ABSTRACT
Poliovirus infection is initiated by attachment to a receptor on the cell surface called Pvr or CD155. At physiological temperatures, the receptor catalyzes an irreversible expansion of the virus to form an expanded form of the capsid called the 135S particle. This expansion results in the externalization of the myristoylated capsid protein VP4 and the N-terminal extension of the capsid protein VP1, both of which become inserted into the cell membrane. Structures of the expanded forms of poliovirus and of several related viruses have recently been reported. However, until now, it has been unclear how receptor binding triggers viral expansion at physiological temperature. Here, we report poliovirus in complex with an enzymatically partially deglycosylated form of the 3-domain ectodomain of Pvr at a 4-Å resolution, as determined by cryo-electron microscopy. The interaction of the receptor with the virus in this structure is reminiscent of the interactions of Pvr with its natural ligands. At a low temperature, the receptor induces very few changes in the structure of the virus, with the largest changes occurring within the footprint of the receptor, and in a loop of the internal protein VP4. Changes in the vicinity of the receptor include the displacement of a natural lipid ligand (called “pocket factor”), demonstrating that the loss of this ligand, alone, is not sufficient to induce particle expansion. Finally, analogies with naturally occurring ligand binding in the nectin family suggest which specific structural rearrangements in the virus-receptor complex could help to trigger the irreversible expansion of the capsid.
IMPORTANCE The cell-surface receptor (Pvr) catalyzes a large structural change in the virus that exposes membrane-binding protein chains. We fitted known atomic models of the virus and Pvr into three-dimensional experimental maps of the receptor-virus complex. The molecular interactions we see between poliovirus and its receptor are reminiscent of the nectin family, by involving the burying of otherwise-exposed hydrophobic groups. Importantly, poliovirus expansion is regulated by the binding of a lipid molecule within the viral capsid. We show that receptor binding either causes this molecule to be expelled or requires it, but that its loss is not sufficient to trigger irreversible expansion. Based on our model, we propose testable hypotheses to explain how the viral shell becomes destabilized, leading to RNA uncoating. These findings give us a better understanding of how poliovirus has evolved to exploit a natural process of its host to penetrate the membrane barrier.
INTRODUCTION
Poliovirus is the type member of the enterovirus genus of the Picornavirus family, and the causative agent of poliomyelitis. The virus is comprised of 60 copies of each of four capsid proteins (VP1, VP2, VP3, and VP4), which form an icosahedrally symmetric capsid that encloses a unique positive-sense single-stranded RNA genome (1, 2). Poliovirus infection is initiated when the virus attaches to a specific receptor, called Pvr (poliovirus receptor) or CD155. The receptor is a cellular glycoprotein with three extracellular immunoglobulin-like domains, a transmembrane domain, and either of two splice-variant C-terminal cytoplasmic domains (3). The protein is nectin-like (Necl) and a member of the nectin superfamily (2, 4). The receptor has been shown to bind to the virus in two modes, a lower-affinity mode (Kd ∼ 1 μM), which dominates at low temperature, and a higher-affinity mode (Kd ∼ 0.1 μM), which dominates at ambient temperature (5).
Attachment of poliovirus to its receptor at physiological temperature catalyzes a conformational alteration of the virus(6–8). The expanded virus particle shows significant changes in its sedimentation constant (135S versus 160S for the mature virion) and its antigenicity (H antigenic versus N antigenic for the virion) (6, 7). During expansion, 60 copies of two polypeptide segments, each about 70 residues long, move from the inner surface of the capsid to the virus exterior: VP4 (which is myristoylated at its N terminus [9]) and the N-terminal extension of VP1 (which is predicted to form an amphipathic helix) (10). These polypeptides insert into membranes, both in vitro (10, 11) and during infection (12), and are thought to form pores in the membranes of endocytic vesicles that permit the transfer of the viral RNA genome into the cytoplasm (12–15), where replication occurs.
The structure of the virus is known at high resolution (16–18) (Fig. 1A). The major capsid proteins (VP1, VP2, and VP3) share a eight-stranded beta sandwich topology (Fig. 1B), but the loops that connect the strands of the beta barrel have different lengths and conformations, and their long N-terminal extensions fold across the inner surface of the capsid (16). The hydrophobic core of VP1 contains a binding site for a fatty acid-like ligand called the “pocket factor” (17). This site also binds a family of capsid-binding drugs that stabilize the capsid, preventing the 135S particle from forming (19–23). Pocket factor binding has been proposed to be a key regulator of viral stability and of the receptor-induced viral expansion that occurs during infection (17).
FIG 1.
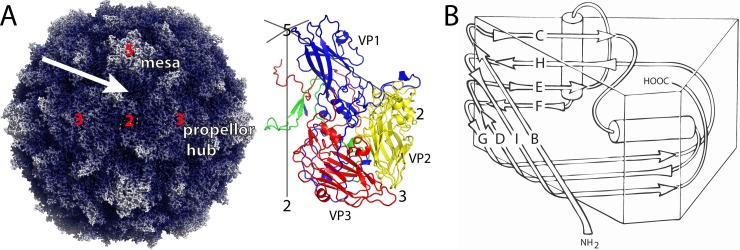
Overview of the poliovirus capsid. (A) Surface rendering of the capsid (PDB 1HXS) (left). Darker hues show regions closer to the center of the particle. Protomer (right) is formed of one subunit each of VP1 (blue), VP2 (yellow), VP3 (red), and VP4 (green). Icosahedral symmetry axes are labeled with numbers, and prominent capsid features are labeled. “Mesas” are formed by five copies of VP1 with the narrow edges of the beta barrels clustered near 5-fold axes. The narrow edge is to the left in panel B. “Propellers” are formed by three copies each of VP2 and VP3 alternating around 3-fold axes. “Canyons” are depressions surrounding the mesas, and “saddle surfaces” cross each 2-fold axis. The quasi-3-fold axis (arrow) is the center of the 5-3-3 asymmetric triangle and the site of Pvr binding. (B) Each of the major capsid proteins has the “jelly-roll” topology of a wedge-shaped eight-stranded antiparallel beta barrel. Beta strands are named for the letters B to I. Loops are named using two letters designating the beta strands that the loop connect.
The outer surface of poliovirus is dominated by star-shaped mesas at each 5-fold axis and by three-bladed propellers surrounding each 3-fold axis (Fig. 1). Each 5-fold mesa is formed by the beta barrels of VP1, and the hubs of the propellers are formed by the beta barrels of VP2 and VP3. Each of the projecting propeller tips is formed by the large EF loop of VP2 (i.e., loops that connect the E and F strands of the VP2 beta barrel) and is supported by a long GH loop of VP1 and by the C-terminal extensions of VP1 and VP2. Neighboring mesas and propellers are separated from one another by the deep depression (“canyon”) that surrounds each mesa, and neighboring propellers are separated from one another by the saddle-shaped depression that crosses each 2-fold axis. In mature virions, the inner surface of the protein shell is decorated by an elaborate stabilizing network formed by the myristoylated protein VP4 and by the intertwined N-terminal extensions of VP1, VP2, and VP3 (24, 25).
As the externalization of RNA, VP4, and the N terminus of VP1 are central to the infection mechanism their routes of exit through the capsid are of keen interest. The structures of expanded forms of poliovirus, and of several related viruses, have recently been reported (26–30). These structures demonstrate that expansion of the capsid produces large holes at the 2-fold axes and near the quasi-3-fold axes. The high-resolution structures of 135S particles of poliovirus (30) and coxsackievirus A16 (27) both demonstrate that the N terminus of VP1 has become externalized at the quasi-3-fold and that the hydrophobic core of VP1 has lost its bound “pocket factor.” In addition, low-resolution structures of poliovirus particles caught in the act of externalizing the viral genome (both with and without membrane present) show that RNA release occurs at a site near the junction of the 2-fold and quasi-3-fold holes (31). These findings contradicted previous models that proposed that the viral peptides and the RNA would exit via a channel at the particle 5-fold axes.
The structure of the virus-receptor complex has not yet been solved by X-ray crystallography, but there have been a number of reports describing the structure of the complex, as determined by cryo-electron microscopy (cryo-EM) at low resolution (32–38). The earliest reports used homology models for the receptor, since no crystal structure was available for Pvr. These reconstructions confirmed early proposals that the receptor would bind within the canyon and demonstrated that only its N-terminal domain would contact the virus directly. However, the use of homology models and the low resolution of the structures prevented accurate descriptions of the virus-receptor interaction in detail, and published models had obvious problems, including serious steric clashes between the virus and docked receptor. A later report described the 3.5-Å crystal structure of a two-domain construct of the receptor that had been mutated to prevent glycosylation (37, 39). That model was fitted to ∼9-Å-resolution cryo-EM reconstructions of the virus-receptor complex for each of the three poliovirus serotypes. Unfortunately, this two-domain structure was of poor quality (high Rfree, and improbable main chain torsions, including seven disallowed cis peptide bonds and six nonplanar peptides), and we never have been able to use it to derive an acceptable model in our high-resolution reconstructions of the complex.
More recently, two high-resolution crystal structures were published that included glycosylated versions of domain 1 (D1) of Pvr. One structure shows the binding of D1 to the immune regulator TIGIT (40). The other is a homodimer of the entire three-domain ectodomain (41). Although the two new structures are similar to each other, they differ from the earlier low-resolution two-domain structure, particularly in D1 (37). The crystal structures show that Pvr displays significant flexibility in the conformation of several loops that are involved in partner interactions. Importantly, these structures also show that the interaction of Pvr with its natural ligands is driven by the need to bury otherwise-exposed aromatic side chains in hydrophobic intermolecular interfaces.
We describe here a cryo-EM reconstruction of a complex between poliovirus and the enzymatically partially deglycosylated form of the Pvr ectodomain at 4-Å resolution. Using this structure, we fitted and refined high-resolution atomic models of the virus and of the three-domain structure of the ectodomain of Pvr based on these reconstructions, and obtained a near-atomic model for the virus-receptor interaction. This new model is consistent with previous mutational data, has chemically plausible contacts between the virus and D1 of the receptor and displays a striking shape complementarity between the virus and D1 of the receptor. Surprisingly, although the virion structure is minimally changed, receptor binding results in the expulsion of the “pocket factor.” Clear analogies can be drawn between the way that poliovirus binds to its receptor and the way that nectins bind to one another. The structure suggests plausible models for (i) how receptor interaction promotes the loss of pocket factor and (ii) how receptor interaction at physiological temperature triggers expansion and other subsequent structural changes that occur early in infection.
MATERIALS AND METHODS
Virus and Pvr preparation.
Poliovirus (serotype 1, Mahoney strain) was grown in suspended HeLa cells and purified by differential centrifugation and CsCl density-gradient fractionation, as described previously (42, 43). Soluble, glycosylated Pvr (sPvr, sCD155), residues 1 to 337 (the ectodomain), with 6 additional histidine residues at the C terminus, was expressed in the 293-T human epithelial kidney cell line and purified by chromatography with Ni-agarose and Q-Sepharose columns, as described previously (5). sPvr was enzymatically deglycosylated using endoglycosylase H, yielding sdPvr (Peter Kwong, unpublished data). The sPvr was provided by V. Racaniello, and sdPvr was provided by V. Racaniello and P. Kwong.
Cryo-EM and three-dimensional reconstruction.
Complexes of poliovirus with sPvr were prepared and imaged as described previously (32). Complexes of poliovirus with sdPvr were prepared similarly. Briefly, samples of the virus (0.9 mg/ml in phosphate-buffered saline) and sdPvr (1 mg/ml in a buffer composed of 2.5 mM Tris, 350 mM NaCl, 0.02% NaN3, 5 mM CaCl2, and 1 mM MnCl2 [pH 7.0]) were mixed at 4°C, producing a 300-fold molar excess of sdPvr. The complexes were then loaded onto glow-discharged holey grids and plunge frozen in liquid ethane. To minimize aggregation, virus and receptor were mixed and plunge frozen within 2 min.
Specimens of poliovirus-sPvr and poliovirus-sdPvr complexes were imaged, and three-dimensional reconstructions were computed to moderate resolutions as described previously (see the supplemental material). Specimens of poliovirus-sdPvr were imaged and reconstructed to near-atomic resolution in the following manner. On a Polara G2 (FEI, Hillsboro, OR) equipped with a K2 Summit camera (Gatan, Pleasanton, CA) micrographs were recorded at 300 kV in super-resolution mode, using dose fractionation over 25 frames. The first two frames were discarded as they showed the most drift, and the remaining 23 were aligned and summed using “dosef” (44). A representative micrograph is shown in Fig. S1A in the supplemental material.
The poliovirus-sdPvr particles were selected and excised using EMAN2 (45). An initial two-dimensional classification of the particles was carried out using Relion (46) to remove particles of low quality. This was followed by a three-dimensional classification procedure to sort the particles into six different classes, using a model of poliovirus that was Fourier filtered to 40-Å resolution as a reference. Two of the six classes (including 9,248 particles and corresponding to 99.7% of the population) appeared to be similar to one another and were retained and combined into a single data set. The full parameter refinement was carried out using GeFrealign (47), gradually extending the upper resolution limit to 4 Å. In each iteration, the refinement was restricted to include resolution shells that showed a high Fourier shell correlation (typically, 0.7 or better). Sections through the final 4-Å map are shown in Fig. S1B to G in the supplemental material.
The 4-Å-resolution electron density map for the complex of poliovirus with the enzymatically deglycosylated three-domain Pvr constructs was deposited in the Electron Microscopy Data Bank (EMDB) as EMD-6147 and -6148 (reconstructions as written by GeFrealign and the sharpened version, respectively). Previously unpublished 9-Å-resolution maps for sPvr (EMD-6242) and sdPvr (EMD-6243), which were determined before the availability of the K2 detector used in the present study (see the supplemental material), have also been deposited.
To facilitate map interpretation, the final electron density map was “sharpened” (i.e., increasing the average Fourier amplitudes at high resolution) using a B-factor of −60 Å2 in “embfactor” (48). For convenience, density values in the sharpened map were then increased radially using a hyperbolic function fit in order to make the density features near the outside of the capsid more nearly equal to those on the inside. In practice, this helped to resolve most of the individual polypeptide chains from one another, which facilitated fitting of the atomic models of poliovirus and D1 (see below). In contrast, D2 and D3 were fitted as a single rigid body to a “dampened” version of the experimental map, which was prepared using a B-factor of 300 Å2.
Model building and refinement.
Atomic models were initially constructed to fit the 9-Å sPvr and sdPvr EM density maps, starting with seven rigid-body domains obtained from prior crystallographic studies. The model included three independently refined domains of Pvr (D1, D2, and D3), obtained from 4FQP.pdb (41), plus poliovirus capsid proteins VP1, VP2, VP3, and VP4, obtained from 1HXS.pdb (49). Together, VP1, VP2, VP3, and VP4 constitute the biological protomer. The initial model was rigid-body refined, with all surrounding symmetry-related neighbors generated (for a total of 26 polypeptide chains), and icosahedral symmetry was strongly enforced. Later, individual local discrepancies between the atomic model and the 4-Å-resolution map were repaired by repeated cycles of model-rebuilding (using COOT [50] and SPDBV [51]) and stereochemically restrained atomic refinement (using Refmac5 [52–54]). D1, D2, and D3 were estimated to be 0.5 occupied in the higher resolution map (versus 1.0 in our older 9- and 22-Å [32] maps).
The refinement of D1 and poliovirus in the model was well behaved, despite the limited resolution, provided that parameter shifts were damped to 1 to 3% of default values, and hundreds of cycles were run each time. Lower-than-normal weights were used to penalize bad nonbonded contacts, and higher-than-normal weights were assigned to Fourier phase agreement, planarity, and single-bond torsion restraints. After refinement, we observed that the atomic model was chemically plausible and fit the map and that the hydrogen-bonding network of the original models was largely preserved. D2 and D3 (whose positions are clearly indicated by a moderate-resolution density envelope, Fig. 2C and D) were modeled as a rigid body and assigned a high temperature factor (B = 300) in the model, to avoid overfitting to noise.
FIG 2.
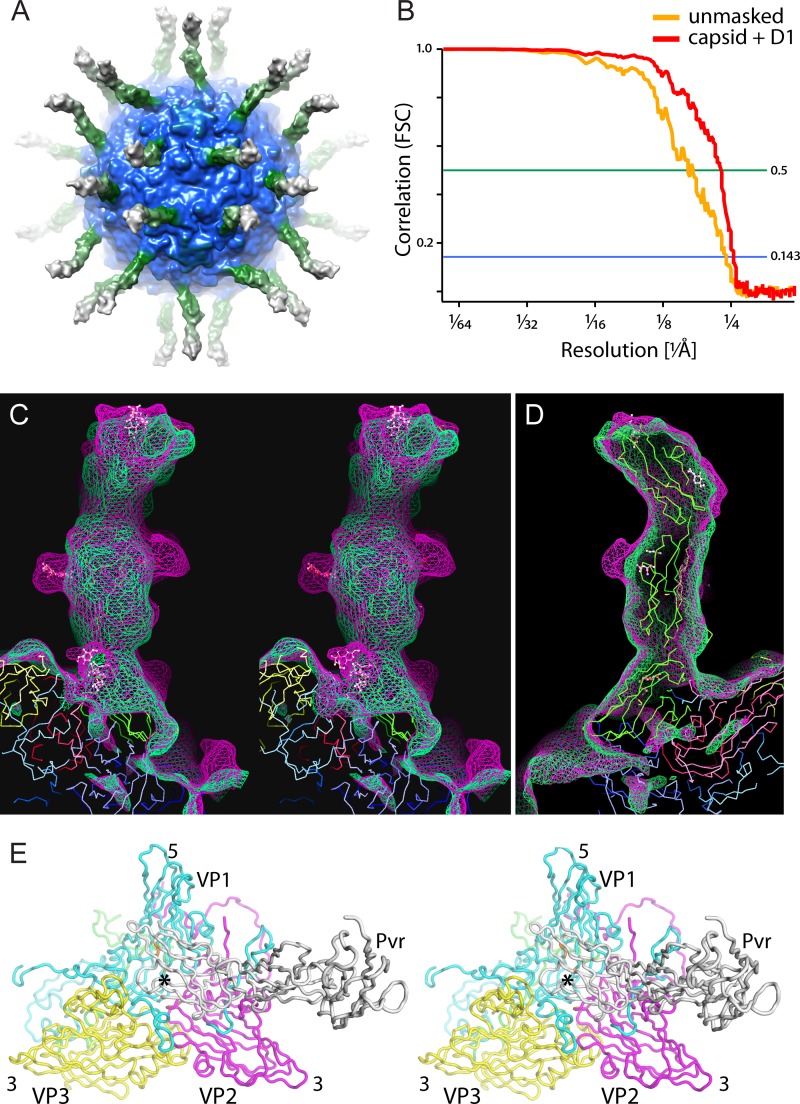
Pvr bound to poliovirus. (A) The high-resolution reconstruction of the Pvr-poliovirus complex is shown as an isocontour surface and colored by radius. Here, it was mildly Gaussian blurred and set to low contour for clarity. (B) The Fourier shell correlation curves show agreement between half-set reconstructions to 4.2-Å resolution and to 3.9-Å resolution when the poorly ordered regions (RNA in poliovirus and D2 and D3 of Pvr) are masked. (C and D) Reconstructions at 9-Å resolution show that the enzymatically deglycosylated (green mesh) and glycosylated (magenta mesh) receptors bind to the virus in similar ways. (C) In stereo, an overlay of the two maps shows excellent agreement between the two density envelopes, except at the glycosylation sites. (D) Refined atomic coordinates of the Pvr monomer fit snugly into these lower-resolution maps. (E) In stereo, backbone traces of the refined model show how Pvr binds at the center of the 5-3-3 icosahedral triangle (at the so-called “quasi-3-fold axis,” marked with an asterisk), making contact with all three major capsid proteins. VP1, VP2, VP3, and Pvr are shown in cyan, yellow, magenta, and white, respectively.
All model refinements were carried out in reciprocal space, using the Fourier transform of a representative portion of the reconstruction as a reference (55) and giving Fourier phase agreement a high weight. As described previously (30), an electron density corresponding to the 26-chain model (with neighbors included) was copied from the reconstruction into a rectangular box, masked with a generous model-based mask, and Fourier transformed to produce the refinement standard. Steps of the refinement were carried out using Unix C-shell scripts that made extensive use of software from the Bsoft (56) and CCP4 (57) program packages. Notably, least-squares superposition was used at the beginning of each refinement cycle to require certain portions of the atomic model to be consistent with their rigid-body standards. Such restraints were applied to all of the atoms, when refining to fit the 9-Å-resolution sPvr and sdPvr maps (see the supplemental material), and to all of the atoms of D2 and D3, when fitting the 4-Å-resolution sdPvr map due to the relatively poorer quality of density in those areas.
Except as noted below, most of the main chain was correctly positioned by automatic all-atom refinement in Refmac5, while a large number of side chain rotomers, throughout the structure, required manual adjustment either to fit the density or to make their chemical contacts plausible. Coordinates for the poliovirus complex with deglycosylated receptor, at a 4-Å resolution, were deposited as PDB 3J8F. For comparison, coordinates for the fit into the previously unpublished 9-Å sPvr and sdPvr maps have also been deposited (3J9F). Figures were prepared using CHIMERA (58), Coot (50), and PyMOL (59).
Database depositions.
The following maps have been deposited into the EMDB: 9-Å sPvr, EMD-6242; 9-Å sdPvr, EMD-6243; 4-Å sdPvr, EMD-6147; and 4-Å sdPvr (sharpened), EMD-6148. The corresponding coordinate depositions in the PDB are as follows: rigid body fit sdPvr, 3J9F; and model of the 4-Å map of sdPvr, 3J8F.
RESULTS
Resolution of the reconstructions of virus-receptor complex.
The resolution of the high-resolution poliovirus-sdPvr reconstruction (Fig. 2A and Fig. 4) was estimated by calculating a Fourier shell correlation (FSC) between two halves of the data set. The resulting FSC plots (Fig. 2B) indicated agreement between half-set reconstructions to a resolution of 6.2 and 4.2 Å, according to the 0.5 and 0.143 correlation criteria, respectively. When the half-set reconstructions were masked to include only the capsid and domain 1 of Pvr, the FSC values improved to 4.4 and 3.9 Å for the 0.5 and 0.143 correlation criteria, respectively. Domains from crystal structures of Pvr were fitted into 9-Å reconstructions of both the glycosylated and the enzymatically deglycosylated Pvr-poliovirus complexes (Fig. 2C and D). Manual model building and restrained refinement were then used to achieve an optimal fit to the 4-Å-resolution map (Fig. 2E).
FIG 4.
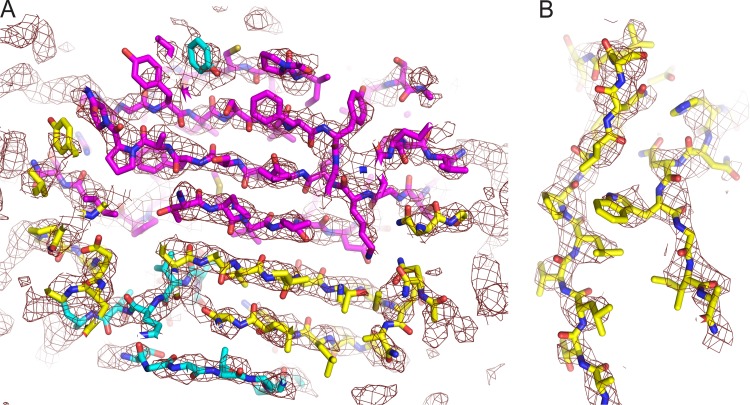
Quality of the electron density map at 4-Å resolution. In the capsid and D1, the position of the main chain is obvious, and there are often indications for the positions of side chains. (A) Seven-stranded beta sheet that is formed by C, H, E, and F strands of VP3 (magenta), the amino-terminal hairpin of VP2 (yellow), and a segment of the N-terminal extension of VP1 (cyan). (B) Side chain density is evident for beta strands G (left strand) and H (right strand), on opposite sides of the VP2 beta barrel.
Site of binding.
As with previous reconstructions, the receptor binds to the center of the 5-3-3 icosahedral triangle, directly over the quasi-3-fold axis (Fig. 2A and E). The 5-3-3 triangle is the icosahedrally unique surface with a 5-fold axis and two 3-fold axes at its corners. It is convenient to describe regions of this “5-3-3 triangle” using compass directions, with the 5-fold corner designated “north,” and the 2-fold axis designated “south.” In the 5-3-3 asymmetric unit, VP1 and VP2 from one protomer lie on the “west” side and a VP3 subunit from an adjacent protomer lies on the “east” side (Fig. 2E; cf. Fig. 1A, right). Interestingly, Pvr binds at the intersection of two protomers. As in our previous poliovirus-sPvr reconstruction (32), density is clearly present for each of the three domains of the receptor (Fig. 2A, C, and D). The best-resolved portion of the receptor is the D1 domain, which is the only domain to contact the virus. The positions of the carbohydrates on Pvr are readily apparent when partially deglycosylated (sdPvr) and fully glycosylated (sPvr) maps are compared (Fig. 2C and D). In our model, residues eligible for glycosylation were found at the glycosylation sites, and this served as a check on fitting of our model.
The contact “footprint” of the receptor on the virus surface.
Domain 1 of Pvr inserts into the canyon and forms an archway, standing on two discrete, well-separated legs (Fig. 3A, B, D, and E). One contact area lies along the east wall of the canyon, running approximately in a north-south direction along the C-terminal extension of VP1. This copy of the C-terminal extension belongs to the VP1 beta barrel lying outside and to the right of the 5-3-3 icosahedral triangle. Along that path, additional viral structures are contacted, including the C terminus of VP3 (in the northeast corner) and the knob-like insertion in the B beta strand of VP3 (in the southeast corner) (Fig. 3A and see Fig. S4 in the supplemental material). The other, somewhat longer, contact area lies on the western side of the binding site, running in a north-south direction along the H beta strand and GH loop of VP1. Along that path, additional contacts are made with the EF loop, BC loop, and C beta strand of VP1 (on the northwestern wall of the canyon). In the southwest, D1 contacts with the GH loops of VP1 and VP3, and with the EF loop of VP2 (Fig. 3A). On the western side, all of the VP1 polypeptide segments belong to the copy of VP1 whose beta barrel forms the Northern third of the 5-3-3 triangle (Fig. 3A).
FIG 3.
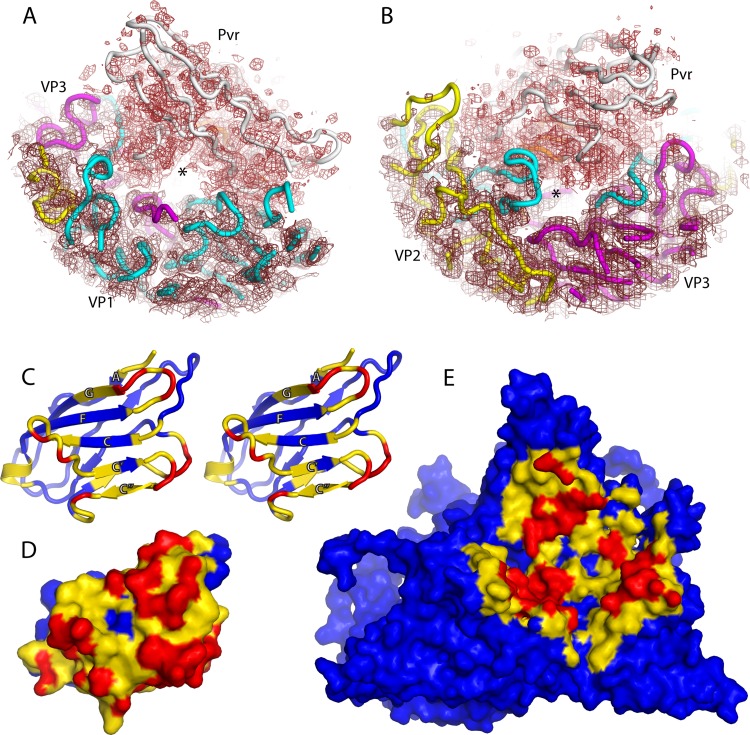
Contacts between the Pvr and poliovirus surfaces create a solvent-filled tunnel. In panels (A) and (B), slices through the experimental density map (mesh) are shown superimposed on backbone traces of VP1 (cyan), VP2 (yellow), VP3 (magenta) and Pvr (white). The tunnel, which appears as a clear area in the center, is apparent when viewed either from the 2-fold axis (B) or the 5-fold axis (A). Panels C-E are calculated from the atomic models, and are color-coded according to contact distance: closer than 8.5 Å (yellow) or 4.5 Å (red). (C) is a stereo ribbon representation of the atomic model, while (D) and (E) are van der Waals surfaces. (C) and (D) show how D1 is covered by poliovirus. (E) Poliovirus outer surface, as covered by D1. The area shown is the 5-3-3 icosahedral triangle with 5-fold axis at the top, and a quasi-3-fold at its center. In panels D and E, note that the ceiling and floor of the tunnel are each visible as a continuous yellow and blue stripe that separates the red close-contact patches.
The locations of the contact areas have been apparent since the earliest structural studies (32, 35, 38), but the detailed interactions with receptor have become clear only recently (37). In each patch, there are loops or terminal extensions that are known to shift, rearrange, or disorder during the 160S-to-135S and 160S-to-80S transitions of poliovirus and related enteroviruses (26, 29, 32, 37, 60). Such changes should either destabilize the native virus structure or facilitate Pvr release, depending on when they occur. Notably, the set of rearranged polypeptides includes both the long C-terminal extension and the long GH loop of VP1, which lie along the eastern and western binding patches, respectively. Several of the residues that make intermolecular contacts have previously been identified as mutations that have an impact on the uncoating process (61–64) (see Table S1 in the supplemental material).
Solvent tunnel.
The receptor forms a high archway that is the roof of a long solvent-filled tunnel that runs in a north-to-south direction between poliovirus and Pvr. The tunnel, in front and back views, is easily visible in the density map, when contoured at an appropriate level (Fig. 3A and B). The existence of a solvent channel within a protein complex is noteworthy, since solvent-filled cavities commonly form binding sites. Indeed, we propose that structural rearrangements that result in viral polypeptides being inserted into this tunnel may be key to triggering viral expansion to the 135S state (see below).
Construction of the atomic model.
As described in Materials and Methods, the atomic models for poliovirus and Pvr were derived starting with mature 160S poliovirions (1HXS.pdb) and Pvr-Pvr homodimers (4FQP.pdb), respectively. After an initial rigid body fit, manual rebuilding was repeatedly alternated with a full atomic refinement, with 21 icosahedrally related neighbor chains present, and stereochemical restraints applied, using the Fourier transform of a portion of the 4-Å-resolution density map as a refinement standard. The resulting model includes VP1 (residues 20 to 302), VP2 (residues 8 to 272), VP3 (residues 1 to 235), VP4 (residues 1 to 69, including N-terminal myristoylation), D1 (residues 28 to 141), D2 (residues 142 to 242), and D3 (residues 243 to 333), plus a total of 20 sugar moieties.
Quality of fit to density.
Qualitatively, the fit of the refined atomic model to the density in the reconstructions is excellent (see Fig. 2C and D). In the 4-Å-resolution map of the sdPvr complex (Fig. 4), the quality of the map varies by radius. In the poliovirus capsid and in D1, the correspondence between the density and the main chain trace is unambiguous, and the docked model is clearly similar in shape to the density. Individual beta strands are usually resolved from one another (Fig. 4A). This is not to imply that the main chain could have been completely traced de novo at the present resolution but rather that the docked model resolves all chain tracing ambiguities and identified those few polypeptide segments that required rebuilding (see below). Most of the large side chains are obvious, and the density map provides an indication for the correct choice of rotomer for many of the smaller side chains, as well (Fig. 4B). Note, however, that the strength of the density varies dramatically, so that no single choice of contour level works well throughout the map. Indeed, to see that the map is similar in shape to the model, the contour level in the higher-radius portions of D1 needs to be reduced by a factor of ∼3, relative to the inner surface of the viral capsid, and ∼1.5, relative to the outer surface. These differences are likely to be due both to the reduced occupancy of the entire receptor (which we estimate to be ∼0.5) and to the greater impact of orientation errors at high radius. Nevertheless, the densities for poliovirus and D1 were sufficiently good that stereochemically restrained atomic refinement behaved well.
For D2 and D3, when visualized in the desharpened map, the location of the solvent boundary is apparent. However, in the sharpened map, the density along the main chain is fragmented and poorly correlated with the known structure. As a result, the density for D2 and D3 was too poor to support atomic refinement, so copies of domains from the crystallographic model (41) were positioned as a rigid body, instead. Flexibility in the D1-D2 junction may have contributed to lower resolution in D2 and D3, along with orientation uncertainties having a greater effect at higher radius.
In the desharpened and the 9-Å maps, the sites of glycosylation are obvious (Fig. 2C). These sites, which remain partially glycosylated after the enzymatic deglycosylation step, correspond almost perfectly to the crystallographically determined positions of the sugar moieties (as seen in 4FQP.pdb). The sole exception involves two spatially close glycosylation sites on one side of D2 (Asn218 and Asn237), where sugars are clearly present, but positioned in a different way. Note that the positions and orientations of domains 2 and 3 and the location of the carbohydrate are completely consistent with 9-Å reconstructions of complexes of the virus with the fully glycosylated (9.6 Å) and enzymatically deglycosylated (8.8 Å) ectodomain (Fig. 2C).
Statistics of fit.
The quality of the overall fit can be put on a quantitative basis by comparing the phases from the Fourier transform of model-based electron density to the phases from the map. The Fourier-amplitude-weighted average of the cosine of the phase discrepancy is analogous to a crystallographic figure-of-merit (with 1.0 indicating perfect phase agreement and 0.0 indicating a complete lack of correlation). At a 3.7-Å resolution that statistic was 0.556. That statistic demonstrated better-than-random phase agreement in the highest resolution shell, phase improvement over the course of the refinement, and convergence at the end. In the last cycle of stereochemically restrained refinement of the 26-chain atomic model, Refmac5 reported an R-factor of 0.45 (0.54 in the outermost shell) versus 581,893 pseudoreflections in the range of 97 to 3.7 Å, with a “figure-of-merit” of 0.73. Note that these numbers are primarily intended to monitor convergence, since their exact values depend on the choice of masked density that is included in the refinement standard. The final atomic model showed an acceptable level of errors in bond lengths and angles of 0.010 Å and 2.08 degrees compared to idealized stereochemical standards.
Interactions between poliovirus and Pvr.
As noted above, D1 forms an archway, standing on two well-separated legs (Fig. 3A and B). Within each contact patch, the receptor makes extensive contacts with the virus surface. In the eastern patch, which lies along the east wall of the canyon, the contacts involve residues in the CC′, C″D, and EF loops of D1 (Fig. 3C). In the western patch, which runs in a north-south direction along the H beta strand and GH loop of VP1, the contacts in Pvr involve the FG, C′C″, and DE loops and C″ and G beta strands of D1. The contacts on the virus surface are consistent with previous mutational data. A detailed description of the contact regions and mutations, is presented in the supplemental material (see Table S1 in the supplemental material).
How Pvr changes when bound to poliovirus.
If the three domains of Pvr crystal structure 4FQP are refined independently into our density maps (either 9 or 4 Å), the overall arrangement of domains changes very little. (Note that this map-fitting calculation is not the same one that results in our high-resolution model, where D2 and D3 are constrained.) Thus, least-squares superposition of the three-domain construct (using LSQKAB [65]) produces an root mean square deviation (RMSD) in alpha carbon positions of 1.84 Å. If only D1 (residues 28 to 142) is used as the basis for comparison, the RMSD is only 1.02 Å (within the error of the refinement), with a maximum displacement of only 2.89 Å (occurring at Ser72). This occurs in the CC′ loop of D1 (residues 68 to 74). Note that when all D1 atoms are included in the calculation, the RMSD rises to 1.76 Å, which suggests that much of the conformational adaptation involves side chain movement. Shifts in individual alpha carbon positions that are due to Pvr binding are plotted in Fig. S2 in the supplemental material.
The superposition of the crystallographic and cryo-EM versions of D1 onto one another, perforce, has the effect of making the discrepancy between D2 copies larger, and D3 copies larger still. Once the D1 copies have been superimposed, the copies of D2 differ primarily by a 12° rotation, pivoting around the D1-D2 junction. About half of this rotation twists D2 around its 40-Å-long major axis, while the other half rotates D2 around the interdomain vector between residues 142 and 143. (The vector between 142 and 143 is roughly perpendicular to the page when the two D2 glycosylation sites project left and right.) The latter rotation is primarily responsible for maximal shifts at the distal (i.e., C-terminal) end of D2 of ∼4 Å. Notably, this rotation of D2 also brings residues 197 and 198 (from the tip of the DE loop of D2) into close contact with Val-115 (from the EF loop of D1), which involves a shift of ∼3 Å, thereby creating a direct contact that is lacking in the 4FQP crystal structure. We surmise that forming this contact helps to prevent the D1-D2 interface from being more flexible, which caused D1 and D2 to appear rod-like in lower-resolution cryo-EM reconstructions (32, 37). Because of this limited flexibility, it is especially noticeable at the present 4-Å-resolution that the D2 density becomes appreciably worse than D1. These differences do not involve residues that are in contact with the virus and may be a simple consequence of crystal packing forces in the crystal structure or differences in the extent of glycosylation.
Direct comparisons between the 4FQP crystal structure and the poliovirus-Pvr complex (see Fig. S2 in the supplemental material) are also useful for establishing which loops of D1 are either the most flexible, or conformationally adaptable upon ligand binding. This is potentially an important issue for a protein with multiple binding partners. Aside from the CC′ loop of D1, which was mentioned above as the site of the largest change, there are concerted localized differences that shift alpha carbon positions by as much as 1.3 to 1.8 Å. These main chain shifts are seen in residues 52 to 59 (in the long BC loop), residues 78 to 84 (in the C′C″ loop), residues 126 to 129 (in the FG loop), and in the N terminus of the construct (residues 28 to 31). All of the named polypeptide segments participate directly in poliovirus binding (see above), except for the BC loop of D1. The BC loop is solvent exposed in the poliovirus complex, but instead binds to the D2-D3 junction of a symmetry-related neighbor in the 4FQP crystal structure, which accounts for the difference in this loop's conformation.
A similar pattern of conserved structure and flexibility in D1 is also evident when the D1 in the Pvr-Pvr homodimer is compared to the structure of D1 in its complex with TIGIT. Thus, the D1 structure in its complex with TIGIT (40) (3UDW.pdb, chain D) superimposes closely with D1 from the D1-D3 homodimer of Pvr (41) (4FQP, chain A), yielding RMS differences of only 0.88 Å among alpha carbons and 1.54 Å among all atoms. The largest loop changes occur in the CC′ and C″D loops (residues 80 to 82 and residues 87 to 90, respectively). The single largest main chain difference (∼3.5 Å) occurs in the CC′ loop and is due to binding contacts with TIGIT in the D1/TIGIT complex. In poliovirus, either version of the CC′ loop could be accommodated in the model without producing bad contacts. It is clear from the above comparisons that the changes in Pvr due to binding its partners are surprisingly small but that the largest of these changes involve direct contacts to its ligand.
How poliovirus changes when bound to Pvr.
When the refined poliovirus coordinates in the sdPvr complex are compared to the starting crystal structure, the two coordinate sets agree very closely (see Fig. S2 in the supplemental material). This is true even though the comparison involves coordinates in the icosahedral frame of reference, without permitting agreement to improve by allowing individual proteins to move. In particular, the RMS difference in alpha carbon coordinates can be reduced to 0.64 Å by choosing a scale factor similar to that of the 1HXS model.
Changes in VP4.
The largest differences between the models (up to 9.9 Å in alpha carbon displacement) involves a loop of VP4 (residues 11 to 23) on the inner surface of the capsid that is not well ordered. The 1HXS.pdb model includes this loop, built to fit to weak density features in its electron density map, but individual atomic temperature factors in the loop have refined to ∼100 Å2, reflecting disorder. When we discovered that the original loop conformation is inconsistent with our present cryo-EM map, we chose to build a better-fitting alternative conformation into the weak density in the 4-Å reconstruction. Although most of the VP4 loop still occupies the same binding site, formed by VP1 and symmetry-related copies of VP4, one portion of the loop, including a Tyr side chain, is now located farther from the capsid and appears to contact the RNA.
Changes in the doorstop and EF loop of VP1.
Aside from VP4, the largest differences in the poliovirus capsid (Fig. 5A) at residues 233 to 236 of VP1, which shifts by >7 Å, relative to the 1HXS model (a region we refer to as the “doorstop” [see below]). These residues are located within the “canyon,” at the carboxyl end of the GH loop of VP1, and just adjacent to the “pore,” where the hydrophilic end of bound “pocket factor” emerges from the upper surface of the VP1 beta barrel in mature poliovirions.
FIG 5.
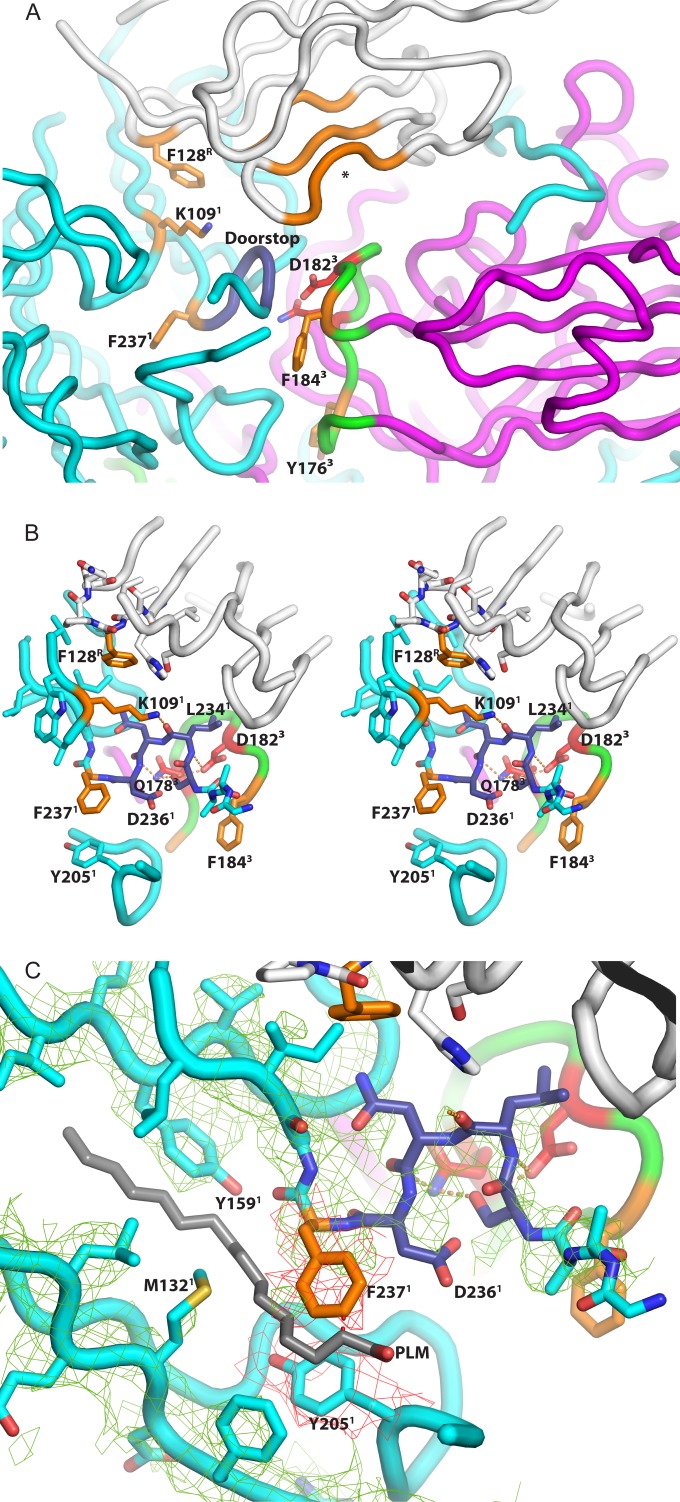
Amino acids that may be relevant to the mechanism of Pvr-catalyzed poliovirus expansion. (A) Main chain traces from selected portions of the poliovirus-Pvr binding site are shown as tubes and colored white (Pvr), cyan (VP1), or magenta (VP3). The key side chains are highlighted in orange or red, and their positions are shown relative to the VP1 doorstop (residues 233 to 236), which is dark blue, and held in a “super-up” conformation by a network of hydrogen bonds. Note the unfilled nectin-family binding site for aromatic side chains, which is colored orange and marked with an asterisk. Polypeptides omitted for clarity include VP2 and the GH loop of VP1 (the truncation is evident below the doorstop), which would both be located in the foreground. The network (seen in more detail in panel B in stereo) includes Lys109 of VP1 (which helps to form the binding pocket for Phe128 of Pvr) and Gln178 and Asp182 of VP3 (red), which belong to the GH loop of VP3 (green). Other side chains that help to form the Phe128-binding pocket are shown but not highlighted. In panel C, a close-up view, the conformation of the VP1 main chain is shown superimposed on the actual electron density from the 4-Å-resolution map (green mesh), and on the hypothetical position of palmitate “pocket factor” (gray). Observe that the position of the Phe-237 side chain (orange model and red mesh) would clash with pocket factor, if it were still present. In panel A, which provides context, note the central clear area (solvent tunnel), which has the GH loop of VP3 (green) at its floor, and the nectin-family aromatic-binding pocket of Pvr (orange tubes) on its ceiling. At physiological temperatures, Pvr binding would trigger expansion of the viral capsid, and the GH loop of VP3 (green) would rearrange to extend outward, toward the nectin pocket. We propose that otherwise-reversible expansion could be trapped by the binding of aromatic side chains (Phe184 and/or Tyr176 of VP3, orange). The superscript “1,” “3,” or “R” at the end of each label denotes the chain to which the residue belongs (1, VP1; 3, VP3; R, Pvr).
It should be emphasized that the conformational change in the “doorstop” itself is not novel or remarkable, since residues 233 to 236 of VP1 have previously been seen in an “up” conformation (e.g., 2PLV) and a “down” conformation (e.g., 1HXS), or a mixture of the two, in poliovirus crystal structures in the past. Indeed, because the “down” conformation would obstruct the “pore” leading to the pocket factor binding site in the hydrophobic core of VP1, it previously has been proposed that the up/open conformation is required for pocket factor to exit, and the exit of pocket factor has been proposed to be a precondition for expansion to the 135S state (28). Figure S3 in the supplemental material shows superimposed main chain traces of VP1 residues 229 to 242 from 2PLV, 1HXS, and the poliovirus-Pvr complex. Comparison between 2PLV and 1HXS shows that the doorstop residues can ordinarily change position without affecting the neighboring structure significantly. In contrast, the constrained conformation of the doorstop in the poliovirus-sdPvr complex forces the Phe237 side chain significantly more deeply into the middle of the VP1 beta barrel.
Pocket factor is missing.
It should be noted that the conformation of the doorstop in the virus receptor complex is an exaggerated version of the previously observed “up” conformation. This “super up” conformation results in a pronounced inward shift of residues 236 to 237 by 1.2 to 1.4 Å. As the density map makes clear (Fig. 5C), this shift “ratchets” the side chain of Phe237 into the center of the VP1 beta barrel into a position that would conflict with the “pocket factor” molecule (palmitic acid, in the case of 1HXS.pdb), if it were present. Simultaneously, residues 203 to 205, from the G beta strand, close to the amino end of the GH loop, are also shifted by almost 1 Å, with the side chain of Tyr205 moving into a position that would conflict with “pocket factor.”
Recent crystal structures of the 135S particle of CAV16 (27) and of 80S particles from EV71 (28) and rhinovirus (26) showed that pocket factor is missing from the middle of the VP1 beta barrel in expanded virus. What was not known until now was the stage at which pocket factor loss occurs. When present, pocket factor is thought to stabilize the virus, thus regulating the 160S-to-135S transition (17, 23, 66). Lack of pocket factor in 80S particles apparently caused the two beta sheets of the VP1 beta barrel to move closer together than they are in mature virus (28), which plausibly could weaken the lateral association between capsid proteins.
The changes in the doorstop are also correlated with localized changes in the EF loop of VP1, particularly around residues 162 to 165 (which move by up to 2 Å) and residues 174 to 177 (which move by up to 1.4 Å). The importance of the EF loop being able to move is also suggested by mutational studies (61, 67) showing that the mutation of Val160 of VP1 to Ile compensates for receptor defects. This residue is located well below the virus surface, deeply buried in the interface between the EF loop and the CHEF sheet that it is attached to. Given the buried location of Val160, it was postulated that this residue was likely to occur at the site of changes in intramolecular contacts during uncoating (67). The changes that we now see in the EF loop of VP1 upon Pvr binding are consistent with that proposal. Collectively, these structural changes convince us that, contrary to our expectations, the binding of Pvr to poliovirus, even at low temperatures, is sufficient to result in the loss of pocket factor but not sufficient to trigger viral expansion. We depict the changes that occur from an energetic perspective using a schematic of the reaction coordinate (Fig. 6).
FIG 6.
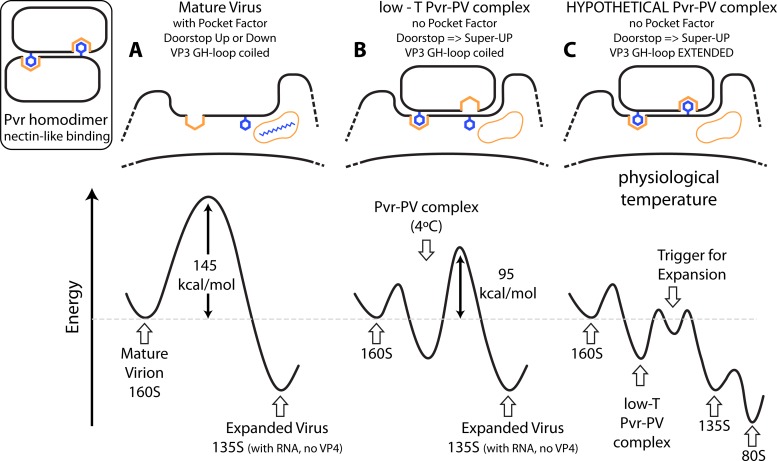
Mechanism of receptor-induced poliovirus expansion. The triggering mechanism that we envision may be clearer in the context of the energy landscape that governs the conversion of mature 160S poliovirions into expanded empty capsids and is mediated by a nectin-like binding interaction (inset). (A) In the absence of receptor, the hydrophobic core of the VP1 beta barrel is usually occupied by pocket factor, and the energy barrier to conversion is large: 145 kcal/mol (8). The GH loop of VP3, represented by the downward facing Phe residue, is coiled. The vacant four-sided orange pleat and the orange pocket containing the blue molecule are hydrophobic sites for nectin binding and pocket factor, respectively. Elevating the temperature to the melting temperature of RNA is required to trigger expansion, to externalize the internal polypeptide segments, and to allow RNA to exit the capsid. (B) In contrast, the initial attachment of Pvr to poliovirus stabilizes an arrangement in which pocket factor is precluded from remaining bound, and the doorstop is held in the “super up” position. At low temperatures, this initial binding mode is stable and corresponds to the complex that we have reconstructed (see Fig. 3 to 5). Phe-128 of Pvr (left downward protruding residue) is buried in a pocket that is formed on the virus surface, which is typical of the natural binding interactions of the nectin family (41) (represented here by the Pvr dimer), but the aromatic-binding pocket of Pvr is uncharacteristically empty. After formation of this initial complex, only 95 kcal/mol is required to trigger expansion (8). (C) Hypothetically, at physiological temperature, the empty Pvr pocket becomes occupied by the rearranged GH loop of VP3. This action promotes the icosahedrally symmetric expansion of the capsid and opens holes at the quasi-3-fold axes, which facilitates the exiting of internal polypeptide segments and creates favorable binding sites for the 60 exposed N termini of VP1 (as seen in the structures of 135S particles [27, 30]). Collectively, these structural changes would reduce the activation barrier to formation of 135S particles. In the presence of membranes (or the absence of divalent cations), 135S particles are unstable, leading to spontaneous loss of RNA and the formation of 80S empty capsids (energy landscape, right).
Changes near the 5-fold axis.
The other relatively large changes that we see in VP1 involve residues 145 to 148 (up to 1.6 Å) and 248 (1.1 Å), which are involved in the contacts of VP1 with its 5-fold-related neighbors, close to the 5-fold axis. Although subtle, these changes may be precursor to the large umbrella-like changes in the expanded 135S particle that cause the VP1 beta barrels to rise, flatten, and separate from one another upon Pvr-triggered viral expansion (29, 30, 68). Although most of the conformational changes in VP1 that were noted in this section are small (1 to 2 Å), they should be understood in the context that typical changes are much smaller. Once we exclude the residues of VP1 that are named above, the RMS change in alpha carbon coordinates in VP1 is reduced to only 0.57 Å, which is comparable to 0.57 and 0.46 Å for VP2 and VP3.
Changes in VP2.
In VP2, the largest changes are seen in the large VP2 EF loop at its uppermost tip (residues 166 to 168, 1.3 to 1.5 Å) and in the HI loop (residues 240 to 243, up to 1.9 Å), which is exposed on the outer surface of the virus, very close to the 3-fold axis. In the 135S structure (30), conformational changes were reported at both of these sites, including extensive disordering of the EF loop after its supporting structures had moved away.
Strained peptide bond.
Smaller changes are seen in one portion of the VP2 EF loop, at residues 140 to 143 (up to ∼1 Å). The peptide bond between residues Thr140 and Met141 is notable for being in a strained conformation, with a highly unfavorable left-handed helical conformation. This arrangement is stabilized by hydrogen bonding to a conserved Asp285 side chain from the C-terminal extension of VP1. Evidence for the existence of the strained bond is strong, as it was observed in the icosahedrally averaged 2.2-Å-resolution poliovirus map of type 1 poliovirus (1HXS.pdb), and it has independently been observed in the crystal structures of type 2 and type 3 poliovirus (1EAH [18] and 1PVC [17], respectively). The importance of the strained polypeptide conformation in VP2 is further suggested by the fact that mutations in residues 140, 142, and 144 (which lie along the edge of the receptor footprint) all affected receptor function (61, 63).
In the poliovirus-Pvr complex, the strained peptide bond is located between two neighboring (5-fold-related) Pvr binding sites. Residues on either side of the strained peptide bond (Thr138 and His142) are hydrogen bonded to backbone oxygens from the DE loop of D1 (at Gly100 and Leu99, respectively). We postulate that the unusual strained peptide bond is a likely site for eventual conformational changes during the 160S-to-135S expansion (including the disordering of parts of the VP2 EF loop and VP1 C terminus), since its conformation looks precarious and easy to destabilize.
DISCUSSION
Reciprocal hydrophobic interactions in nectin-ligand complexes.
As elegantly demonstrated by Harrison et al. (41), Pvr and its nectin family share a characteristic mode of binding during homodimerization. Thus, each copy of the receptor has a projecting FG loop at one corner of its beta sheet, with an exposed Phe-Pro motif at its tip (residues 128 to 129 in Pvr) (Fig. 5B). The solvent exposure of the aromatic residues in (hypothetical) isolated monomers obviously would be unfavorable, so there is a strong tendency for the projecting aromatic side chain to become buried in a hydrophobic pocket (see the inset in Fig. 6). In the nectin dimers (including the Pvr homodimer) this exposed aromatic side chain is buried deeply a hydrophobic pocket (or pleat) on an outward-facing side of the C, C′, and C″ beta strands, involving residues 63 to 65, residues 77 to 79, and residues 82 to 85, respectively. The leading edge of the phenyl group contacts the main chain of the beta strands, while the sides of the aromatic side chain are surrounded by contacts with the aliphatic portions of those same residues (Fig. 5A).
Analogously, in the Pvr interaction with TIGIT (40) a quasidimeric interaction is seen, wherein TIGIT provides a Tyr-Pro motif (residues 113 to 114), instead of Phe-Pro, in the FG loop at one corner of the binding site. To complete the analogy, TIGIT provides a fold in the beta sheet, including residues 68 to 70 and residues 73 to 76, as a binding site for Phe128 of Pvr. Clearly, significant binding energy comes from burying these aromatic side chains that otherwise would be solvent exposed in a hypothetical monomer.
Role of nectin-like interactions in poliovirus binding.
The burying of aromatic side chains plays an important role in the natural binding interactions of the nectin family (40, 41). Given that fact, we pay close attention to what the corresponding residues do in the poliovirus complex with Pvr. Just as in the Pvr-Pvr dimer and the Pvr-TIGIT complex, Phe128 from the FG loop of D1 plays a key role in the binding of poliovirus. In the complex with poliovirus, Phe128 lies in contact with the C strand of the VP1 beta barrel, close to the position of Trp108 of VP1. The role of VP1 Lys109 is noteworthy, as the aliphatic portion of the lysine side chain wraps around Phe128 to protect it. At the same time, the epsilon-amino group of lysine is hydrogen bonded either to the Asn235 side chain carboxamide group or to the main chain oxygen of Leu234 in the “doorstop,” depending on the rotomer (Fig. 5B). Either hydrogen bond would help to stabilize a “super up” arrangement of the doorstop. As discussed above, this arrangement differs somewhat from either the “up” or the “down” conformation seen in native virus (in 2PLV and 1HXS, respectively) when pocket factor is present. Notably, these changes in the “doorstop” move the side chain of VP1 Phe237 into a position that is normally occupied by the pocket factor and is clearly inconsistent with the presence of ligand in the pocket. (Indeed, the most convincing proof that pocket factor is missing in the Pvr-virus complex is the positions of the side chains of Phe237 and Tyr205 in the VP1 hydrophobic pocket.)
Conceivably, receptor binding could induce the release of pocket factor. Alternatively, pocket factor release may occur spontaneously and reversibly in the time frame of receptor binding, and receptor binding then traps the virus in a state that prevents pocket factor from returning. In this regard it may be worth noting that some rhinoviruses appear to have lost pocket factor during purification (which is consistent with the reversible binding of pocket factor) (69, 70). In addition, in some rhinoviruses (71–73) (and for poliovirus binding at low temperature [74]), capsid-stabilizing ligands that displace pocket factor can block receptor binding (consistent with the idea that pocket occupancy and receptor binding are mutually exclusive). Thus, the release of pocket factor must either be a consequence of receptor binding or a precondition for it.
It had long been postulated that release of pocket factor is required for receptor-induced conformational changes during viral entry (17, 75, 76), a postulate supported by hints in our previously published low-resolution map, and recently it has been shown that pocket factor is missing in 135S (poliovirus, CAV16) (27, 30) and 80S (EV71, HRV2) (26, 28) particles. Nevertheless, the lack of pocket factor in the virus-receptor complex comes as somewhat of a surprise, since it had generally been thought that release of pocket factor serves as a trigger for the virus-to-135S transition. In contrast, our result demonstrates that pocket factor loss is not sufficient to trigger the 135S rearrangement at a low temperature (even though it might prime the virus for subsequent rearrangements).
What virus residues bind to the hydrophobic pleat in D1?
In the Pvr-poliovirus complex, the hydrophobic site in D1 lies on the under surface of D1, facing the outer surface of poliovirus, but not touching it, held aloft in an arch (Fig. 5A). Importantly, the hydrophobic binding site and the viral surface are separated by a solvent-filled tunnel, which is evident in the density map, starting near the 2-fold hole and running northward toward the 5-fold axis (Fig. 3A). The hydrophobic (aromatic-binding) site of D1 forms the roof of this tunnel. In protein complexes, such cavities are often binding sites, and we postulate that the filling of this cavity will eventually serve as the trigger for expansion (Fig. 6C).
Interestingly, the floor of the tunnel, just below the nectin-specific hydrophobic binding site, is formed by the GH loop of VP3, in its native conformation (see Fig. S4A in the supplemental material). Thus, in mature virus, the GH loop winds around itself, lying roughly parallel to the surface, and forming a plug that blocks what would otherwise be a hole at the quasi-3-fold (the place where VP1, VP2, and VP3 meet at the center of the 5-3-3 triangle [16–18]). In all known high-resolution structures of expanded picornavirus particles (i.e., 80S particles from EV71 [28] and 135S particles from coxsackievirus A16 [27] and poliovirus [30]), the stem of the GH loop of VP3 rearranges to form an extended two-stranded beta sheet and reorients to project in a more nearly radial direction away from the viral surface. Together with shifts in the beta barrels, this conformational change in the GH loop of VP3 opens the quasi-3-fold hole, thereby creating a passageway for the membrane-binding N terminus of VP1 to exit from the virus particle interior. Eventually, the exposed VP1 N terminus will anchor against the GH loop of VP3 (27), and its exposure will permit the virus to interact with membranes (10–12), which is an essential precondition for endocytosis (11, 15) and for RNA delivery (77). It should be noted that the transition from the native state to the extended conformation of the GH loop of VP3 would require at least local conformational rearrangements of neighboring portions of the structure.
Amino acids in the virus that might bind in the nectin pocket.
In the poliovirus-Pvr complex, the GH loop of VP3, which rearranges during viral expansion, includes two aromatic side chains (Phe184 and Tyr176). Phe184 belongs to a well-conserved Asp-x-Phe/Tyr-Thr/Ser sequence motif, which is common to almost all enteroviruses. Similarly, Tyr176 belongs to a conserved Thr/Ser-x-Tyr/Phe-Arg motif, where “x” is usually polar. In all of the polioviruses, these two sequence motifs are separated by exactly four amino acids, although a spacer in the loop of as many as 10 amino acids is seen in other picornaviruses that do not use Pvr.
Of these two choices, we favor Phe184 as the more likely candidate for nectin-like binding. This is because the Tyr176 side chain faces away from the nectin pocket in native virus, as well as in the extended GH loop conformations seen in expanded virus (27, 28). However, Tyr176 cannot be ruled out entirely, since a rotation of the extended GH loop around its long axis could allow the Tyr176 side chain to face the pocket. Similarly, Phe237 of VP1 cannot be ruled out entirely as the pocket occupant, though a massive (and previously unseen) shift and reorganization of the VP1 doorstop region would be required to bring it into position.
Key residues that connect the GH loop of VP3 to the “doorstop.”
Pursuing an analogy with the natural binding of nectins, we have focused on D1 Phe128 (whose binding stabilizes a “super-up” conformation in the “doorstop”) and Phe184 and Tyr 176 of the VP3 GH loop (which appear poised to bind the aromatic-binding pocket on the underside of D1). Given the potential importance of these two sites, attention should be drawn to two key side chains that clearly tie the VP3 GH loop to the VP1 “doorstop,” in its unusual “super-up” conformation. Thus (see Fig. 5B), the side chain carboxylate of VP3 Asp182 binds to the main chain nitrogen in the VP1 Ser233-Leu234 peptide bond. At the same time, the side chain carboxamide group of VP3 Gln178 binds to the main chain oxygen in the Asn235-Asp236 peptide bond. In contrast, in native virus (1HXS.pdb), the “down” conformation is stabilized, with Asp182 and Gln178 binding to the Ser233 and Asp236 side chains, rather than the main chain. Clearly, it is relevant that the Gln178 mutation previously has been shown to affect both virus stability (17, 78) and receptor binding (62) and likely plays a role in regulating the 135S transition. These interactions (seen at low temperature) are clearly involved in stabilizing the receptor-bound conformation and (at higher temperatures) might also connect the rearrangement of the VP3 GH loop to the disordering of the VP1 GH loop.
What protects VP3-Phe184 from solvent in the 160S structure?
The availability of an aromatic binding pocket could be catalytic for the 160S-to-135S transition only if the aromatic side chain (i) would be exposed to solvent, in the absence of Pvr, and (ii) would be sheltered from solvent in the mature virion. Indeed, this would help to explain why native virus is so resistant to uncoating when Pvr is not present. In mature virus, and in the present complex, VP3-Phe184 is protected from solvent exposure by the two stems of the GH loop of VP1 (Fig. 5A). Thus, the Phe184 side chain abuts laterally against the Tyr209 and Ser213 side chains of VP1 (which belong to the amino end of the GH loop), and it is covered by the Ala231-Ala232-Ser233 main chain (which belongs to the carboxyl end). In contrast, once the 135S transition is complete, the GH loop of VP3 has been extended, portions of the GH loop of VP1 have rearranged, and Phe184 is no longer protected (30).
Conceivably, the transition between the two states could begin with the rearrangement of the GH loop of VP1. Indeed, it could be relevant that the Ala-Ala-Ser (VP1 231-233) sequence is a direct contact site for D1 in the poliovirus-Pvr complex (Fig. 5 and see Table S1 in the supplemental material), along with numerous other residues in the GH loop (as described above). However, it is not obvious how those contacts could help the GH loop to rearrange, since the low-temperature poliovirus-Pvr complex appears to be quite stable.
Alternatively, “breathing” motions of the capsid at physiological temperature could cause the transient separation of beta barrels. Then, the rearrangement of the GH loops of VP1 and VP3 might be the effects, rather than the cause, of virus expansion. Such motions could be completely reversible, unless the C-C′-C″ pleat of D1 happened to be waiting to trap them.
Receptor binding and energy.
The action of poliovirus receptor as a catalyst for conformational change is perhaps best understood by examining the energies required for viral expansion (see Fig. 6). In the absence of Pvr, the activation energy for conversion to the 135S state is 145 kcal/mol (8). Adding Pvr to the mixture results in an activation energy of 95 kcal/mol—a decrease of 50 kcal/mol in the energy of the activated complex (8). This present cryo-EM structure represents a semistable state at higher energy than the 80S particle, but the low temperature at which the experiments were carried out does not provide enough thermal energy to surmount the activation energy barrier leading to expansion. Under physiological conditions, however, we hypothesize that further rearrangements of the capsid, stabilized by the underside of the receptor, lead to the irreversible change to the 135S particle.
Breathing and trapping.
It is well established that native poliovirus, above room temperature (but not at low temperatures) undergoes a reversible “breathing” motion that causes the transient externalization of VP4 and of the N-terminal extension of VP1. During these breathing motions, both VP4 and the N terminus of VP1 can become “trapped” on the outside of the virus, using specific antibodies or Fabs (79), and the resulting complexes can be visualized as EM reconstructions (80). Importantly, these reconstructions are icosahedrally symmetric, showing that trapping a few polypeptides in transit through holes in the capsid suffices to keep the entire virion in an expanded state. Just as important, viral expansion was reversible, once the trapping Fab molecules were released (79).
By analogy we propose a model in which physiological “breathing” motions result in the transient expansion of the capsid and the uncoiling of the GH loop of VP3 (Fig. 6). In the poliovirus-Pvr complex, we see the key hydrophobic residue of the receptor, Phe128, interacting with VP1, but the VP3-GH loop remains coiled (Fig. 6B). At physiological temperatures, however, the exposed VP3-GH loop would permit the insertion of Phe184 (or Tyr176) into the hydrophobic binding site in the roof of the tunnel (Fig. 3A and 5A), lowering the activation energy for the transition state for the 160S-to-135S conformational change (Fig. 6C). The thermal energy available at physiological temperatures drives the irreversible expansion of the particle, resulting in the displacement of the receptor and the externalization of VP4 and the N terminus of VP1. Interactions of the N-terminal extension of VP1 with the GH loops of VP3 and VP1 then lock the particle into the expanded state, making the process irreversible. Thus, the receptor would catalyze the 160S-to-135S conversion in a two-step process: first, locking the “doorstop” in the “super-up” conformation and displacing pocket factor (which primes the particle for additional changes but is not sufficient to trigger irreversible expansion), and second, stabilizing an otherwise transient state in which the particle is expanded and the GH loop of VP3 is exposed.
Kinetics of Pvr binding by poliovirus.
As reported previously (5), the receptor has been shown to bind to the virus in two modes: a lower-affinity mode (Kd ∼ 1 μM), which dominates at low temperatures, and a higher-affinity mode (Kd ∼ 0.1 μM), which dominates at ambient temperatures. In the past, we and others have postulated that the two binding modes are structurally distinct, with one representing a weak-binding complex and the other representing a tight-binding complex. However, the current structure (together with a consideration of the on-rates and off-rates for each binding mode) suggests an alternative model. Thus, for the low-affinity mode, kon is 3.6 × 103/Ms and koff is 2.4 × 10−3/s. For the high-affinity mode, kon is 3.2 × 104/Ms and koff is 3.3 × 10−3/s. Neither on-rate approaches the diffusion limit, but the on-rate at higher temperatures is 10-fold faster than it is at lower temperatures. Slow on-rates are generally thought to indicate that a slow conformational change is required for binding. In this case, the conformational changes that are associated with pocket factor release are plausible candidates for the kinetic slow step. Correspondingly, the observed temperature dependence of binding affinity may solely be due to an increase in the speed of pocket factor dissociation (and its associated structural changes) as the temperature rises. If this is the case, then the two binding modes may be structurally indistinguishable.
Generality of the model.
Finally, it should be noted that many aspects of the models we have presented here may be generally relevant to other enteroviruses. Thus, two distinct binding modes have also been observed in rhinoviruses (38) and for coxsackievirus B3 (81) (both of which also use canyon-binding immunoglobulin superfamily members as their receptors). The doorstop region has been identified as a key area for initiating structural transitions in the “trigger-effector” model proposed for the initiation of expansion of EV71, and there are striking similarities between the structures of the products of receptor-induced expansion in a wide range of enteroviruses. These include poliovirus (30) and rhinoviruses (26) (both species C enteroviruses), EV71 (28) and CAV16 (27) (both species A enteroviruses), and coxsackie B3 virus (82) (a species B enterovirus). The similarities include rearrangements of the GH loop of VP1 that expose previously buried aromatic residues, changes in the doorstop, loss of pocket factor, and separation of the EF loop of VP1 from the remainder of the VP1 beta barrel. These similarities strongly suggest that several distinct species of the enterovirus genus share a machinery for inducing the conformational changes that are associated with genome release and membrane translocation. It remains to be seen whether these similarities extend to more distantly related members of the picornavirus family.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Kwong and Vincent Racaniello for a generous supply of the Pvr.
M.S. was sponsored by the Alexander von Humboldt Foundation. This study was supported in part by the grant NIH-AI020566 to J.M.H. and R15AI084085 from the National Institutes of Health to D.M.B. This study was also supported by University of Utah and BYU institutional funds (to D.M.B.) and the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (D.M.B. and N.C.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03101-14.
REFERENCES
- 1.Racaniello VR. 2001. Picornaviridae: the viruses and their replication, p 685–722. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed, vol 1 Lippincott/The Williams & Wilkins Co, New York, NY. [Google Scholar]
- 2.Biederer T. 2006. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics 87:139–150. doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Mendelsohn CL, Wimmer E, Racaniello VR. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda W, Kakunaga S, Itoh S, Shingai T, Takekuni K, Satoh K, Inoue Y, Hamaguchi A, Morimoto K, Takeuchi M, Imai T, Takai Y. 2003. Tage4/Nectin-like molecule-5 heterophilically trans-interacts with cell adhesion molecule Nectin-3 and enhances cell migration. J Biol Chem 278:28167–28172. doi: 10.1074/jbc.M303586200. [DOI] [PubMed] [Google Scholar]
- 5.McDermott BM, Rux AH, Eisenberg RJ, Cohen GH, Racaniello VR. 2000. Two distinct binding affinities of poliovirus for its cellular receptor. J Biol Chem 275:23089–23096. doi: 10.1074/jbc.M002146200. [DOI] [PubMed] [Google Scholar]
- 6.Fenwick ML, Cooper PD. 1962. Early interactions between poliovirus and ERK cells. Some observations on the nature and significance of the rejected particles. Virology 18:212–223. [DOI] [PubMed] [Google Scholar]
- 7.De Sena J, Mandel B. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554–566. [DOI] [PubMed] [Google Scholar]
- 8.Tsang SK, McDermott BM, Racaniello VR, Hogle JM. 2001. A kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J Virol 75:4984–4989. doi: 10.1128/JVI.75.11.4984-4989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow M, Newman JFE, Filman D, Hogle JM, Rowlands DJ, Brown F. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- 10.Fricks CE, Hogle JM. 1990. Cell-induced conformational change of poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol 64:1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuthill TJ, Bubeck D, Rowlands DJ, Hogle JM. 2006. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J Virol 80:172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danthi P, Tosteson M, Li QH, Chow M. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J Virol 77:5266–5274. doi: 10.1128/JVI.77.9.5266-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosteson MT, Chow M. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol 71:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosteson MT, Wang H, Naumov A, Chow M. 2004. Poliovirus binding to its receptor in lipid bilayers results in particle-specific, temperature-sensitive channels. J Gen Virol 85:1581–1589. doi: 10.1099/vir.0.19745-0. [DOI] [PubMed] [Google Scholar]
- 15.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. 2007. Imaging poliovirus entry in live cells. PLoS Biol 5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogle JM, Chow M, Filman DJ. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 17.Filman DJ, Syed R, Chow M, Macadam AJ, Minor PD, Hogle JM. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J 8:1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lentz KN, Smith AD, Geisler SC, Cox S, Buontempo P, Skelton A, DeMartino J, Rozhon E, Schwartz J, Girijavallabhan V, O'Connell J, Arnold E. 1997. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure 5:961–978. doi: 10.1016/S0969-2126(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 19.Badger J, Minor I, Oliveira MA, Smith TJ, Rossmann MG. 1989. Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses. Proteins 6:1–19. doi: 10.1002/prot.340060102. [DOI] [PubMed] [Google Scholar]
- 20.Badger J, Minor I, Kremer MJ, Olivera MA, Smith TJ, Griffith JP, Guerin DMA, Krishnaswamy S, Luo M, Rossmann MG, McKinlay MA, Diana GD, Dutko FJ, Fancher M, Rueckert RR, Heinz BA. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc Natl Acad Sci U S A 85:3304–3308. doi: 10.1073/pnas.85.10.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiremath CN, Grant RA, Filman DJ, Hogle JM. 1995. Binding of the antiviral drug WIN51711 to the Sabin strain of type 3 poliovirus: structural comparison with the drug binding in rhinovirus 14. Acta Crystallogr D 51:473–489. doi: 10.1107/S090744499401084X. [DOI] [PubMed] [Google Scholar]
- 22.Grant RA, Hiremath C, Filman DJ, Syed R, Andries K, Hogle JM. 1994. Structures of poliovirus complexes with antiviral drugs: implications for viral stability and drug design. Curr Biol 4:784–797. [DOI] [PubMed] [Google Scholar]
- 23.Tsang SK, Danthi P, Chow M, Hogle JM. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J Mol Biol 296:335–340. doi: 10.1006/jmbi.1999.3483. [DOI] [PubMed] [Google Scholar]
- 24.Levy H, Bostina M, Filman DJ, Hogle JM. 2010. Cell entry: a biochemical and structural perspective, p 87–107. In Ehrenfeld E, Domingo E, Roos R (ed), The picornaviruses. ASM Press, Washington, DC. [Google Scholar]
- 25.Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. 2010. Picornaviruses. Curr Top Microbiol Immunol 343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog 8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol 84:4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butan C, Filman DJ, Hogle JM. 2014. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol 88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol 85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belnap DM, McDermott BM Jr, Filman DJ, Cheng N, Trus BL, Zuccola HJ, Racaniello VR, Hogle JM, Steven AC. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc Natl Acad Sci U S A 97:73–78. doi: 10.1073/pnas.97.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bubeck D, Filman DJ, Hogle JM. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat Struct Mol Biol 12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bostina M, Bubeck D, Schwartz C, Nicastro D, Filman DJ, Hogle JM. 2007. Single particle cryoelectron tomography characterization of the structure and structural variability of poliovirus-receptor-membrane complex at 30A resolution. J Struct Biol 160:200–210. doi: 10.1016/j.jsb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Bowman VD, Mueller S, Bator CM, Bella J, Peng X, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. 2000. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci U S A 97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y, Mueller S, Chipman PR, Bator CM, Peng X, Bowman VD, Mukhopadhyay S, Wimmer E, Kuhn RJ, Rossmann MG. 2003. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol 77:4827–4835. doi: 10.1128/JVI.77.8.4827-4835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci U S A 105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing L, Tjarnlund K, Lindqvist B, Kaplan GG, Feigelstock D, Cheng RH, Casasnovas JM. 2000. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J 19:1207–1216. doi: 10.1093/emboj/19.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholls A. 1992. GRASP: graphical representation and analysis of surface properties. Columbia University, New York, NY. [Google Scholar]
- 40.Stengel KF, Harden-Bowles K, Yu X, Rouge L, Yin J, Comps-Agrar L, Wiesmann C, Bazan JF, Eaton DL, Grogan JL. 2012. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc Natl Acad Sci U S A 109:5399–5404. doi: 10.1073/pnas.1120606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison OJ, Vendome J, Brasch J, Jin X, Hong S, Katsamba PS, Ahlsen G, Troyanovsky RB, Troyanovsky SM, Honig B, Shapiro L. 2012. Nectin ectodomain structures reveal a canonical adhesive interface. Nat Struct Mol Biol 19:906–915. doi: 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rueckert RR, Pallansch MA. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol 78:315–325. [PubMed] [Google Scholar]
- 43.Yeates TO, Jacobson DH, Martin A, Wychowski C, Girard M, Filman DJ, Hogle JM. 1991. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J 10:2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Grigorieff N, Cheng Y. 2010. GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors. J Struct Biol 172:407–412. doi: 10.1016/j.jsb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez JJ, Luque D, Caston JR, Carrascosa JL. 2008. Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol 164:170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Miller ST, Hogle JM, Filman DJ. 2001. Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 Å resolution. J Mol Biol 307:499–512. doi: 10.1006/jmbi.2001.4485. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 52.Nicholls RA, Long F, Murshudov GN. 2012. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr 68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 55.Jacobson DH, Hogle JM, Filman DJ. 1996. A pseudo-cell based approach to efficient crystallographic refinement of viruses. Acta Crystallogr D Biol Crystallogr 52:693–711. doi: 10.1107/S0907444996001060. [DOI] [PubMed] [Google Scholar]
- 56.Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Schrodinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1 Schrodinger LLC, New York, NY. [Google Scholar]
- 60.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol 79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colston EM, Racaniello VR. 1995. Poliovirus variants selected on mutant receptor-expressing cells identify capsid residues that expand receptor recognition. J Virol 69:4823–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colston E, Racaniello VR. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J 13:5855–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao S, Racaniello V. 1997. Allele-specific adaptation of poliovirus VP1 B-C loop variants to mutant cell receptors. J Virol 71:9770–9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harber J, Bernhardt G, Lu HH, Sgro JY, Wimmer E. 1995. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology 214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 65.Kabsch W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr A 32:922–923. doi: 10.1107/S0567739476001873. [DOI] [Google Scholar]
- 66.Katpally U, Smith TJ. 2007. Pocket factors are unlikely to play a major role in the life cycle of human rhinovirus. J Virol 81:6307–6315. doi: 10.1128/JVI.00441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wien MW, Curry S, Filman DJ, Hogle JM. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat Struct Biol 4:666–674. doi: 10.1038/nsb0897-666. [DOI] [PubMed] [Google Scholar]
- 68.Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, Hogle JM. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J Virol 74:1342–1354. doi: 10.1128/JVI.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim SS, Smith TJ, Chapman MS, Rossmann MC, Pevear DC, Dutko FJ, Felock PJ, Diana GD, McKinlay MA. 1989. Crystal structure of human rhinovirus serotype 1A (HRV1A). J Mol Biol 210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 70.Zhao R, Pevear DC, Kremer MJ, Giranda VL, Kofron JA, Kuhn RJ, Rossmann MG. 1996. Human rhinovirus 3 at 3.0 Å resolution. Structure 4:1205–1220. doi: 10.1016/S0969-2126(96)00128-1. [DOI] [PubMed] [Google Scholar]
- 71.Pevear DC, Fancher MJ, Felock PJ, Rossmann MG, Miller MS, Diana G, Treasurywala AM, McKinlay MA, Dutko FJ. 1989. Conformation change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J Virol 63:2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heinz BA, Shepard DA, Rueckert RR. 1990. Escape mutant analysis of a drug-binding site can be used to map functions in the rhinovirus capsid, p 173–186. In Laver WG, Air GM (ed), Use of X-ray crystallography in the desing of antiviral agents. Academic Press Inc, San Diego, CA. [Google Scholar]
- 73.Shepard DA, Heinz BA, Rueckert RR. 1993. WIN 52035-2 inhibits both attachment and eclipse of human rhinovirus 14. J Virol 67:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dove AW, Racaniello VR. 2000. An antiviral compound that blocks structural transitions of poliovirus prevents receptor binding at low temperatures. J Virol 74:3929–3931. doi: 10.1128/JVI.74.8.3929-3931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oliveira MA, Zhao R, Lee WM, Kremer MJ, Minor I, Rueckert RR, Diana GD, Pevear DC, Dutko FJ, McKinlay MA, et al. 1993. The structure of human rhinovirus 16. Structure 1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 76.Mosser AG, Rueckert RR. 1993. WIN 51711-dependent mutants of poliovirus type 3: evidence that virions decay after release from cells unless drug is present. J Virol 67:1246–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. 2013. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol 87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macadam AJ, Arnold C, Howlett J, John A, Marsden S, Taffs F, Reeve P, Hamada N, Wareham K, Almond J, Cammack N, Minor PD. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology 172:408–414. doi: 10.1016/0042-6822(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 79.Li Q, Yafal AG, Lee YM-H, Hogle J, Chow M. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J Virol 68:3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin J, Lee LY, Roivainen M, Filman DJ, Hogle JM, Belnap DM. 2012. Structure of the Fab-labeled “breathing” state of native poliovirus. J Virol 86:5959–5962. doi: 10.1128/JVI.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carson SD. 2014. Kinetic models for receptor-catalyzed conversion of coxsackievirus b3 to A-particles. J Virol 88:11568–11575. doi: 10.1128/JVI.01790-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. 2014. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol 88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.