ABSTRACT
Stress granules (SGs) are protein-mRNA aggregates that are formed in response to environmental stresses, resulting in translational inhibition. SGs are generally believed to play an antiviral role and are manipulated by many viruses, including various alphaviruses. GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1) is a key component and commonly used marker of SGs. Its homolog G3BP2 is a less extensively studied SG component. Here, we demonstrate that Chikungunya virus (CHIKV) infection induces cytoplasmic G3BP1- and G3BP2-containing granules that differ from bona fide SGs in terms of morphology, composition, and behavior. For several Old World alphaviruses it has been shown that nonstructural protein 3 (nsP3) interacts with G3BPs, presumably to inhibit SG formation, and we have confirmed this interaction in CHIKV-infected cells. Surprisingly, CHIKV also relied on G3BPs for efficient replication, as simultaneous depletion of G3BP1 and G3BP2 reduced viral RNA levels, CHIKV protein expression, and viral progeny titers. The G3BPs colocalized with CHIKV nsP2 and nsP3 in cytoplasmic foci, but no colocalization with nsP1, nsP4, or dsRNA was observed. Furthermore, G3BPs could not be detected in a cellular fraction enriched for CHIKV replication/transcription complexes, suggesting that they are not directly involved in CHIKV RNA synthesis. Depletion of G3BPs did not affect viral entry, translation of incoming genomes, or nonstructural polyprotein processing but resulted in severely reduced levels of negative-stranded (and consequently also positive-stranded) RNA. This suggests a role for the G3BPs in the switch from translation to genome amplification, although the exact mechanism by which they act remains to be explored.
IMPORTANCE Chikungunya virus (CHIKV) causes a severe polyarthritis that has affected millions of people since its reemergence in 2004. The lack of approved vaccines or therapeutic options and the ongoing explosive outbreak in the Caribbean underline the importance of better understanding CHIKV replication. Stress granules (SGs) are cytoplasmic protein-mRNA aggregates formed in response to various stresses, including viral infection. The RNA-binding proteins G3BP1 and G3BP2 are essential SG components. SG formation and the resulting translational inhibition are generally considered an antiviral response, and many viruses manipulate or block this process. Late in infection, we and others have observed CHIKV nonstructural protein 3 in cytoplasmic G3BP1- and G3BP2-containing granules. These virally induced foci differed from true SGs and did not appear to represent replication complexes. Surprisingly, we found that G3BP1 and G3BP2 were also needed for efficient CHIKV replication, likely by facilitating the switch from translation to genome amplification early in infection.
INTRODUCTION
Chikungunya virus (CHIKV) is a reemerging arbovirus that is currently causing a large outbreak in the Caribbean, affecting 41 countries and territories with ∼1 million suspected and ∼22,500 confirmed cases (http://www.cdc.gov/chikungunya/geo/americas.html). CHIKV will likely continue to spread throughout the Americas, as competent vectors are present in many countries in the region, including parts of the United States. The magnitude of this recent outbreak and the fact that CHIKV might soon be endemic in many parts of the world stress the need for a deeper understanding of this important human pathogen.
In the past decade, there has been an increasing interest in stress granules (SGs) and their interplay with the replication of a variety of viruses (reviewed in references 1 and 2). SGs are cytoplasmic ribonucleoprotein condensations formed in eukaryotic cells in response to environmental stress, and their appearance is linked to inhibition of translation (3). SGs contain stalled translation preinitiation complexes and are characterized by the presence of cellular mRNAs, translation initiation factors (e.g., eIF3 and eIF4B), the small ribosomal subunit, and RNA-binding proteins such as T-cell-restricted intracellular antigen 1 (TIA-1), TIA-1-related protein (TIAR), and GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1) (4–6). Environmental stress is sensed by double-stranded RNA (dsRNA)-dependent protein kinase (PKR) (7), PKR-like endoplasmic reticulum kinase (PERK) (8), general control nonderepressible 2 (GCN2) kinase (9), or heme-regulated inhibitor kinase (HRI) (10). Upon their activation, these kinases phosphorylate the α-subunit of eukaryotic translation initiation factor 2 (eIF2α), which, in turn, leads to dephosphorylation of G3BP1 and enables G3BP1 multimerization and subsequent SG formation (5). RNA-binding proteins TIA-1 and TIAR play a similar role in SG formation (11). The G3BP1 homolog G3BP2 is relatively poorly characterized, but it also localizes to SGs (12, 13). The G3BPs share a conserved acidic domain, a nuclear transport factor 2-like domain, a number of SH3 domain binding motifs, an arginine/glycine-rich box, and an RNA recognition motif. The last two elements are associated with RNA binding. The G3BPs may be partly functionally redundant, but there are also functional differences. For example, G3BP1 has phosphorylation-dependent endoribonuclease activity, which has not been reported for G3BP2 (14). Furthermore, only G3BP2 binds the N-terminal domain of IκBα (15), and differences in the number of SH3 domain binding motifs (PxxP) suggest that G3BP1 and G3BP2 differ in their interactions with other proteins.
RNA viruses commonly induce SG formation via dsRNA replication intermediates that are sensed by PKR, although in some cases activation of PERK by ER stress is involved (reviewed in reference 1). Alphaviruses also induce the formation of SGs or aggregates resembling those, as G3BP1-containing foci have been observed in cells infected with Semliki Forest virus (SFV) and Sindbis virus (SINV) or transfected with a CHIKV replicon (16–19). Several laboratories have demonstrated colocalization and coimmunoprecipitation of alphavirus nsP2, nsP3, or nsP4 with G3BPs, and it was therefore speculated that these host proteins might be associated with the alphavirus replication machinery (16, 18, 20–22). The interaction of nsP3 with G3BP1 has been studied extensively (18, 20, 21, 23, 24), and the induction of SGs by SFV has also been characterized in detail (17, 19, 23). SFV induces SGs early in infection; these are disassembled around 8 h postinfection (p.i.) following recruitment of G3BPs by nsP3 (17). Colocalization of G3BPs with SFV nsP1 and dsRNA suggests that they are recruited to replication and transcription complexes (RTCs) (17). Fros et al. showed that expression of CHIKV nsP3 was sufficient to induce G3BP-containing foci (16). The C-terminal repeat of SFV nsP3 that allows G3BP binding is conserved among Old World alphaviruses and was demonstrated to be responsible for the interaction between CHIKV nsP3 and both G3BPs as well (23), although earlier studies suggested the involvement of another nsP3 domain (16). In general, SG formation is considered an antiviral response that limits translation, and it has been hypothesized that alphaviruses prevent SG formation through the nsP3-mediated sequestering of G3BPs into cytoplasmic granules (16, 17). Despite the reported CHIKV nsP3-G3BP1 interaction and the SG-like aggregates that have been described to occur in cells transfected with a CHIKV replicon (16), it remains unclear whether such aggregates are formed in the context of a complete CHIKV infection and what the role of these SG-like G3BP aggregates might be. We therefore set out to elucidate the role of SGs, in particular that of the G3BPs, in the CHIKV replicative cycle. Rather than focusing only on G3BP1, we also analyzed the role of the often-overlooked G3BP2 and assessed the impact of their combined knockdown on CHIKV replication. We show that late in the CHIKV replication cycle, the bulk of G3BP1 and G3BP2 is not associated with the viral RTCs but sequestered in nsP3-G3BP aggregates, likely to prevent the formation of bona fide SGs. Surprisingly, we discovered that the G3BPs, in particular G3BP2, also have a proviral function early in CHIKV replication, as their knockdown delayed the accumulation of negative-stranded RNA. The G3BPs appear to play a regulatory role in the switch from genome translation to negative-strand RNA synthesis, perhaps by clearing ribosomes from the viral RNA, thus enabling it to serve as a template for RNA synthesis.
MATERIALS AND METHODS
Cells, viruses, and virus titration.
Vero E6 and 293/ACE2 cells (25) were cultured and infected with CHIKV Leiden Synthetic 3 (LS3) (GenBank accession number KC149888) or enhanced green fluorescent protein (eGFP)-expressing reporter virus CHIKV LS3-GFP (GenBank accession number KC149887) as described previously (26). CHIKV LS3 is an infectious clone-derived virus with a genome sequence based on the consensus sequence of “recent A226V strains,” with growth kinetics and other properties similar to those of natural isolates (26). All experiments were performed with this virus unless indicated otherwise. CHIKV variants expressing Renilla luciferase either as an nsP3 fusion (ICRES1-P3Rluc) or from a duplicated subgenomic RNA (sgRNA) promoter (ICRES1-2SG-Rluc) were generated by standard cloning techniques, based on previously described constructs (27). The ICRES1 CHIKV variants were based on the sequence of natural isolate LR2006-OPY1. A replication-deficient variant (ICRES1-P3Rluc-nsP4-GAA) was created by mutating the GDD motif of nsP4 (amino acids [aa] 465 to 467) to GAA using QuikChange site-directed mutagenesis. CHIKV strain ITA07-RA1 (GenBank accession number EU244823) was isolated during the 2007 outbreak in Italy (26). Virus titers were determined by plaque assay on Vero E6 cells in six-well clusters in medium containing 1.2% Avicel RC-581 (FMC BioPolymer) as described previously (26). All work with live CHIKV was performed inside biosafety cabinets in a biosafety level 3 facility. CHIKV LR2006-OPY1-nsP4/FLAG, which was used for colocalization studies, encodes a 3× FLAG-tagged nsP4 (unpublished data). SINV-GFP was created by cloning eGFP into the previously described SINV MRE16 infectious clone (28) using standard techniques (details available upon request). CVB3-GFP (29) was a kind gift from Frank van Kuppeveld (Utrecht University, the Netherlands).
In vitro transcription and RNA transfection.
In vitro transcription and RNA transfection were essentially performed as described previously (26). The ICRES1-P3Rluc and ICRES1-2SG-Rluc plasmids were linearized with NotI and transcribed using the mMessage mMachine SP6 kit (Ambion).
Indirect immunofluorescence microscopy.
For visualization of SGs, Vero E6 cells were grown on coverslips and treated for 60 min with 0.5 mM sodium arsenite (Sigma). Disassembly of SGs was induced by treatment with 100 μg/ml of cycloheximide (CHX) for 30 min. Cells were processed for indirect immunofluorescence microscopy as described previously (26). Double-stranded RNA was detected using mouse monoclonal antibody J2 (English & Scientific Consulting; 10010500). Mouse anti-G3BP1 (BD Transduction Laboratories; 61126) or rabbit anti-G3BP1 (Aviva; ARP37713) was used to detect G3BP1. G3BP2 was detected with a rabbit antiserum (12) generously provided by Christer Larsson (Lund University, Sweden) or using a commercially available antibody (Assay Biotech; C18193-2). Other SG components were visualized using antibodies against eIF3α (Santa Cruz; sc-16377), TIA-1 (Santa Cruz; sc-1751), TIAR (Santa Cruz; sc-1749), p-eIF2α (Cell Signaling; 9721), or PABP (Santa Cruz, sc-32318). Primary antibodies were detected with Cy3- or Alexa 488-conjugated secondary antibodies (Jackson/Life Technologies). Nuclei were stained with Hoechst 33342. The coverslips were analyzed using a Leica TCS SP5 confocal microscope and LAS AF Lite software (Leica).
To assess colocalization of G3BPs with CHIKV nsPs, Vero cells were grown on coverslips and infected with CHIKV LR2006-OPY1-nsP4/FLAG at a multiplicity of infection (MOI) of 5. At 6 h p.i. cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and permeabilized with ice-cold methanol. The nsPs were visualized using mouse anti-FLAG, rabbit anti-o'nyong-nyong nsP1, anti-CHIKV helicase (nsP2), and anti-CHIKV nsP3 sera. Primary antibodies were detected with Alexa 488- or Alexa 568-conjugated secondary antibodies. The coverslips were analyzed using a Zeiss LSM 710 confocal microscope.
Western blot analysis.
Western blot analysis was performed basically as described previously (26) using the primary antibodies listed in the previous paragraph. In addition, rabbit antisera against CHIKV nsP1 and nsP4 (raised against bacterially expressed full-length recombinant proteins), nsP2 (aa 453 to 798), and nsP3 (aa 1 to 320) were used. Rabbit antiserum against CHIKV E2 (30) was kindly provided by Gorben Pijlman (Wageningen University, the Netherlands).
Cellular fractionation experiments.
The subcellular fractionation and isolation of active replication complexes from CHIKV-infected Vero E6 cells have been described elsewhere (31). Briefly, CHIKV LS3-infected cells were harvested at 6 h p.i. by trypsinization and lysed using a Dounce homogenizer. Unlysed cells, debris, and nuclei were pelleted by centrifugation at 1,000 × g, and the resulting postnuclear supernatant (PNS) was further fractionated in a 15,000 × g pellet (P15) and supernatant (S15).
RNA interference.
ON-TARGETplus SMARTpool small interfering RNAs (siRNAs) targeting G3BP1 (L-012099-00), G3BP2 (L-015329-01), the corresponding G3BP2-specific deconvoluted pool, and scrambled (non-targeting pool) control siRNAs (D-001810-10) were obtained from Dharmacon. C911 mutant siRNAs (32) and a nontargeting control siRNA were custom made by Sigma. 293/ACE2 cells were transfected with a final concentration of 50 to 100 nM siRNA using DharmaFECT1 (Dharmacon) according to the manufacturer's instructions. Cells that were transfected with two siRNA pools, targeting both G3BPs, received in total the same amount of siRNA and transfection reagent as the cells transfected with one SMARTpool. At 48 h posttransfection (p.t.), cells were infected with CHIKV or harvested to determine the silencing efficiency by Western blotting. To quantify the replication of the eGFP-expressing reporter virus, siRNA-transfected cells in 96-well clusters were infected with CHIKV LS3-GFP. Cells were fixed with 3% paraformaldehyde in PBS at 16 to 24 h p.i., depending on the MOI that was used. In parallel, the viability of siRNA-transfected cells was assessed at 48 h p.t. using the CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega). Absorbance and eGFP expression were quantified using a Berthold Mithras LB 940 96-well plate reader.
Rescue experiments with an siRNA-resistant G3BP2 expression plasmid.
G3BP2 expression plasmid pCMV-FLAG-G3BP2 was created by cloning the G3BP2 coding sequence obtained from MRC5 cDNA into p3xFLAG-CMV-10 (Sigma). Transcripts from the resulting construct are resistant to G3BP2 siRNAs 2 and 4, which target the 3′ untranslated region (UTR) of the natural G3BP2 mRNA. During rescue experiments the G3BP2 expression plasmid or the empty vector was cotransfected with the G3BP2 siRNAs using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. These cells were infected with CHIKV at 24 h p.t.
RNA isolation, gel electrophoresis, and in-gel hybridization.
CHIKV RNA isolation using acid phenol, denaturing gel electrophoresis, and detection by in-gel hybridization with 32P-labeled oligonucleotides specific for positive or negative-strand RNA were performed as described previously (26).
qRT-PCR.
An internally controlled multiplex quantitative TaqMan real-time PCR (qRT-PCR) was used to determine the copy number of CHIKV genomic RNA (probe in nsP1-coding region) and total RNA (probe in E1-coding region). Briefly, forward (CTAGCTATAAAACTAAUAGAGCAGGAAATTG) and reverse (GACTTTTCCTGCGGCAGATGC) primers and a probe (Texas red-TCCGACATCATCCTCCTTGCTGGCG-black hole quencher 2 [BHQ2]) in the nsP1-encoding region were used in combination with a set of primers and 6-carboxyfluorescein (FAM)-labeled probe specific for the E1-coding region that has been described previously (33). Samples were analyzed using the SensiFast Probe (Bioline) or TaqMan Fast Virus 1-Step (ABI) qRT-PCR kit and a CFX384 Touch Real-Time PCR detection system (Bio-Rad) according to the manufacturers' instructions. Serial dilutions of in vitro-transcribed RNA were used as standards for copy number determination, and the cellular PGK1 mRNA was used as an internal control in multiplex reactions.
Luciferase assays.
293/ACE2 cells (5 × 103 per well) were seeded in 96-well plates and transfected with siRNAs (described above). After 48 h, the cells were infected with ICRES1-P3Rluc or ICRES1-2SG-Rluc at an MOI of 5. Alternatively, the cells were transfected with 100 ng of in vitro-transcribed full-length viral RNA (ICRES1-P3Rluc, ICRES1-2SG-Rluc, or ICRES1-P3Rluc-nsP4-GAA) using Lipofectamine 2000. At the desired time points, cells were lysed in passive lysis buffer (Promega) and luciferase substrate was added according to the manufacturer's instructions. Luciferase activity was measured in a GloMax 96 microplate luminometer (Promega).
Metabolic labeling and immunoprecipitation.
Proteins synthesized in infected 293/ACE2 cells were labeled with [35S]methionine and [35S]cysteine ([35S]Met/Cys) as described previously (26). Cells were lysed in Laemmli sample buffer, and antibody binding was carried out overnight at 4°C in AVIP buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholine) containing a final concentration of 0.5% SDS. Immune complexes were pulled down using a 1:1 mixture of protein A/G-Sepharose beads (GE Healthcare). Beads were washed three times in AVIP buffer before elution by boiling in Laemmli sample buffer for 5 min. Eluted proteins were separated in 10% polyacrylamide gels, and detection was done by autoradiography with phosphorimager screens and a Typhoon-9410 scanner (GE Healthcare).
Data analysis.
Band intensities were quantified using Quantity One v4.5.1 (Bio-Rad) or ImageQuant TL software (GE Healthcare). Two or three independent experiments were quantified (one representative experiment is shown in figures). Statistical significance was calculated using a two-tailed Student t test in GraphPad Prism 5 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RESULTS
CHIKV replication induces G3BP-containing foci that resemble SGs.
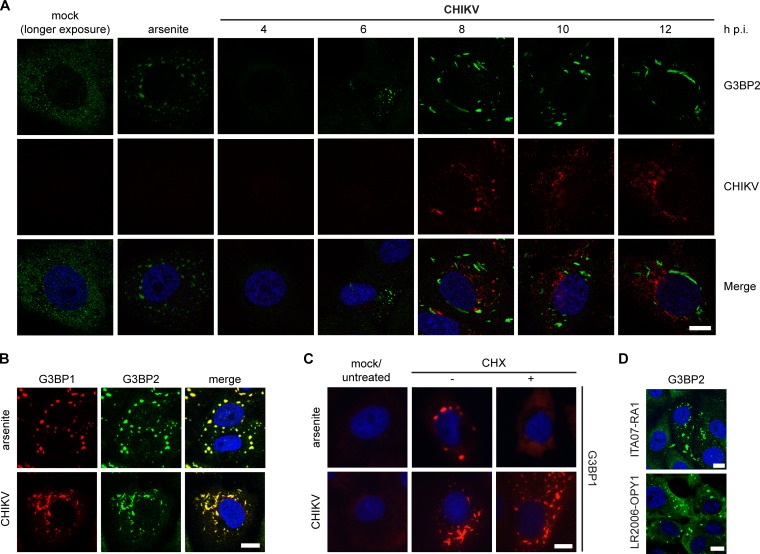
A variety of viruses, including alphaviruses, induce the formation of SGs or SG-like cytoplasmic granules. To investigate whether CHIKV infection induces SGs, Vero E6 cells were infected and the localization of the SG marker G3BP2 was monitored (Fig. 1A). Arsenite, commonly used to induce SGs via oxidative stress, was employed as a positive control. In uninfected cells, G3BP2 displayed a diffuse cytoplasmic distribution, whereas in CHIKV-infected cells, G3BP2-containing foci appeared by 6 to 8 h p.i., continued to grow in size until ∼10 h p.i., and remained present thereafter. Similar observations were made for G3BP1 (data not shown), and costaining for G3BP1 and G3BP2 revealed that the two proteins localize to the same puncta (Fig. 1B). These G3BP puncta did not possess the typical rounded morphology of arsenite- or heat shock-induced SGs but had a more rod-like appearance (Fig. 1A to C). Genuine SGs are dispersed upon cycloheximide (CHX) treatment, which stabilizes polysomes and prevents their disassembly, a crucial step in SG formation. As expected, arsenite-induced G3BP1 puncta readily dispersed upon CHX treatment. However, the CHIKV-induced G3BP1 puncta were not affected by CHX treatment (Fig. 1C). Identical observations were made when the granules were stained for G3BP2 (data not shown). To exclude the possibility that the induction of G3BP-containing puncta was a unique feature of the infectious clone-derived strain LS3, which is based on the consensus sequence of several CHIKV strains, we also analyzed cells infected with other strains. Natural isolate CHIKV ITA07-RA1 and CHIKV LR2006-OPY1-nsP4/FLAG, which is based on a clinical isolate from La Reunion, induced similar G3BP2-containing granules (Fig. 1D), suggesting that this is a general CHIKV property.
FIG 1.
Induction of G3BP-containing foci by CHIKV replication. (A) Vero E6 cells were infected with CHIKV (MOI, 5), fixed at the indicated time points postinfection, and immunostained for G3BP2 and CHIKV. A longer exposure of mock-infected cells is shown to visualize diffuse G3BP2 staining. (B) CHIKV-induced granules (MOI, 5; 8 h p.i.) were costained for G3BP1 and G3BP2. (C) Vero E6 cells were infected with CHIKV (MOI, 5; analyzed at 8 h p.i.) or treated with arsenite (0.5 mM for 1 h) to induce SG formation and subsequently incubated with CHX (100 μg/ml for 30 min), followed by immunostaining for G3BP1. (D) G3BP2 immunostaining of Vero cells that were infected with CHIKV ITA07-RA1 or LR2006-OPY1-nsP4/FLAG (MOI, 5) and fixed at 6 h p.i. Scale bar: 10 μm.
The composition of CHIKV-induced granules differs from that of genuine SGs.
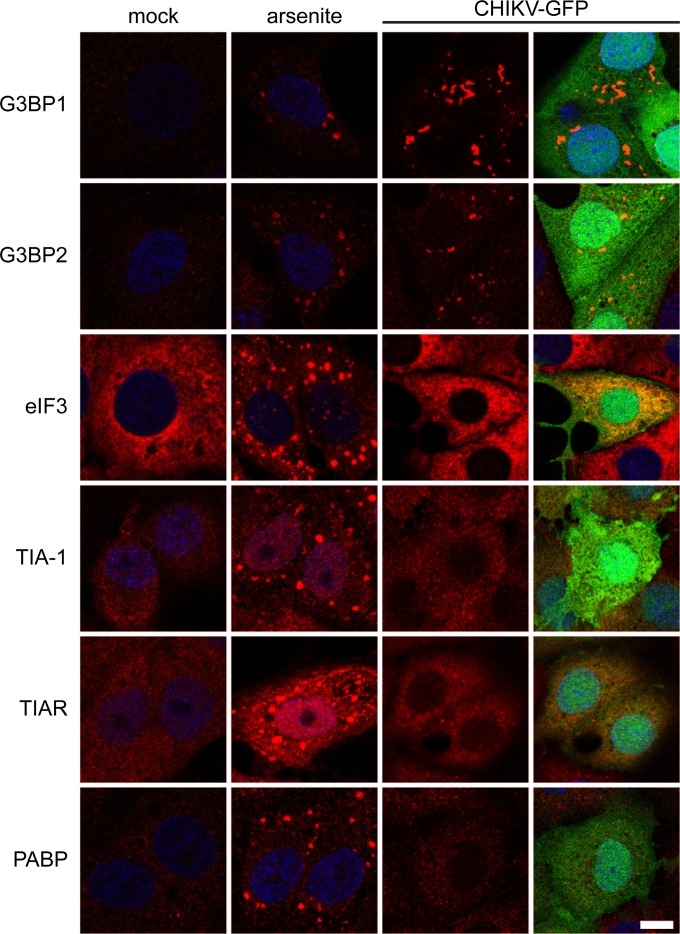
The composition of the CHIKV-induced granules was examined by immunostaining for several SG markers. In arsenite-induced SGs, G3BP1, G3BP2, TIA-1, TIAR, PABP, and eIF3 could readily be detected (Fig. 2). However, the CHIKV-induced granules were labeled only for G3BP1 and G3BP2, demonstrating that they differ not only in morphology and response to CHX treatment but also in protein composition.
FIG 2.
Composition of arsenite-induced SGs and CHIKV-induced granules. Vero E6 cells were treated with arsenite or infected with CHIKV-GFP (MOI, 5; fixed at 8 h p.i.). After fixation, the localization of the SG markers indicated to the left of each row was analyzed by immunofluorescence microscopy. The rightmost column shows the overlays of the signals of the SG marker and eGFP. Scale bar: 10 μm.
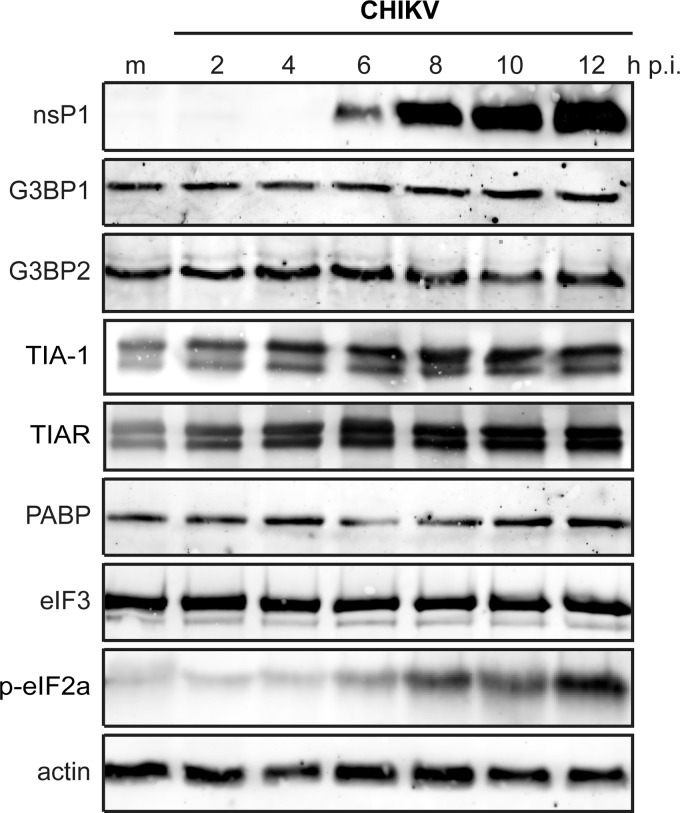
Some viruses, e.g., poliovirus (34), block SG formation by cleaving key SG components, like G3BP1. To assess whether the CHIKV-induced granules lack SG components due to their degradation or downregulation, the expression level of a number of SG markers in CHIKV-infected cells was determined. Western blot analysis showed that G3BP1 and G3BP2 protein levels did not change during the course of CHIKV infection (Fig. 3). Also, the expression levels of eIF3 and PABP remained unchanged during CHIKV infection. As expected, the level of eIF2α phosphorylation increased strongly in CHIKV-infected cells between 6 and 12 h p.i. (Fig. 3). The expression level of TIA-1 and TIAR proteins increased ∼2-fold within 6 h p.i. (Fig. 3).
FIG 3.
Levels of stress granule proteins during CHIKV infection. 293/ACE2 cells were infected with CHIKV (MOI, 5), and whole-cell lysates were prepared at the indicated time points postinfection. The expression level of the indicated proteins was determined by Western blotting. Mock-infected cells (m) were included as a negative control, and actin was used as a loading control.
G3BPs colocalize with CHIKV nsP2 and nsP3 but not with nsP1, nsP4, or dsRNA.
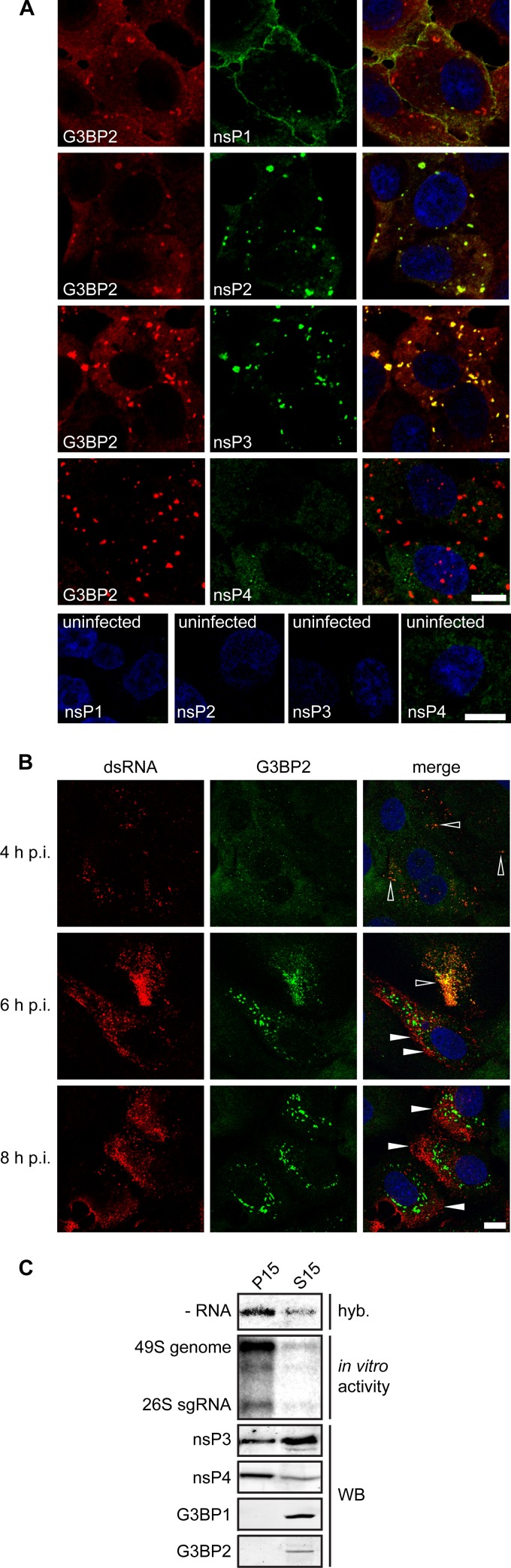
To investigate if the G3BP granules represent CHIKV replication complexes, we analyzed the possible colocalization of G3BP2 with CHIKV nsPs and dsRNA. Vero cells were infected with CHIKV at an MOI of 5, fixed at 6 h p.i., and immunostained for G3BP2 and CHIKV nsPs. This staining confirmed the previously reported nsP3-G3BP2 colocalization in cytoplasmatic granules (Fig. 4A). In addition, we observed nsP2-G3BP2 colocalization in similar foci. Strikingly, no colocalization of G3BP2 with nsP1 or nsP4 could be detected (Fig. 4A). Next, we analyzed the distribution of dsRNA and G3BP2 in CHIKV-infected cells (Fig. 4B). Only early in infection (4 h p.i.), when the dsRNA and G3BP2 signals were barely detectable, did there appear to be some colocalization of G3BP2 and dsRNA. At 6 h p.i. there was a very limited overlap between the dsRNA- and G3BP2-containing puncta, and at 8 h p.i. most of the dsRNA and G3BP2 signals were clearly not colocalizing (Fig. 4B).
FIG 4.
Localization of CHIKV nonstructural proteins, dsRNA, and G3BP. (A) Vero cells were infected with a CHIKV encoding nsP4-FLAG (LR2006-OPY1- nsP4/FLAG) at an MOI of 5, fixed at 6 h p.i., and stained for G3BP2, nsP1, nsP2, nsP3, and nsP4/FLAG. The bottom part of panel A shows the staining of uninfected cells, with the antibodies indicated in each frame. (B) Vero E6 cells were infected with CHIKV (MOI, 5), fixed at the indicated time points p.i., and stained for G3BP2 and dsRNA. Open arrowheads indicate colocalization of dsRNA with G3BP2, whereas closed arrowheads indicate some examples of nonoverlapping signals. Scale bar: 10 μm. (C) Distribution of G3BPs and CHIKV nsPs between the “heavy membrane” fraction P15 and “cytoplasmic” fraction S15. Vero E6 cells were infected with CHIKV (MOI, 5) and then harvested, lysed, and subjected to subcellular fractionation at 6 h p.i. The presence of CHIKV negative-stranded RNA was determined by hybridization with a specific probe (hyb). The RNA synthesizing activity was assessed with an in vitro assay in which the incorporation of [32P]CTP into CHIKV RNA was determined (31). The levels of CHIKV nsP3, nsP4, G3BP1, and G3BP2 in the P15 and S15 fractions were determined by Western blotting (WB).
We did not observe complete colocalization between the nsP1 and nsP4 foci and the dsRNA puncta, which makes it impossible to unequivocally pinpoint the intracellular location of the RTC. This is likely because only a fraction of nsP1 and nsP4 is located within RTCs. Nonetheless, regardless of which of these markers most accurately identifies the CHIKV RTCs, clearly none of them colocalized with the G3BPs, suggesting that the G3BP granules did not represent the RTCs. All 4 nsPs are needed to form the RTC, but nsP2 and nsP3 also have other functions and different intracellular localizations outside the membrane-associated RTC. Thus, it is likely that the previously reported nsP3-G3BP interaction occurs with the pool of nsP3 that is not associated with the RTC.
G3BPs are not associated with membrane-bound CHIKV replication complexes.
To independently confirm that the G3BP granules observed in our immunofluorescence microscopy analysis were not the (membrane-associated) CHIKV RTCs, subcellular fractionation experiments were performed. CHIKV-infected cells were fractionated into a crude membrane fraction (P15) and a cytosolic fraction (S15). The P15 fraction contained 80 to 90% of the in vitro RNA synthesizing activity and was enriched in negative-strand RNA and nsP4, suggesting that it contained the majority of the membrane-associated RTCs (Fig. 4C). The bulk of nsP3 was found in the cytosolic S15 fraction. G3BP1 and G3BP2 were detected exclusively in the S15 fraction, suggesting that they are not associated with the RTCs. This supports the idea that the pool of nsP3 that associates with G3BPs differs from the one that is part of the RTC. The subcellular distribution of G3BPs in mock-infected cells was similar to that in CHIKV-infected cells (data not shown).
Depletion of G3BPs inhibits CHIKV replication.
To further study the role of G3BP1 and G3BP2 in CHIKV replication, we assessed the effect of their knockdown on viral replication (Fig. 5). It was previously shown that G3BP expression is controlled by an apparent feedback mechanism that results in the upregulation of one G3BP when the expression of the other is reduced (35). These observations suggest that the proteins are functionally linked, and therefore, the impact of simultaneous knockdown of G3BP1 and G3BP2 was also examined.
FIG 5.
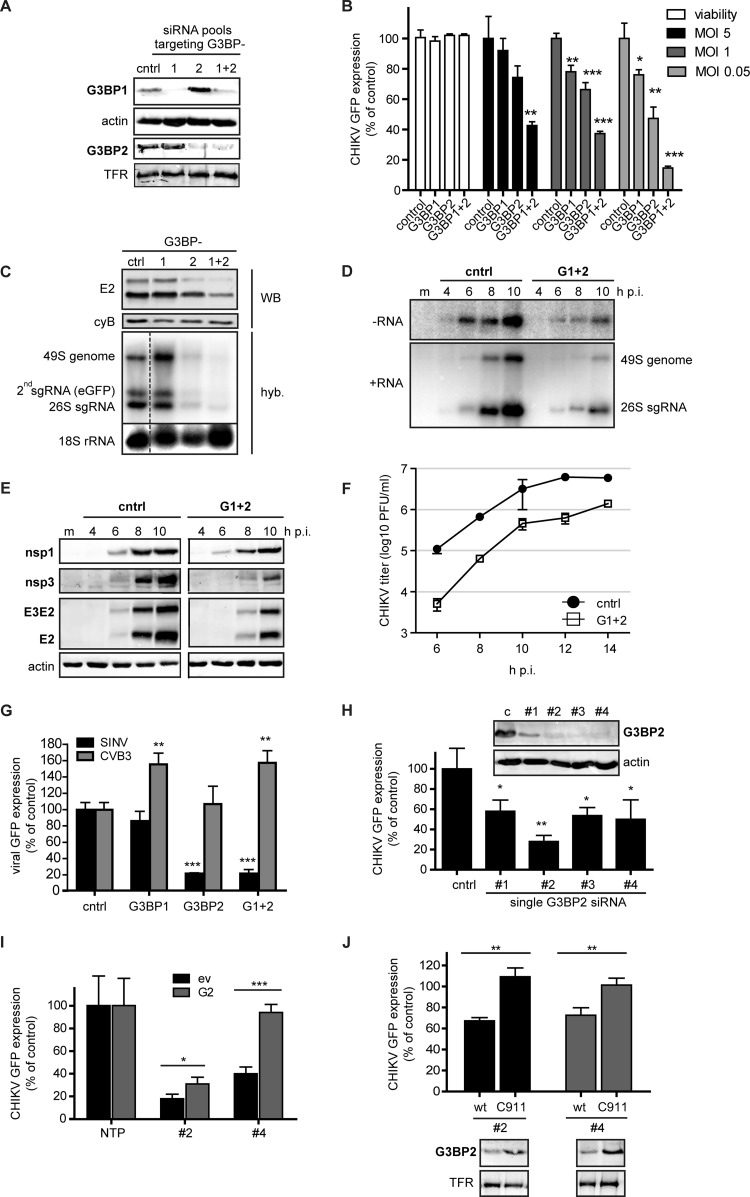
Effect of siRNA-mediated depletion of G3BP1 and G3BP2 on CHIKV replication. (A) Western blot analysis of the protein levels of G3BP1 and G3BP2 in 293/ACE2 cells transfected with control siRNAs or those targeting G3BP1, G3BP2, or both. The transferrin receptor (TFR) and actin were used as loading controls. (B) Cell viability and CHIKV-driven eGFP expression in cells that were depleted of G3BP1 and G3BP2 and subsequently infected with reporter virus CHIKV LS3-GFP at an MOI of 5 (black bars), 1 (dark gray bars), or 0.05 (light gray bars). Cell viability was determined at 48 h p.t. (white bars), and eGFP expression was quantified at 16, 20, or 24 h p.i., depending on the MOI used. (C) G3BP-depleted, CHIKV-infected cells (MOI, 0.05) were analyzed for E2 expression (by Western blotting using cyclophilin B as a loading control) and positive-strand RNA levels (by in-gel hybridization [hyb]) at 24 h p.i. (D) In-gel hybridization analysis of CHIKV RNA levels with probes specific for negative-strand (−RNA) or positive-strand (+RNA) RNA in G3BP-depleted 293/ACE2 cells. The cells were infected with CHIKV (MOI, 5) 48 h after siRNA transfection, and total RNA was isolated at the indicated time points. cntrl, control. (E) Western blot analysis of CHIKV protein expression levels after G3BP depletion. 293/ACE2 cells were transfected with control or G3BP-specific siRNAs and 48 h later infected with CHIKV (MOI, 5). Cell lysates for Western blot analysis were harvested at the indicated time points and analyzed for the viral proteins indicated, using actin as a loading control. (F) Infectious progeny titers of CHIKV-infected cells (MOI, 5) that were transfected with control or G3BP-specific siRNAs. (G) G3BP-depleted 293/ACE2 cells were infected with SINV-GFP or CVB3-GFP (MOI, 1) and fixed at 16 or 10 h p.i., respectively. The level of GFP expressed by the viruses was normalized to infected cells transfected with control siRNAs. (H) 293/ACE2 cells were transfected with 50 nM individual siRNA duplexes (deconvoluted pool) and infected with CHIKV-GFP (MOI, 1) 2 days later, followed by quantification of GFP expression at 20 h p.i. The (remaining) level of G3BP2 expression was determined by Western blotting. (I) Analysis of GFP expressed by CHIKV in cells depleted for G3BP2 with siRNA 2 or 4 and cotransfected with a plasmid encoding siRNA-resistant G3BP2 (G2) or an empty vector (ev). (J) Cells were transfected with C911 mutant siRNAs (C911) or the corresponding G3BP2-targeting siRNAs (wt), followed 24 h later by infection with CHIKV-GFP (MOI, 1) and quantification of GFP expression at 20 h p.i. GFP levels were normalized to cells transfected with a nontargeting control siRNA (100%). G3BP2 knockdown efficiency at 24 h p.t. was determined by Western blotting.
Transfection of cells with the G3BP1-specific siRNA pool resulted in an ∼90% reduction in G3BP1 expression and an ∼2-fold increase in G3BP2 levels. The G3BP2 siRNA treatment achieved an ∼80% reduction of G3BP2 levels, accompanied by an ∼2-fold increase in G3BP1 expression (Fig. 5A). After treatment with a combination of G3BP1- plus G3BP2-targeting siRNAs, cells displayed about 40% and 30% of their original levels of expression of G3BP1 and G3BP2, respectively. The siRNA-transfected cells were infected with a CHIK reporter virus at MOIs of 0.05, 1, and 5, and cells were fixed at 24, 20, and 16 h p.i., respectively. The eGFP expression was quantified, and cell viability assays performed in parallel showed no negative effect of G3BP depletion (Fig. 5B). Despite efficient knockdown, G3BP1 depletion had little effect on CHIKV replication. G3BP2 depletion reduced eGFP levels in cells infected at an MOI of 0.05 by ∼55%, and the combined depletion of G3BP1 and G3BP2 reduced eGFP levels even further (∼80%). The effect of G3BP depletion was less pronounced in cells infected at an MOI of 1 or 5 (Fig. 5B). Because silencing of G3BP1 alone had little effect, and because the simultaneous depletion of both G3BPs exerted a stronger effect on CHIKV replication than depletion of G3BP2 alone (Fig. 5C), all subsequent knockdown experiments were done with siRNA pools that targeted G3BP1 and G3BP2 simultaneously. The combined depletion of G3BP1 and G3BP2 resulted in severely reduced negative-strand RNA levels, which were about 85 to 90% lower (6 to 8 h p.i.) than in control cells (Fig. 5D). Consequently, positive-stranded RNA levels were also affected (80 to 85% lower than in control cells).
The strongly reduced genomic RNA levels in G3BP-depleted cells resulted in a reduction of nsP and E2 levels (Fig. 5E). It is noteworthy that G3BP depletion affected the accumulation of nsP3 more strongly than the other nsPs, as the amount of protein could not be quantified at 6 h p.i. and was ∼80% lower at 8 h p.i. than in control cells. Viral progeny titers from G3BP-depleted cells were approximately 1 log lower at 8 h p.i. (Fig. 5F). In conclusion, G3BP2 depletion caused an ∼2-h delay in the accumulation of viral RNA and proteins and the production of infectious progeny, while a less pronounced effect was observed at later time points, suggesting that the G3BPs play a role early in the CHIKV replication cycle.
The sensitivity of CHIKV replication to G3BP depletion is striking, as SINV was previously reported to replicate slightly better in G3BP-depleted cells (35). We therefore also infected G3BP-depleted cells with a SINV reporter virus (SINV-GFP) to investigate the effect of G3BP depletion. As observed for CHIKV, simultaneous depletion of the two G3BPs also inhibited SINV replication in our experimental setup (Fig. 5G). Depletion of G3BP1 alone barely affected SINV replication, whereas depletion of G3BP2 alone strongly affected SINV replication (Fig. 5G).
To ensure that the observed inhibition of CHIKV (and SINV) replication was not due to a general negative effect of G3BP2 depletion on cellular homeostasis, we infected G3BP-depleted cells with an unrelated reporter virus: the picornavirus coxsackie B3 virus (CVB3-GFP). CVB3 is not expected to be negatively affected by G3BP2 depletion, as its protease has been reported to cleave G3BP1 (36) and possibly also G3BP2. Indeed, G3BP2 depletion had no effect on GFP expression by CVB3, and depletion of G3BP1 alone or both G3BPs simultaneously even enhanced CVB3-GFP replication (Fig. 5G). This demonstrated that CVB3 replication was not negatively affected in G3BP-depleted cells, suggesting that the knockdown did not lead to serious negative effects on cellular physiology.
Another issue that needs to be addressed when using siRNAs is the possibility that the observed phenotype is due to off-target effects. Therefore, the pools of four G3BP siRNAs were deconvoluted and the siRNA duplexes were tested individually. As anticipated based on the results obtained with the G3BP1 pool, none of the four single G3BP1 siRNAs had a strong effect on CHIKV replication, despite the fact that 3 out of 4 siRNAs (1 to 3) significantly reduced G3BP1 levels (data not shown). Transfection of 3 out of the 4 individual G3BP2 siRNAs (2, 3, and 4) resulted in a strong reduction in G3BP2 expression (Fig. 5H), whereas transfection of siRNA 1 was somewhat less effective in reducing G3BP2 protein levels. Single siRNA duplexes 2 to 4 reduced CHIKV replication to various extents, but all single siRNAs reduced GFP expression by ∼50% or more (Fig. 5H). It should be noted that a larger effect was not expected, as depletion of G3BP2 alone with the SMARTpool resulted in a similar reduction in GFP expression (Fig. 5B and C). The correlation between the level of CHIKV replication and remaining G3BP expression indicates that the siRNA-mediated inhibition is unlikely due to off-target effects. This is further supported by the fact that expression of an siRNA-resistant form of G3BP2 restored CHIKV replication to ∼30 to 90% of that in control cells (Fig. 5I). The rescue of CHIKV replication by G3BP2 overexpression was more efficient in cells in which G3BP2 was depleted with siRNA 4 than in those transfected with siRNA 2. This suggests that besides G3BP2 depletion, siRNA 2 also caused some off-target effects. To further exclude potential off-target effects, we employed C911 mutant siRNAs (32), in which the “targeting” residues 9 to 11 are mutated. These mutant siRNAs should no longer induce knockdown of the target while still causing the same off-target effects as the corresponding targeting siRNA. We designed and tested C911 mutant siRNA corresponding to G3BP2 single siRNAs 2 and 4 (Fig. 5J). These custom-synthesized targeting siRNAs did not deplete G3BP2 protein levels to the same extent as the original (modified) Dharmacon siRNAs and only reduced CHIKV replication by ∼30%. However, no reduction in CHIKV-driven eGFP expression was observed in cells transfected with the corresponding C911 mutant siRNAs, indicating that the inhibition of CHIKV replication was due to G3BP2 depletion rather than off-target effects of the siRNAs (Fig. 5J).
G3BP levels do not influence nonstructural polyprotein processing.
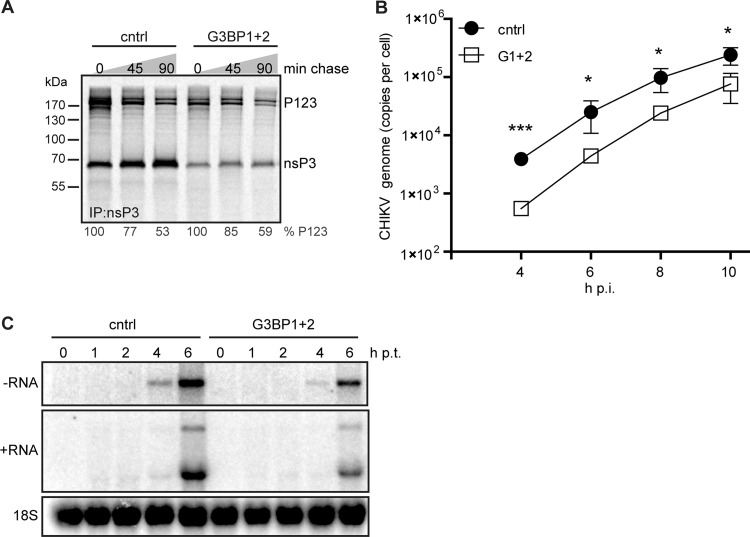
We noticed that—especially early in infection—G3BP depletion affected the level of structural proteins (E2) more than that of nsPs (Fig. 5E). The expression of structural proteins is dependent on sgRNA synthesis, which for alphaviruses is controlled by the extent to which nsP123 is proteolytically processed (37). SFV mutants lacking the G3BP-binding domain exhibited delayed polyprotein processing, resulting in the appearance of an extra uncleaved processing intermediate (17). Therefore, we assessed CHIKV polyprotein processing in G3BP-depleted cells. Pulse-chase metabolic labeling with [35S]Met/Cys, followed by immunoprecipitation of nsP3, showed no differences in the kinetics of processing of the P123 precursor when G3BP-depleted and control cells were compared (Fig. 6A). The total level of viral protein was lower in G3BP-depleted cells at this early time point, but P123 was processed at the same rate and no additional uncleaved intermediates were found, indicating that polyprotein processing was not specifically affected by the absence of G3BP.
FIG 6.
Effect of G3BP depletion on CHIKV nonstructural polyprotein processing, rate of RNA synthesis, and entry. 293/ACE2 cells were transfected with siRNAs targeting the G3BPs and 48 h later infected with CHIKV (MOI, 5) or transfected with in vitro-transcribed CHIKV genomic RNA. (A) Cells were metabolically labeled at 5 h p.i. and chased for 0, 45, or 90 min before lysis and immunoprecipitation. (B) Total RNA from infected cells was isolated at the indicated time points, and CHIKV genome copy numbers were determined using qRT-PCR. (C) G3BP-depleted cells were transfected with 1 μg of viral RNA (in vitro transcript) and harvested at the indicated time points. CHIKV RNA was analyzed using in-gel hybridization using probes specific for negative- or positive-strand RNA. The 18S rRNA was probed as a loading control.
G3BP levels do not affect CHIKV entry or RNA synthesis.
We noticed that viral protein accumulation, as analyzed by Western blotting, appeared to be delayed by ∼2 h in G3BP-depleted cells. Comparison of the kinetics of CHIKV RNA accumulation in control and G3BP-depleted cells by qRT-PCR revealed that RNA production was also delayed by ∼2 h but occurred at the same rate, since a semilog plot of the copy number over time had the same slope as the curve obtained for control cells (Fig. 6B). These results suggested a role for G3BPs early in the CHIKV replication cycle. We therefore investigated whether G3BPs are involved in viral entry by transfecting control and G3BP-depleted cells with in vitro-transcribed CHIKV full-length genomic RNA, a procedure that bypasses virion attachment and entry. The transfection of control cells with CHIKV RNA led to readily detectable amounts of negative-strand RNA by 4 h p.t., after which genome and sgRNA levels increased rapidly in the next 2 h (Fig. 6C). Also in G3BP-depleted cells replication of transfected CHIKV RNA was detected, although negative-strand RNA levels were much lower than in control cells and the accumulation of positive-strand RNA was impaired (Fig. 6C). The observation that bypassing CHIKV entry still resulted in a delayed replication indicates that the G3BPs are involved in an early, but postentry, step of the CHIKV replication cycle. The effect of G3BP depletion in these transfection experiments appeared to be smaller than that observed upon (low-MOI) infection (Fig. 5D). This may have been due to the large amount of RNA transfected into the cells, probably mimicking a very high-MOI infection, which makes the effect of G3BP depletion less pronounced (as shown in Fig. 5B) for reasons discussed below. However, we cannot formally exclude a (minor) additional role for the G3BPs during entry and/or uncoating.
G3BP depletion does not affect translation, but G3BPs appear to regulate the switch to minus-strand synthesis.
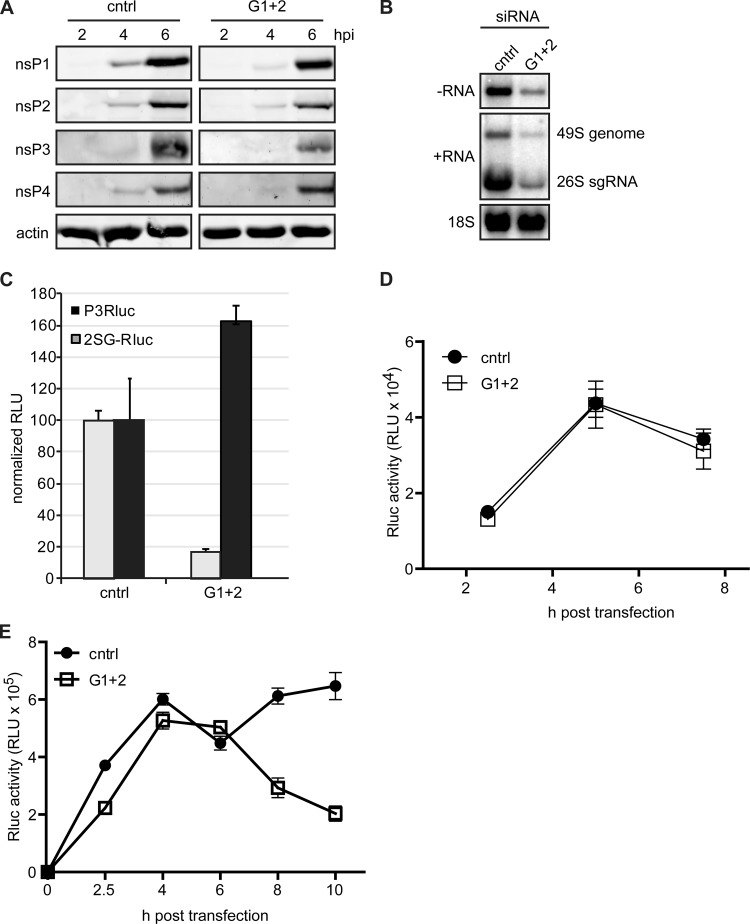
Our experiments indicated that the G3BPs are likely involved in an early step of the CHIKV replication cycle. This could be either translation or early (negative-strand) RNA synthesis. Unfortunately, it is difficult to study translation and genome replication separately, as these processes are interdependent. Incoming viral genome first serves as an mRNA for nsP production and then is copied into the negative-strand template for genome replication, which produces novel positive-strand RNA that, in turn, serves as an mRNA for polyprotein production. However, infection at a very high MOI is thought to provide enough input RNA to render translation largely independent of newly synthesized RNA, enabling analysis of the translation of incoming genomes. To investigate if the incoming CHIKV genome is translated normally in G3BP-depleted cells, these cells were infected at an MOI of 50, and total cell lysates were harvested at the desired time points. Western blot analysis revealed only minor differences in nsP levels between G3BP-depleted and control cells (Fig. 7A). Only the nsP3 level was clearly lower in G3BP-depleted cells. In contrast, the accumulation of CHIKV negative-strand RNA and (consequently also) positive-strand RNA was strongly reduced in G3BP-depleted cells infected at an MOI of 50 and analyzed at 5 h p.i. (Fig. 7B), indicating that the G3BPs are involved in (early) RNA synthesis. To further examine the effect of G3BP depletion on CHIKV translation and RNA synthesis, reporter viruses were used that express Renilla luciferase either as part of the nonstructural polyprotein (fused to nsP3; P3Rluc) or under the control of a duplicated subgenomic promoter (2SG-Rluc). G3BP-depleted and control cells were infected with these reporter viruses (MOI, 5). Quantification of luciferase activity at 8 h p.i. showed a decrease in luciferase expressed from the duplicated subgenomic promoter in G3BP-depleted cells (Fig. 7C). This is in line with the effect of G3BP depletion on eGFP reporter gene expression (from the second sgRNA) that was observed with our reporter virus. Surprisingly, G3BP depletion resulted in an increased luciferase signal from P3Rluc virus, indicating that the translation of genomic RNA was not affected or even slightly enhanced by G3BP depletion. The increase of nsP3-Rluc signal in the luciferase assay contrasts with the apparent nsP3 decrease shown by Western blotting but can be explained by the fact that the degradation signal present in nsP3 (38) was lost in the nsP3-Rluc fusion. Alternatively, a larger amount of nsP3-Rluc could have been solubilized from G3BP-depleted cells compared to control cells, in which the protein is expected to be in G3BP-containing aggregates. Taken together, these data suggest that G3BP depletion does not directly affect translation and might even slightly stimulate nonstructural polyprotein translation.
FIG 7.
Effect of G3BP depletion on translation of genomic RNA and early negative-strand RNA synthesis. (A) Cells transfected with control (cntrl) or G3BP1- and G3BP2-targeting (G1 + 2) siRNAs were infected with CHIKV at an MOI of 50 and harvested at the indicated time points, after which viral protein levels were analyzed by Western blotting. (B) siRNA-treated cells were infected with CHIKV (MOI, 50), and viral RNA was isolated at 5 h p.i. and analyzed by in-gel hybridization. The 18S rRNA was probed as a loading control. (C) G3BP-depleted cells were infected with reporter viruses that express Renilla luciferase either fused to nsP3 (P3Rluc) or from a duplicated subgenomic promoter (2SG-Rluc) at an MOI of 5. Luciferase activity was determined at 8 h p.i. and normalized to the activity in cells transfected with control siRNAs. (D) Replication-deficient CHIKV-P3Rluc RNA was transfected into control and G3BP-depleted cells and luciferase activity was assessed at the indicated time points. (E) Replication-competent CHIKV-P3Rluc RNA was transfected into control and G3BP-depleted cells, followed by measurement of luciferase activity at the indicated time points.
In addition to performing infections at an MOI of 50, we have employed a replication-deficient CHIKV RNA (nsP4 GDD motif mutated to GAA), encoding luciferase fused to nsP3. G3BP-depleted or control cells were transfected with replication-deficient CHIKV nsP3-Rluc RNA, and the luciferase expression was quantified at various time points (Fig. 7D). Translation of this transfected RNA resulted in a peak of luciferase activity around 5 h p.t., followed by a decrease at later time points, likely due to degradation of the nonreplicating RNA and luciferase turnover. The luciferase signals in G3BP-depleted and control cells were very similar, indicating that G3BP depletion did not affect the translation of the CHIKV genome into nonstructural polyproteins (Fig. 7D). We performed a similar experiment with replication-competent CHIKV-nsP3-Rluc RNA, which allowed us to study translation as well as genome amplification. In control cells, an initial peak of luciferase activity was detected at 4 h posttransfection. After a small decrease in luciferase expression at 6 h p.i. (likely due to degradation of mRNA), a further increase in luciferase signal was observed, likely driven by translation of newly synthesized positive-strand RNA (Fig. 7E). Strikingly, this second increase did not occur in G3BP-depleted cells, indicating that the G3BPs play a role not in translation but in the switch from translation of the (incoming) positive-strand RNA to negative-strand synthesis and RNA replication.
DISCUSSION
Many viruses manipulate the formation and dynamics of SGs, likely because their formation results in inhibition of translation. G3BP1 is an extensively studied SG marker, while the related G3BP2 remains less well characterized. These proteins, collectively referred to as G3BPs, are multifunctional RNA-binding proteins that have been implicated in the replication of several RNA viruses (34, 39–42). G3BPs have also been implicated in the alphavirus replication cycle, as they have been identified as binding partners of SINV nsP2, nsP3, and nsP4 (18, 20–22, 35) and SFV nsP3 (17, 19). Replication of a CHIKV replicon was shown to induce G3BP1-containing granules, and the expression of nsP3 alone was sufficient to sequester G3BP1 into granules (16). In addition, CHIKV-induced G3BP1-capsid protein foci have been described (43).
We found that late in infection, CHIKV induced foci that contained both G3BP1 and G3BP2 but that differed from bona fide SGs in morphology, CHX sensitivity, and composition (Fig. 1). These granules are probably similar to those observed in an earlier study using a CHIKV replicon (16). CHIKV-induced granules did not contain other SG markers, like TIA-1, TIAR, eIF3, or PABP, and the nsP3-G3BP aggregates therefore likely block the formation of genuine SGs by sequestering G3BPs. The expression level of these other SG components did not change over the course of infection, with the exception of TIA-1 and TIAR, which even slightly increased (Fig. 3). Therefore, the lack of these proteins in the CHIKV-induced granules was not due to their absence in the infected cell. SFV- and CHIKV replicon-induced G3BP-granules also lack other typical SG markers (16, 17).
SGs and G3BPs are generally thought to exert an antiviral effect on alphavirus replication. Surprisingly, we found by siRNA-mediated depletion that G3BPs were also required for efficient CHIKV replication (Fig. 5). CHIKV replication was affected most strongly when G3BP1 and G3BP2 were depleted simultaneously, which resulted in reduced viral RNA levels, diminished CHIKV protein expression, and an ∼10-fold reduction in progeny titers. G3BP1 is generally considered an antiviral protein, and it was therefore surprising that depletion of G3BP1 did not stimulate CHIKV replication. However, proviral roles have also been described for G3BPs in the replication of respiratory syncytial virus (39) and hepatitis C virus (HCV) (40). The observed inhibition of CHIKV replication in siRNA-transfected cells could be rescued by expressing siRNA-resistant G3BP2, demonstrating that it was not due to off-target effects. This was further corroborated by the use of C911 mutant siRNAs and by demonstrating that coxsackievirus replication was not affected in G3BP2-depleted cells. Our results demonstrate that simply studying the role of G3BP1 in viral replication, without taking G3BP2 into consideration, can lead to the misinterpretation or underestimation of the role of the G3BPs, as these homologous proteins can likely complement each other (in part) but also possess unique properties. This is illustrated by the fact that knockdown of G3BP1 did not affect CHIKV replication, possibly due to the concomitant increase in G3BP2 expression.
Our data show that G3BP2 colocalized with CHIKV nsP2 and nsP3 in cytoplasmic granules but not with nsP1, nsP4, or dsRNA. In addition, subcellular fractionation experiments demonstrated that G3BPs were undetectable in the fraction (P15) enriched for CHIKV RTCs, suggesting that G3BPs are not associated with the active membrane-associated RTCs that can be isolated from infected cells (Fig. 4). SFV induces true SGs very early in infection, which disappear and are replaced by different nsP3-containing structures later in infection (17). For SINV, two types of nsP3-containing granules have been described: one type is likely associated with RTCs, while the other lacks dsRNA (20). Our findings suggest that most CHIKV nsP3 (and interacting G3BPs) is in the second type of granule. In contrast, G3BPs were found to be present in a fraction containing active SFV RTCs in a proteomics analysis of isolated cytopathic vacuoles (44). Which proportion of total cellular G3BPs was present in this fraction, however, was not determined. Clearly, we cannot formally exclude that trace amounts of G3BPs were present in our CHIKV RTC-containing membrane fraction. Unfortunately, technical limitations did not allow us to study the composition and in vitro activity of the early (negative-strand RNA-synthesizing) RTCs. Therefore, it is well possible and even likely (see below) that G3BPs play a role in negative-strand RNA synthesis. This is supported by the fact that G3BP depletion caused a delay in the replication cycle and affected an early postentry step. Translation of viral mRNA and nonstructural polyprotein processing were not impaired in G3BP-depleted cells, suggesting a role for G3BPs in early RNA synthesis. Indeed, infecting G3BP-depleted cells at a very high MOI in order to render viral mRNA translation to a certain extent independent from RNA synthesis showed that negative-strand RNA levels were severely reduced, despite the production of almost normal nsP levels (Fig. 7A and B). Therefore, G3BPs appear to be involved in the switch from translation of the incoming genome to negative-strand RNA synthesis. The G3BPs might clear the viral genome of proteins and/or translating ribosomes that would otherwise interfere with a negative-strand-synthesizing RTC moving in the opposite direction. A similar proviral role has been proposed for G3BPs during HCV replication, in which they were shown to be important during viral genome amplification but not translation (40). By analogy, impairing G3BP-induced CHIKV mRNA clearance would not affect nsP synthesis, which is in line with the observed close-to-normal nsP levels and the slightly enhanced luciferase signal of a recombinant virus expressing an nsP3-Rluc fusion protein in G3BP-depleted cells. A less efficient switch from translation to genome amplification after G3BP depletion would explain the observed reduction of RNA levels and structural protein expression, which is dependent on sgRNA synthesis.
Our findings that G3BP depletion reduced CHIKV replication may appear to disagree with data from an earlier study on SINV (35). Cristea et al. observed enhanced SINV polyprotein expression (similar to what we found for CHIKV), but they also found similar or even slightly (though not statistically significant) increased RNA levels and virion production. It is possible that G3BP2 protein levels were not sufficiently depleted in this earlier study, as only mRNA levels were analyzed, which does not necessarily mean there was a similar reduction in G3BP protein levels. If G3BP2 protein levels were not sufficiently depleted, it would be compatible with our observation that G3BP1 depletion alone had little effect on CHIKV replication. When we analyzed SINV in G3BP2-depleted cells using our own experimental setup, we did observe reduced replication, similar to what we found for CHIKV (Fig. 5G). Of course, the differences between our data and those previously reported by Cristea et al. may also be due to differences in experimental setup or the cell lines used. We have not analyzed the effect of G3BP depletion on SINV in much detail, and it remains possible that CHIKV and SINV respond differently to G3BP depletion. Previous reports have identified at least one other RNA-binding protein that has different effects on CHIKV and SFV replication (44), so a similar difference between SINV and CHIKV would not be unimaginable.
Commonly, G3BP1 is implicated in SG formation, and therefore, its effect on viral replication has often been attributed to this function. However, both G3BPs possess multiple domains and a wide range of other functions unrelated to SG formation, which may (also) be relevant for CHIKV replication. G3BPs are part of the HCV replication complex (40), but it is unlikely that they are essential components of the CHIKV RTC, at least not in the membrane-associated complexes that produce the bulk of the genomic and sgRNA during the later stages of the replication cycle. This is in line with the recently identified interaction between CHIKV nsP3 and G3BPs (16, 23) that we have confirmed in this study (Fig. 4A). This interaction between nsP3 and G3BPs appears to occur not in RTCs (Fig. 4) but in SG-like structures that differed in composition and behavior from traditional SGs. The at-first-sight contradicting pro- and antiviral roles of G3BPs could perhaps be reconciled in a more refined model that would discriminate between early and late events in the replication cycle. Early in infection, nsP3 is present at low levels (and as part of the polyprotein), and it could then recruit G3BPs to the genomic RNA that is being translated, to mediate or support the switch from translation to the synthesis of negative-strand RNA. G3BPs might be involved in clearing ribosomes or proteins from the RNA and/or stabilize the naked viral RNA. Alternatively, G3BPs could be involved in a very early step of RTC formation, although they do not appear to be a major component of, or required for the activity of, the positive-strand RNA-synthesizing RTCs. Later in infection, when negative-strand RNA synthesis ceases, and higher levels of fully processed nsP3 are present, a cytosolic (non-RTC-associated) pool of nsP3 might sequester G3BPs into the aggregates that prevent the formation of true SGs, which could otherwise exert an antiviral effect on the translation of viral mRNAs. This model is supported by the notions that the G3BPs seem to play a (proviral) role only early in CHIKV replication and that at this stage genuine SGs can still be formed in alphavirus-infected cells, as we and others (17) have observed. Indeed, at 4 h p.i. we observed some colocalization of dsRNA and G3BPs, while at 8 h p.i. there clearly was no colocalization of dsRNA with the nsP3- and G3BP2-containing granules.
Since G3BP1 and G3BP2 have so many (sometimes poorly understood) functions (reviewed in reference 45), they might be involved in more steps of the CHIKV replication cycle, besides their proposed role in the translation-replication switch. For example, the G3BPs could be involved in stabilizing viral RNAs via their RNA-binding properties or even in NF-κB signaling or ubiquitin-mediated degradation, in which they have also been implicated (15, 46). NF-κB phosphorylation and protein levels were suggested by Zhang et al. to be affected by G3BP depletion (47), but we could not detect any changes in NF-κB protein levels or intracellular localization (data not shown). Another intriguing observation was that nsP3 levels appeared to be more strongly affected by G3BP depletion than the other nonstructural proteins, suggesting a role for G3BPs in stabilizing nsP3. Future work should assess the additional roles that the G3BPs might play during CHIKV replication.
ACKNOWLEDGMENTS
We thank Emmely Treffers and Adriaan de Wilde for helpful discussions and for sharing their unpublished data. We are grateful to Christer Larsson (Lund University, Sweden) for his generous gift of G3BP2 antiserum.
Part of this research was supported by project IUT20-27 from the Estonian Research Council (E.Z. and A.M.).
REFERENCES
- 1.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol 20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai W-C, Lloyd RE. 2014. Cytoplasmic RNA granules and viral infection. Annu Rev Virol 1:147–170. doi: 10.1146/annurev-virology-031413-085505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kedersha N, Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30:963–969. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 4.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 5.Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams BR. 2001. Signal integration via PKR. Sci STKE 2001:re2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- 8.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 9.Kimball SR. 2001. Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol 26:155–184. doi: 10.1007/978-3-642-56688-2_6. [DOI] [PubMed] [Google Scholar]
- 10.Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 20:6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T, Winslow S, Sunesson L, Hellman U, Larsson C. 2012. PKCalpha binds G3BP2 and regulates stress granule formation following cellular stress. PLoS One 7:e35820. doi: 10.1371/journal.pone.0035820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 14.Tourrière H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. 2001. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prigent M, Barlat I, Langen H, Dargemont C. 2000. IkappaBalpha and IkappaBalpha/NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem 275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 16.Fros JJ, Domeradzka NE, Baggen J, Geertsema C, Flipse J, Vlak JM, Pijlman GP. 2012. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol 86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panas MD, Varjak M, Lulla A, Eng KE, Merits A, Karlsson Hedestam GB, McInerney GM. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem 281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 19.McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol 80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J Virol 81:5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panas MD, Ahola T, McInerney GM. 2014. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 88:5888–5893. doi: 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, Frolova EI. 2013. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol 87:1997–2010. doi: 10.1128/JVI.02853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholte FE, Tas A, Martina BE, Cordioli P, Narayanan K, Makino S, Snijder EJ, van Hemert MJ. 2013. Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1-226V isolates. PLoS One 8:e71047. doi: 10.1371/journal.pone.0071047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohjala L, Utt A, Varjak M, Lulla A, Merits A, Ahola T, Tammela P. 2011. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One 6:e28923. doi: 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myles KM, Pierro DJ, Olson KE. 2003. Deletions in the putative cell receptor-binding domain of Sindbis virus strain MRE16 E2 glycoprotein reduce midgut infectivity in Aedes aegypti. J Virol 77:8872–8881. doi: 10.1128/JVI.77.16.8872-8881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanke KH, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJ. 2009. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol 83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metz SW, Geertsema C, Martina BE, Andrade P, Heldens JG, van Oers MM, Goldbach RW, Vlak JM, Pijlman GP. 2011. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol J 8:353. doi: 10.1186/1743-422X-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albulescu IC, Tas A, Scholte FE, Snijder EJ, van Hemert MJ. 2014. An in vitro assay to study Chikungunya virus RNA synthesis and the mode of action of inhibitors. J Gen Virol 95(Part 12):2683–2692. [DOI] [PubMed] [Google Scholar]
- 32.Buehler E, Chen Y-C, Martin S. 2012. C911: a bench-level control for sequence specific siRNA off-target effects. PLoS One 7:e51942. doi: 10.1371/journal.pone.0051942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastorino B, Bessaud M, Grandadam M, Murri S, Tolou HJ, Peyrefitte CN. 2005. Development of a TaqMan RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J Virol Methods 124:65–71. doi: 10.1016/j.jviromet.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 34.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol 84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P, Han L, Chu F, Jagdeo J, Jan E, Fujita T, Luo H. 2013. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 8:e79546. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68:1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varjak M, Žusinaite E, Merits A. 2010. Novel functions of the alphavirus nonstructural protein nsP3 C-terminal region. J Virol 84:2352–2364. doi: 10.1128/JVI.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE. 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi ZG, Pan TT, Wu XF, Song WH, Wang SS, Xu Y, Rice CM, MacDonald MR, Yuan ZH. 2011. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J Virol 85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward AM, Bidet K, Ang YL, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews JD, Frey TK. 2012. Analysis of subcellular G3BP redistribution during rubella virus infection. J Gen Virol 93:267–274. doi: 10.1099/vir.0.036780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Y, Kielian M. 2013. Imaging of the alphavirus capsid protein during virus replication. J Virol 87:9579–9589. doi: 10.1128/JVI.01299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varjak M, Saul S, Arike L, Lulla A, Peil L, Merits A. 2013. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J Virol 87:10295–10312. doi: 10.1128/JVI.01105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irvine K, Stirling R, Hume D, Kennedy D. 2004. Rasputin, more promiscuous than ever: a review of G3BP. Int J Dev Biol 48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 46.Soncini C, Berdo I, Draetta G. 2001. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Zhang S, He H, Zhao W, Chen J, Shao RG. 2012. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin [sic]-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci 103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]