ABSTRACT
Tumor suppressor p53 has been suggested to be a host restriction factor against HIV-1 replication, but the detailed molecular mechanism has remained elusive for decades. Here, we demonstrate that p53-mediated HIV-1 suppression is attributed to double-stranded RNA (dsRNA)-dependent protein kinase (PKR)-mediated HIV-1 trans-activator (Tat) phosphorylation and inactivation. p53 silencing significantly enhanced HIV-1 replication in infected cells. Ectopic expression of p53 suppressed Tat activity, which was rescued by PKR silencing. In addition, ectopic expression of PKR abolished Tat activity in p53−/− and eIF2αCA cells. Finally, we found that HIV-1 infection activates p53, followed by the induction and activation of PKR. PKR directly interacted with HIV-1 Tat and phosphorylates the first exon of Tat exclusively at five Ser/Thr residues (T23, T40, S46, S62, and S68), which inhibits Tat-mediated provirus transcription in three critical steps: (i) phosphorylation near the arginine-rich motif (ARM) inhibits Tat translocation into the nucleus, (ii) accumulation of Tat phosphorylation abolishes Tat–Tat-responsive region (TAR) binding, and (iii) Tat phosphorylation at T23 and/or T40 obliterates the Tat-cyclin T1 interaction. These five Ser/Thr sites on Tat were highly conserved in HIV-1 strains prevalent in Europe and the United States. Taken together, our findings indicate that p53-derived host restriction of HIV-1 replication is likely attributable, at least in part, to a noncanonical p53/PKR/Tat phosphorylation and inactivation pathway in HIV-1 infection and AIDS pathogenesis.
IMPORTANCE HIV-1-mediated disease progression to AIDS lasts for years to decades after primary infection. Host restriction and associated viral latency have been studied for several decades. p53 has been suggested as an important host restriction factor against HIV-1 replication. However, the detailed molecular mechanism is still unclear. In the present study, we found that the p53-mediated HIV-1 restriction is attributed to a p53/PKR/Tat-inactivation pathway. HIV-1 infection activated p53, which subsequently induced PKR expression and activation. PKR directly phosphorylated Tat exclusively at five specific Ser/Thr residues, which was accompanied by significant suppression of HIV-1 replication. Accumulation of Tat phosphorylation at these sites inhibited Tat function by blocking Tat nuclear localization, Tat binding to TAR, and Tat-cyclin T1 interaction. Our findings provide a better understanding of the p53-derived host restriction mechanism against HIV-1 replication in AIDS pathogenesis and may contribute to further research focusing on the investigation of potential therapeutic targets for HIV-1.
INTRODUCTION
Human immunodeficiency virus type I (HIV-1) encodes a highly conserved trans-activator (Tat) protein, which is essential for viral replication and disease progression (1). Although Tat is a potent trans-activator in HIV-1 replication, HIV-1-mediated disease progression lasts for decades after primary infection (2). Host restriction mechanisms have been discussed in association with latent reservoirs, inappropriate provirus integration, epigenetic silencing, absence of host transcription factors for HIV-1 expression, cellular repressors, and insufficient Tat activity (3–5). However, the host restriction of HIV-1 replication and associated viral latency still remains unclear and is a major barrier in the eradication of AIDS. The tumor suppressor p53 has been suggested as an important factor in host restriction of HIV-1 replication (6–8). HIV-1 infection induces upregulation of p53 in primary CD4+ T cells (9, 10). It was also reported that HIV-1 infection activates p53 target genes, leading to cell apoptosis (11). However, p53-mediated host restriction of HIV-1 replication is not completely understood.
Protein kinase R (PKR) is a well-characterized double-stranded RNA-dependent protein kinase with important roles in type 1 interferon (IFN)-mediated antiviral activity (12). Recently, we reported that PKR is a novel p53 target gene which plays a crucial role in the tumor suppressor function of p53 under genotoxic conditions (13) and induces G2 arrest by triggering Cdc2 degradation (14).
The replicating capacity of HIV-1 strains has been suggested to be closely associated with Tat activity and sequence variations. Tat proteins from African strains Mal and Eli are more active than those from Europe and the United States (15). In addition, the natural amino acid substitution T23N in Tat is predominant in clade A, C, D, and G of HIV-1 strains in Africa that are known to be fast-replicating (16). On the other hand, insufficient Tat activity is accompanied by the establishment of HIV-1 latency (17, 18), and latent infection can be reactivated by exogenous Tat (19). Tat activity is altered by cellular modification factors such as p300, p300/CBP-associating factor (PCAF), Hdm2, Hexim1, and SKIP (20–23). Tat phosphorylation by PKR (24) and CDK2 (25) has also been reported, and phosphorylation of Tat by PKR at three Ser/Thr sites (S62, T64, and S68) was shown to increase Tat–Tat-responsive region (TAR) binding and enhance transcription (26). However, the detailed molecular mechanisms underlying host restriction of HIV-1 replication via p53/PKR-mediated Tat phosphorylation and inactivation after HIV-1 infection are largely unknown.
Here, we found that HIV-1 infection activates p53, which subsequently induces PKR expression, followed by significant inactivation of Tat by PKR-mediated Tat phosphorylation at five specific Ser/Thr residues that are highly conserved in European and American HIV-1 strains, independent of the canonical PKR/eIF2α translational inhibition pathway. PKR-mediated Tat phosphorylation inhibited Tat nuclear localization, Tat binding to TAR, and Tat-cyclin T1 interaction, accompanied by suppression of HIV-1 replication. Our findings indicate that p53 plays an important role in the inhibition of HIV-1 replication via Tat inactivation in patients infected with HIV-1 strains which are predominant in Europe and the United States. This study provides a better understanding of the p53-derived host restriction mechanism against HIV-1 replication in AIDS pathogenesis and may contribute to further research focusing on the investigation of potential therapeutic targets for HIV-1.
MATERIALS AND METHODS
Cells, viruses, plasmids, and antibodies.
HeLa, HEK293, C8166, and Jurkat cells were obtained from American Type Culture Collection (ATCC) or the Korean Type Culture Collection (KTCC). Isogenic HCT116 p53+/+ and p53−/− cells were kindly provided by B. Vogelstein (Johns Hopkins Oncology Center, USA). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or in RPMI 1640 medium, supplemented with 10% fetal bovine serum (FBS) (GIBCO/BRL). Jurkat-Tat (Tat-expressing Jurkat) cells were kindly provided by J. Sodroski (Dana-Farber Cancer Institute, Harvard Medical School, USA). HeLa/ΔPKR and Jurkat/ΔPKR cells, stably transfected with the pSLX-antisense PKR(620-180) plasmid, were selected in the presence of 800 μg/ml G418 and maintained in medium containing 400 μg/ml G418. PKR knockdown (PKRKD) HCT116 (p53+/+ or p53−/−), HeLa, and Jurkat cells, as well as constitutively active eIF2α mutant (eIF2αCA) cells were prepared by transfection with PKR-targeting short hairpin RNA (shRNA) (sh-PKR) or pSLX-eIF2α51A plasmids as reported previously (13). The HIV-1 laboratory strain HXB2, its cDNA clone (pHXB2), and the HIV-1IIIB strain were obtained from the AIDS Research and Reference Reagent Program (ARRRP, NIH, USA) and cultivated as described previously (27). HXB2 cDNAs containing mutant Tat were generated by subcloning the cDNA fragment at NheI/NcoI sites with mutant tat-containing fragments after PCR-based mutagenesis on tat. pcDNA-p53, pcDNA3-(Flag/Myc)-PKR, pcDNA3-Flag-tat, pGEX-5X-1-PKR, and pEGFPc2-cyclin T1 plasmids were constructed as described previously (13, 14), with minor modifications. Plasmid pcDNA-HA-PML-IV (28) was kindly provided by C. Y. Choi (Sungkyunkwan University, Republic of Korea). Tat-expressing plasmids were constructed by cloning wild-type (wt) and mutant (mt) Tat cDNAs (for sequences, see Table S1 in the supplemental material) into the pSV2-Tat (from J. Sodroski), pcDNA3-Flag (Invitrogen), pGEX-5X-1 (Amersham), or pRSETc (Promega) plasmid. Mutant Tats and shRNA/small interfering RNA (siRNA)-resistant silent mutant PKR/p53-expressing plasmids were generated using the QuikChange site-directed mutagenesis kit (Stratagene), and the PCR products were verified by sequencing. “Off-target effects” of each siRNA (si-PKR and si-p53, designed and provided by Invitrogen Stealth) and shRNA (sh-PKR) on HIV-1 replication in infected cells were evaluated with each siRNA/shRNA-resistant expressing plasmids (pcDNA-PKR/m and pcDNA-p53/m, having silent mutations on the target site of each siRNA/shRNA) (data not shown). PCR primers and siRNA/shRNA sequences used for the present study are summarized in Tables S2 and S3 in the supplemental material. Cells were transfected with these expression plasmids using Lipofectamine 2000 (Invitrogen) or by electroporation. Antibodies against p53 (DO-1; Santa Cruz Biotechnology), PKR and phospho-PKR (Cell Signaling), Tat (monoclonal Ab-N3 [Abcam]; rabbit antiserum [AIDS Research and Reference Reagent Program]), phospho-p53 (Cell Signaling), p21waf1/cip1 (Cell Signaling), and Puma (Cell Signaling), anti-Flag monoclonal antibody (MAb) (Cell Signaling), anti-cyclin T1 polyclonal antibody (PAb) (Cell Signaling), and antibodies against HIV-1 p24 (Abcam and Chemicon), β-actin (Sigma), and α-tubulin (Sigma) were used.
Recombinant Tat, eIF2α, and PKR protein expression and purification. (i) His-tagged HIV-1 Tat and human eIF2α protein.
6×His-tagged wild-type (wt) and mutant (mt) (Ala-A and Asp-D) Tat genes and wt and mt eukaryotic initiation factor 2 α subunit (eIF2α) genes were cloned into the pRSETc/pcDNA/pSLX expression plasmid. Escherichia coli BL21 (Stratagene) was transformed with these plasmids and cultured in 2× YTA broth medium (50 μg/ml ampicillin). Recombinant proteins were used for the experiments after purification.
(ii) GST-Tat and GST-PKR fusion proteins.
Glutathione S-transferase (GST)-Tat and GST-PKR recombinant proteins (cloned in the pGEX-5X-1 vector) were prepared as described previously (29, 30), with minor modifications.
Yeast two-hybrid experiments.
Yeast two-hybrid experiments were performed using the Matchmaker LexA two-Hybrid system (Clontech) as described previously (31). PKR was cloned into the DNA-binding domain (BD)-containing pLexA vector (His3 Ampr), and HIV-1 tat or eIF2α gene was cloned into the activation domain (AD)-containing pB42AD vector (Trp1 Ampr) and then transformed into yeast strain EGY48. Positive clones were selected in UHW-auxotrophic minimal agar medium containing 2% glucose, and β-galactosidase (β-gal) expression was examined in UHW-auxotrophic medium supplemented with 2% galactose, 1% laffinose, 80 mg/liter X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and BU salt. Blue colonies indicate direct interactions between the two molecules in vivo.
In vitro kinase assay with recombinant proteins Tat, eIF2α, and PKR.
In vitro kinase assays were performed as described previously (14). Purified recombinant GST-PKR (0.2 μg) was preactivated with poly(I·C) at 30°C for 1 h in the presence or absence of 1 μCi of [γ-32P]ATP and then incubated with 0.5 μg of 6×His(GST)-wt or -mt Tat or eIF2α at 30°C for 1 h or for the time periods described in the figure legends. Each reaction was separated on a 12% or 15% SDS-polyacrylamide gel. Tat/eIF2α phosphorylation was autoradiographed by exposing a dried gel to X-ray film (Eastman Kodak Co.) or by Western blot analysis using anti-phospho-Thr (Cell signaling) and/or anti-phospho-Ser (Zymed Co.) antibodies.
ESI-MS/MS analysis of PKR-treated Tat.
Mass spectrometry (MS) was performed as described previously (14) with minor modifications. Tat bands following in vitro kinase reaction with PKR were gel extracted and digested with trypsin. The tryptic peptides were subjected to liquid chromatography-electrospray ionization-tandem MS (LC-ESI-MS/MS) in a data-dependent scan mode. MS/MS spectra were searched via the Turbo SEQUEST algorithm against a target protein (HIV-1 Tat) database, and the resulting identified phosphopeptides were further validated by manual inspection.
PKR-mediated Tat phosphorylation in vivo.
Flag-tagged wt or mt Tat-expressing plasmids were transfected into PKR-normal (sh-con) or PKRKD (sh-PKR) HEK293 cells or cotransfected with PKR-expressing plasmid (pcDNA-Myc-PKR) into HEK293 cells. Cells were cultured for 48 h, then treated with 50 nM calyculin A (Ser/Thr phosphatase inhibitor; Sigma) before harvest, and lysed with lysis buffer. Portions of cell extracts were treated with 1 U/μl of Lambda protein phosphatase (λPPase) (Sigma) for 1 h at 30°C. Tat phosphorylation was assessed by Western blotting using anti-Flag MAb, anti-phospho-Thr PAb (Cell Signaling), and anti-phospho Ser PAb (Zymed Co.).
HIV-1 replication and titration.
The HIV-1 titer in culture supernatant was assessed with the Alliance HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) kit (Perkin-Elmer Life Sciences). In brief, C8166 cells, human peripheral blood mononuclear cells (PBMCs), or primary CD4+ T cells were transfected with 1 μg of wt or mt Tat-containing HXB2 cDNA by electroporation or infected with HIV-1 variants. Complementation assay was performed by cotransfection with pcDNA-Flag-tat (wt or mt) and HXB2/ΔTat cDNA, After transfection/cotransfection/infection, the HIV-1 titers in the supernatants were assessed using the HIV-1 p24 ELISA kit, and intracellular HIV-1 was analyzed by Western blotting using anti-p24 antibody.
HIV-1 infection of human PBMCs and purified CD4+ T cells.
Human peripheral blood mononuclear cells (PBMCs) obtained from healthy volunteers or human CD4+ T cells isolated from the human PBMCs using the Dynabeads Untouched human CD4 T cell kit (Invitrogen) were cultured for 3 days in RPMI 1640 supplemented with 0.25 μg/ml phytohemagglutinin P (PHA-P), human interleukin 2 (hIL-2) (100 units/ml for CD4+ T cells) and 10% FBS before infection. After infection with the HIV-1IIIB strain at a multiplicity of infection (MOI) of 0.1 to 1, cells were cultured in a medium supplemented with hIL-2 at a final concentration of 100 units/ml. Three to 4 days after infection, the HIV-1 titer in each culture supernatant was measured with the HIV-1 p24 ELISA kit (Perkin-Elmer). HIV-1 p24, p53, and PKR in the infected cells were assessed by Western blotting.
Reporter assays.
The long terminal repeat (LTR)-conjugated luciferase reporter plasmid (pLTR-luc) was constructed by cloning the HIV-1IIIB LTR sequence into the PGL-3 vector (Promega). Luciferase reporter assays were performed as described previously (13) with minor modifications. To assess the effects of p53 and PKR on Tat activity, cells were transfected with 200 ng of pLTR-luc and 100 ng of pcDNA-Flag-tat (or pSV2-tat) together with different amounts of pcDNA-p53 and/or pcDNA-PKR plasmids using Lipofectamine 2000 (Invitrogen). In the reporter assay, pCMV-lacZ (20 ng) was always cotransfected as a transfection control. At 48 h after transfection, the luciferase activity of each sample was measured and normalized to the β-galactosidase activity of each transfection control (20 ng of pCMV-lacZ) using the Bright-Glo luciferase assay system (Promega) and Genios luminometer (Tecan, Austria). Data are represented as means ± standard deviations (SD). The LTR-chloramphenicol acetyltransferase (CAT) reporter system (pU3III-CAT plasmid) was obtained from the ARRRP (NIH, USA). The CAT reporter assay was performed as described previously (27).
RNA retardation assays.
TAT-TAR interaction and retardation assays were performed as described previously (32) with some modifications. 32P-labeled wt TAR RNA (or mutant TAR having 20-GACUCGA-26 mutations at the bulge region) was synthesized by in vitro transcription of pTZ18R-TAR using a commercial T7 RNA polymerase system (NEB) and [α-32P]UTP (Amersham). Phosphorylated Tat protein was prepared by incubating Tat protein with preactivated PKR for the indicated period of time (0 to 120 min) in the presence or absence of [γ-32P]ATP. Tat protein was incubated with 32P-labeled TAR RNA for 15 min in 10 μl of RNA binding buffer (15 mM HEPES-KOH [pH 7.4], 5 mM MgCl2, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin, 1 μM dithiothreitol [DTT], 1 unit of RNasin [Promega]). The TAT-TAR binding assay was also performed with different concentrations of wild-type or phosphor-mimic (D-mt) Tat proteins and 3 pmol of 32P-labeled TAR RNA. The retardation assay was carried out on a 3% native or denaturing (SDS) polyacrylamide gel and visualized by autoradiography.
Immunocytochemistry analyses.
Immunocytochemistry was performed as described previously (13) with minor modifications. Cells were transfected with appropriate expression plasmids or treated with recombinant Tat proteins and then fixed and permeabilized with Cytofix/Cytoperm (BD Bioscience Inc.). Cells were then incubated for 1 h with primary anti-Flag (1/500), antihemagglutinin (anti-HA) (1/500), or anti-Tat antibodies and then incubated with fluorescence (fluorescein isothiocyanate [FITC] or Texas Red)-labeled secondary antibodies (1/500) overnight at room temperature. Fluorescence signals were observed on a fluorescence microscope (Olympus X100) or confocal laser scanning microscope (Zeiss F510).
Co-IP assays.
Coimmunoprecipitation (co-IP) assays were performed as described previously (14) with minor modifications. C8166 cells were transfected with wt or mt Tat-expressing plasmids (pcDNA3-Flag-tat) using Lipofectamine 2000 (Invitrogen). After 24 h, Tat in cell lysates was immunoprecipitated with anti-Flag antibody (M2; Sigma) together with protein A/G agarose beads (Santa Cruz) at 4°C for 5 h. Pellets were washed and assessed by Western blotting. Co-IP of cyclin T1 (CycT1) and Tat was performed as follows. 6×His-Tat was fully phosphorylated by overnight incubation with preactivated PKR in the presence of [γ-32P]ATP and was incubated further with HeLa cell extracts at 4°C for 3 h. The mixtures were immunoprecipitated with anti-CycT1 antibody (Santa Cruz), and Tat was assessed by Western blotting and autoradiography.
GST pulldown experiments.
Reaction mixtures containing GST-Tat (wt or mt) proteins were incubated with 1 mg of HeLa cell nuclear extracts at 4°C for 4 h. Reaction mixtures were incubated for 30 min with 10 μl of glutathione beads (Amersham-Pharmacia Biotech), washed three times with binding buffer, and analyzed by Western blotting assay.
Statistical analysis.
Statistical analysis was performed using Student's t test with GraphPad Instat software. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
NCBI GenBank accession numbers for the major genes and proteins that are mentioned in the text are as follows: p53, XM_008679.2; PKR, NM_002759.3; HIV-1 Tat, the sequence and accession number of each variant mentioned in the manuscript are summarized separately in Table S4 in the supplemental material; eIF2α, J02645.1; TAR, M36464.1.
RESULTS
HIV-1 infection activates p53, which inhibits Tat function.
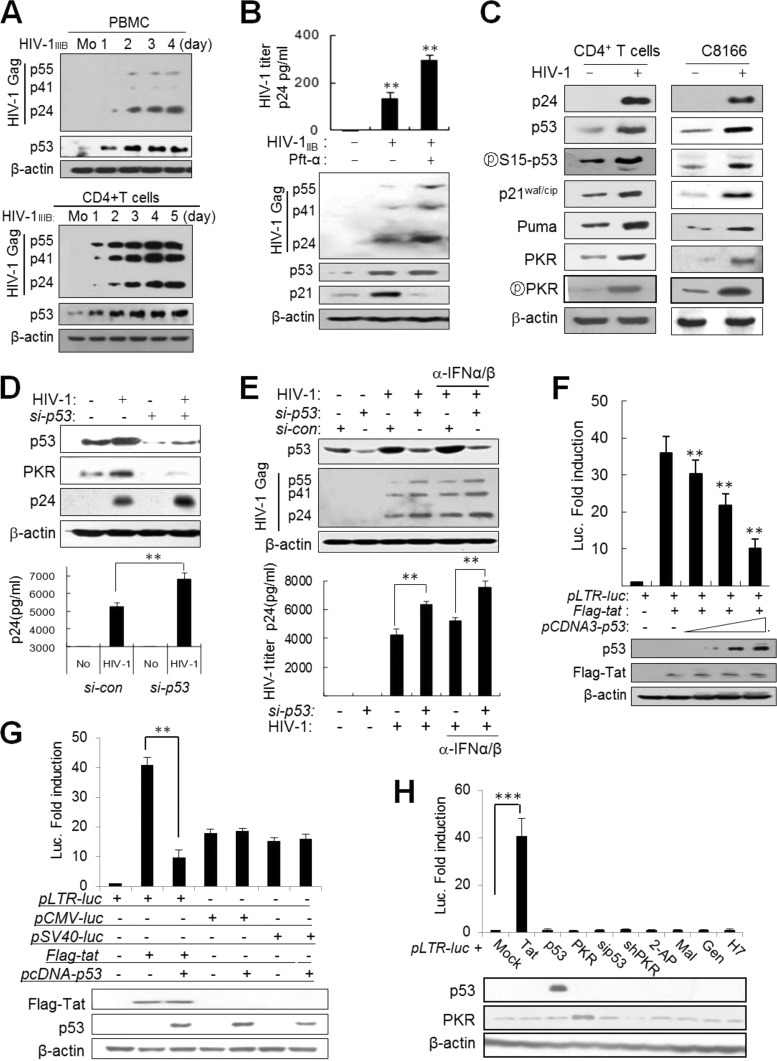
HIV-1 infection increased the levels of p53 in primary peripheral blood mononuclear cells (PBMCs) and purified CD4+ T cells (Fig. 1A). Treatment of HIV-1-infected cells with pifithrin-α (Pft-α, a p53 inhibitor) significantly increased viral replication after infection (Fig. 1B), implying that induced p53 suppresses HIV-1 replication in infected cells. In addition, induced p53 after infection led to the expression of several p53 target genes, including p21, Puma, and PKR, and the expression profile of p53 target genes in primary CD4+ T cells was very similar to that in the C8166 human T cell line (Fig. 1C), indicating that C8166 cells could be substituted for primary CD4+ T cells in the following experiments. HIV-1 replication in C8166 cells was markedly enhanced by p53-knockdown (p53KD) in HIV-1-infected cells (Fig. 1D). It was reported that HIV-1 infection induces the expression of type 1 interferon (IFN), accompanied by upregulation of p53 mRNA in CD4+ T cells (10). To examine the effects of type 1 IFN on p53-mediated HIV-1 suppression, HIV-1 infected p53-normal (p53+/+) and p53KD cells were cultured in the presence of IFN-α/β neutralizing antibodies. As shown in Fig. 1E, the titer of virus was enhanced by blocking type 1 IFN in p53-normal cells, but the enhancement by p53 knockdown was more significant than that by blocking of type 1 IFN (Fig. 1E). These data indicate that p53 induced by HIV-1 infection plays a crucial role in the inhibition of HIV-1 replication in the infected cells. In an HIV-1 long terminal repeat (LTR)-luciferase (LTR-luc) reporter assay, ectopic expression of p53 inhibited Tat activity in a dose-dependent manner with little effect on Tat expression in p53−/− cells (Fig. 1F). In order to assess whether p53-mediated Tat suppression is specific to the HIV-1 LTR promoter, other viral promoters were examined under the same conditions. The cytomegalovirus (CMV) promoter and the simian virus 40 (SV40) promoter were minimally affected by ectopic expression of p53 in luciferase reporter assays (Fig. 1G). Thus, p53-mediated Tat inhibition appeared to be due to suppression of Tat activity rather than to p53-derived general inhibition. The LTR-luc reporter alone without Tat was activated neither by ectopic expression of p53 and PKR nor by transfection of cells with siRNA or treatment of cells with kinase inhibitors (Fig. 1H), indicating that LTR-promoter activation was restricted only to Tat without being affected by the experimental procedures. We next examined whether p53 interacts directly with Tat protein. Direct interaction of p53 and Tat has been controversial (33–35), and we could not find any evidence of a direct interaction between p53 and Tat in coimmunoprecipitation (co-IP) assays or yeast two-hybrid analysis (data not shown). These data suggested that another factor(s) may be involved in p53-mediated Tat suppression rather than direct interactions between the two molecules.
FIG 1.
HIV-1 infection induces the expression of p53, and p53 inhibits HIV-1 replication by suppression of Tat activity through PKR. (A) Preactivated PBMCs (upper panel) and CD4+ T cells (lower panel) were infected with HIV-1IIIB at an MOI of 1 for the indicated periods of time, and the levels of p53 and HIV-1 Gag proteins in infected cells were assessed by Western blotting with anti-p53 and anti-p24 antibodies. Mo, mock infection. (B) Upper panel, HIV-1-infected PBMCs were cultured in the presence or absence of 10 μM pifithrin-α (Pft-α) (a p53 inhibitor). Four days after infection, titers of virus in culture supernatants were assessed by p24 ELISA and are represented as means ± SD (n = 3). **, P < 0.01. Lower panel, HIV-1 Gag proteins in infected cells were assessed by Western blotting. (C) The expression of p53 and p53 target genes was assessed 4 days after infection of CD4+ T cells with HIV-1IIIB and 40 h postinfection of C8166 cells with HIV-1 (HXBc2) at an MOI of 1. (D) HIV-1 replication was assessed in p53-normal and p53KD (si-p53) (p53 siRNA; Cell Signaling) C8166 cells 48 h after infection (MOI of 1) from cell extracts by Western blotting (upper panel) and from culture supernatants using a p24 ELISA kit (represented as means ± SD; n = 3) (lower panel). **, P < 0.01 for untreated versus treated cells. (E) Effects of p53 and type 1 interferon (IFN) on HIV-1 replication and PKR expression. Upper panel, p53-normal (si-con) and p53KD (si-p53) C8166 cells were infected with HIV-1 (HXB2) at an MOI of 1 in the presence or absence of 5 μg/ml anti-IFN-α/β neutralizing antibodies (BioLegend Inc.). After 48 h, cell extracts were assessed by Western blotting with anti-HIV-1 Gag antibodies. Lower panel, the titers of HIV-1 in culture supernatants were assessed in triplicates at 48 h postinfection using a p24 ELISA kit (Perkin-Elmer) and are represented as means ± SD (n = 3). **, P < 0.01 versus control siRNA (si-con). (F) HCT116 p53−/− cells were transfected with pLTR-luc (200 ng), pcDNA-Flag-tat (100 ng), and increasing amounts of pcDNA3-p53 (1, 2.5, and 5 μg), together with 20 ng of pCMV-lacZ as a transfection control. Luciferase activity was assessed 2 days after transfection and expressed after normalization to β-galactosidase activity. Data are represented as means ± SD (n = 4). Protein levels were assessed by Western blotting after normalization to β-actin. **, P < 0.01. (G) pLTR-luc, pCMV-luc, and pSV40-luc reporter plasmids were cotransfected with the indicated Tat or p53 plasmid combinations into HeLa cells together with pLTR-luc as a transfection control. Luciferase activity was expressed after normalization with the β-galactosidase activity of each reaction. Protein levels were assessed by Western blotting after normalization with β-actin. **, P < 0.01. (H) HeLa cells were cotransfected with pLTR-luc and other expression vectors together with pCMV-lacZ as a transfection control. In addition, HeLa cells transfected with pLTR-luc and p pCMV-lacZ were treated with each kinase inhibitor as shown. Luciferase activity was assessed. ***, P < 0.001.
PKR plays a crucial role in p53-mediated Tat suppression.
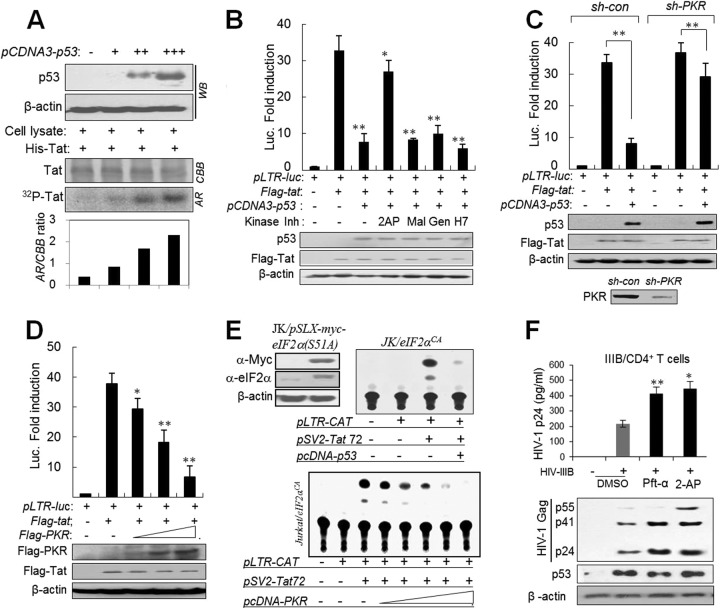
Next we investigated which factors are involved in p53-mediated Tat suppression. We found that a recombinant Tat protein was phosphorylated by p53-expressing cell lysates in a dose-dependent manner (Fig. 2A). We examined which protein kinases downstream of p53 were involved in Tat phosphorylation and inactivation. Among several kinase inhibitors, 2-aminopurine (2-AP) (a potent PKR inhibitor) restored Tat function from the p53-mediated Tat suppression in LTR-luciferase reporter assays (Fig. 2B), suggesting that PKR is likely involved in p53-mediated Tat suppression. Although 2-AP is a well-established PKR inhibitor, its nonspecific inhibition cannot be completely excluded. Therefore, PKR involvement was further verified by significant attenuation of p53-mediated Tat suppression upon PKR silencing (Fig. 2C). In addition, ectopic overexpression of PKR suppressed Tat function in a dose-dependent manner in p53−/− cells (Fig. 2D). To examine the association of PKR-mediated Tat suppression with eIF2α phosphorylation, PKR or p53 was cotransfected to Jurkat/eIF2αCA cells (bearing eIF2α-S51A, a constitutively active form of eIF2α) with pSV2-Tat72. As shown in Fig. 2E, p53/PKR-mediated Tat suppression was not attenuated in eIF2αCA cells, suggesting that p53/PKR-mediated Tat suppression is not associated with the canonical PKR/eIF2α translational inhibition pathway. These results were similarly repeated in human primary CD4+ T cells isolated from human PBMCs. HIV-1IIIB replication was significantly enhanced in CD4+ T cells when infected cells were treated with 2-AP (Fig. 2F). Taken together, our results imply that p53-mediated HIV-1 suppression is likely attributable to the p53/PKR-mediated Tat inactivation after HIV-1 infection.
FIG 2.
PKR plays a crucial role in p53-mediated Tat suppression. (A) p53-expressing cell lysates phosphorylate HIV-1 Tat. HCT116 p53−/− cells were transfected with increasing amounts of pCDNA-p53 (1, 3, and 5 μg). Two days after transfection, p53 expression was assessed by Western blotting (upper panel), and the cell lysates were incubated with recombinant Tat proteins (6×His-Tat) in the presence of [γ-32P]ATP. The reaction mixtures were immunoprecipitated with anti-His MAb and then applied to an SDS-polyacrylamide gel. The gel was stained with Coomassie brilliant blue and autoradiographed (lower panel). (B) HCT116 p53−/− cells were transfected with pLTR-luc (200 ng), pcDNA-Flag-tat (100 ng), and pcDNA3-p53 (5 μg), together with 20 ng of pCMV-lacZ as a transfection control. One day after transfection, cells were treated with the protein kinase inhibitor 2-AP (2 mM 2-aminopurine; PKR inhibitor), Mal (30 μM Mallotoxin; PKC inhibitor), Gen (20 μM Genestein; PTK inhibitor), or H7 (5 μM H-7; PKA and PKC inhibitor). After 24 h, luciferase reporter assays were performed. Values are means ± SD (n = 4). *, P < 0.05, **, P < 0.01. (C) PKR-normal (sh-con) and PKRKD (sh-PKR) HCT116 p53−/− cells were transfected as described above, and luciferase activity was assessed. Values are means ± SD (n = 4). **, P < 0.01. (D) HCT116 p53−/− cells were transfected with increasing concentrations of pcDNA-Flag-PKR, and luciferase activity was assessed. Values are means ± SD (n = 4). *, P < 0.05; **, P < 0.01. (E) Ectopic expression of p53 and PKR inhibits Tat activity in eIF2αCA (pSLX-myc-eIF2α-S51A-transformed) Jurkat cells (JK/eIF2αCA). Myc-eIF2α expression was examined in JK/eIF2αCA cells by Western blotting (upper left). JK/eIF2αCA cells were transfected with pLTR-CAT and pSV2-tat72 together with pCDNA-p53 plasmids as indicated (upper right). JK/eIF2αCA cells were cotransfected with pLRT-CAT and pSV2-Tat72 together with increasing concentrations of pCDNA-PKR plasmid (lower). Intracellular CAT activity was assessed 2 days after transfection. (F) Human CD4+ T cells purified from the PBMCs were infected with HIV-1IIIB at an MOI of 1 for 1 h and then cultured in the presence or absence of 10 μM Pft-α or 2 mM 2-AP (PKR inhibitor) for 4 days. HIV-1 titer in the culture supernatants (upper panel) and intracellular Gag proteins (lower panel) were assessed by p24 ELISA and Western blotting, respectively, and results are represented as means ± SD (n = 3). *, P < 0.05; **, P < 0.01 (untreated versus treated cells). All luciferase data in panels B to G were normalized to the β-galactosidase activity of each transfection control (20 ng of pCMV-lacZ), and expression levels of Tat, p53, and PKR were examined by Western blotting.
PKR interacts with Tat and phosphorylates Ser/Thr residues on the first exon of Tat.
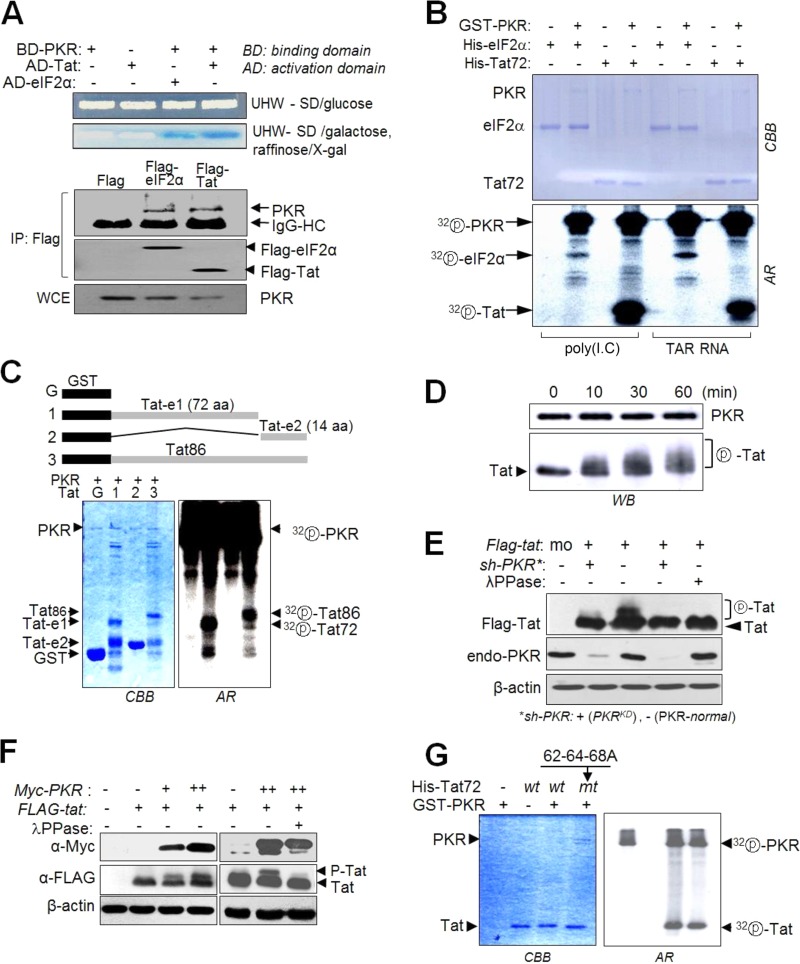
PKR directly interacted with HIV-1 Tat in yeast two-hybrid and co-IP assays (Fig. 3A). In an in vitro kinase assay, Tat was more strongly phosphorylated by PKR than eIF2α (Fig. 3B). Full-length Tat and the first exon part of Tat were phosphorylated to a similar extent by PKR, while the second exon part of Tat alone was not phosphorylated at all under the same conditions (Fig. 3C). The kinetics of Tat phosphorylation by PKR revealed that the Tat band shifted up to multiple bands over time during in vitro kinase assays with recombinant PKR and Tat protein (Fig. 3D), suggesting that Tat is likely to have multiple PKR phosphorylation sites. The Tat bands were clearly shifted in PKR-normal cells, while these shifted band patterns were not observed in PKRKD (sh-PKR) cells (Fig. 3E). Furthermore, the shifted Tat bands in PKR-normal cells disappeared upon additional treatment with phosphatase (λPPase) (Fig. 3E). The intensity of the shifted Tat band increased upon ectopic expression of PKR (Fig. 3F). These data indicated that the shifted Tat bands were attributable to Tat phosphorylation by PKR. Phosphorylation of Tat by PKR on residues 62, 64, and 68 was previously reported in an in vitro biochemical study (24). In the present study, however, S62A-T64A-S68A mutant (mt) Tat was also equivalently phosphorylated by PKR (Fig. 3G, top), indicating that S62-T64-S68 are not the major sites on Tat for phosphorylation by PKR.
FIG 3.
PKR phosphorylates the first exon of HIV-1 Tat. (A) Upper panel, yeast two-hybrid experiments with PKR and Tat as described in Materials and Methods. Lower panel, endogenous PKR was assessed by Co-IP with anti-Flag Ab in C8166 cells transfected with pcDNA-Flag-tat/eIF2α. (B) Recombinant GST-PKR protein (0.5 μg), preactivated with poly(I·C) or TAR RNA for 1 h, was incubated with 0.5 μg of recombinant His-eIF2α or His-Tat proteins in the presence of 1 μCi of [γ-32P]ATP, and phosphorylation of Tat and eIF2α was examined by autoradiography after SDS-PAGE. (C) Recombinant GST-Tat proteins encoding the first exon (72 aa), second exon (14 aa), or full-length Tat (86 aa) were incubated with preactivated PKR in the presence of [γ-32P]ATP, separated on a 12% polyacrylamide gel, and then assessed by autoradiography. (D) Preactivated PKR (GST-PKR) was incubated with column-purified His-Tat72 for 10 to 60 min at 30°C. Reaction mixtures were separated by 15% SDS-PAGE and then subjected to Western blot analysis. (E) PKRKD (sh-PKR+) and PKR-normal (−) HEK293 cells were transfected with pcDNA3-Flag-tat. Cells were treated with 50 nM calyculin A (a phosphatase inhibitor) for 1 h prior to harvest, and then cell extracts were assessed by Western blotting. Parts of samples were additionally treated for 1 h with λPPase (1 U/μl) before SDS-PAGE. (F) PKR-mediated Tat phosphorylation in vivo. HEK293 cells were cotransfected with pCDNA-Flag-tat and different concentrations of pCDNA-Myc-PKR. Two days after transfection, cells were treated with 50 nM calyculin A for 1 h prior to harvest, and cell extracts were separated by 15% SDS-PAGE and then subjected to Western blot analysis with anti-Flag, anti-Myc, and anti-β-actin antibodies. One of the samples was treated with λPPase prior to the Western blot assay. (G) wt or 62-64-68A mt Tat (72 aa) was incubated with preactivated PKR in the presence of [γ-32P]ATP. The reaction products were separated on a 12% SDS-polyacrylamide gel and then examined by autoradiography.
PKR phosphorylates Tat on five Ser/Thr residues, mainly at T23, T40, and S46.
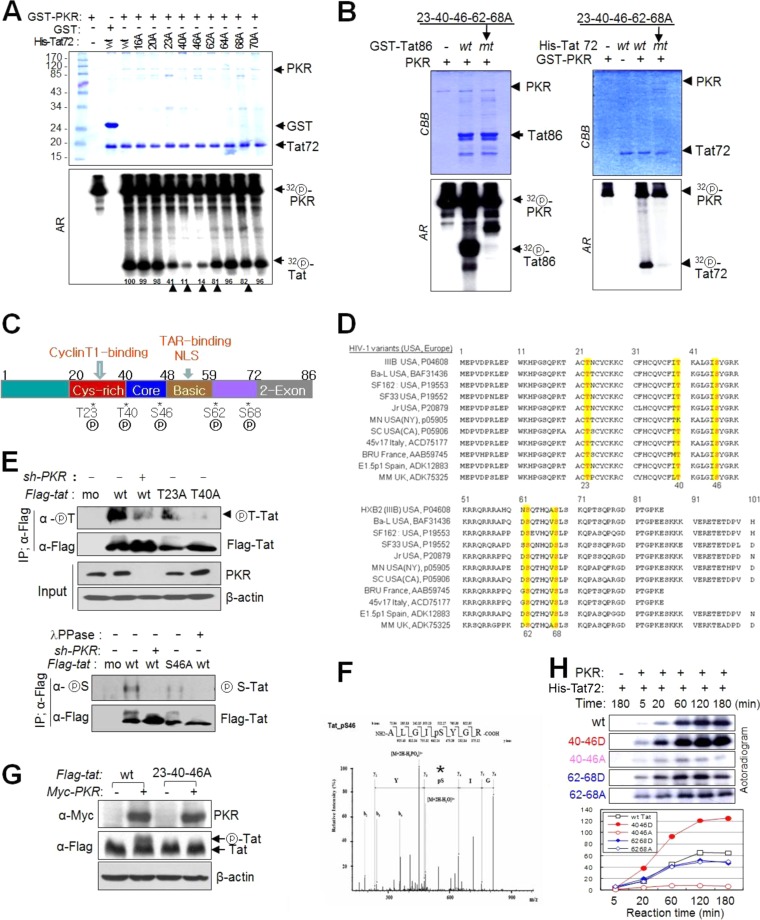
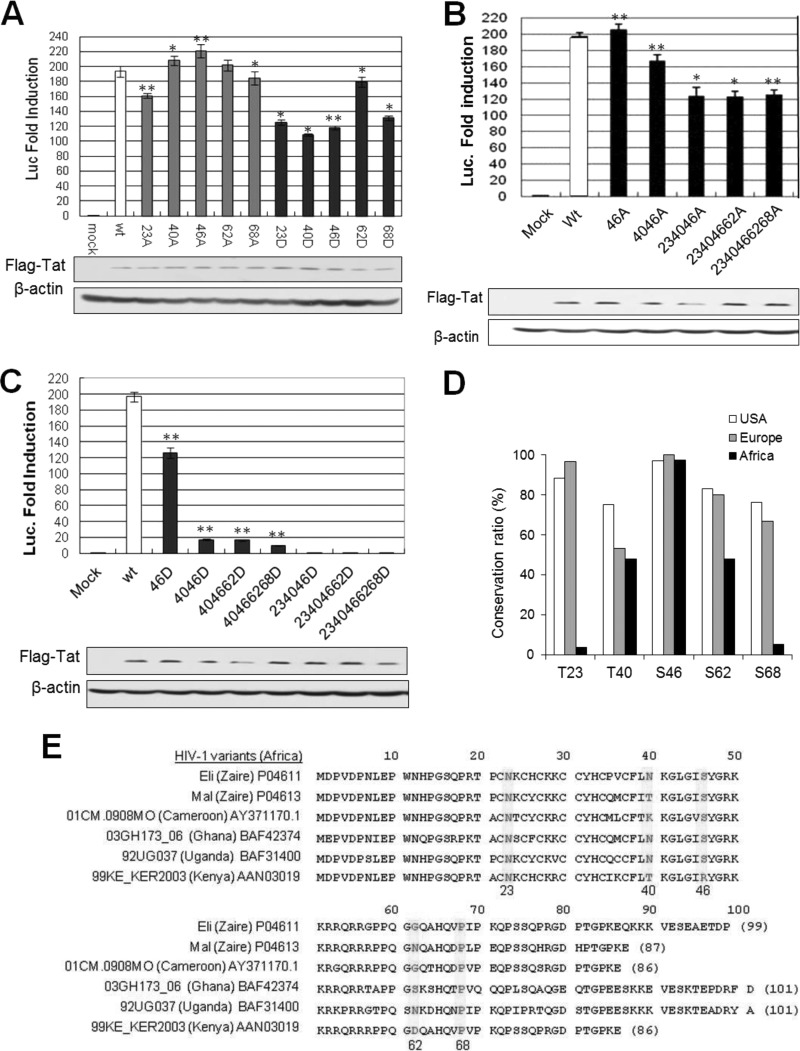
To determine the target sites for Tat phosphorylation by PKR, we constructed Tat mutants with Ala substitutions at nine Ser/Thr residues and performed in vitro kinase assays with PKR. Tat phosphorylation by PKR was markedly attenuated by the substitution T23A, T40A, or S46A, weakly attenuated by S62A or S68A, and not attenuated at all by T64A compared with wt Tat (Fig. 4A). When the five Ser/Thr residues in the first exon were replaced by Ala, not only the first exon (Tat72; Fig. 4B, right panels) but also full-length Tats (Tat86; Fig. 4B, left panels) were not phosphorylated by PKR, suggesting that PKR phosphorylates exclusively the 5 Ser/Thr sites in the first exon of Tat. These 5 phosphorylation sites on the Tat functional motif (Fig. 4C) were found to be highly conserved in HIV-1 strains such as Ba-L, Bru, Oyi, JR, and IIIB, which are predominant in the United States and Europe (Fig. 4D; see Table S4 in the supplemental material). In order to demonstrate intracellular phosphorylation of Tat on these Ser/Thr sites, we tried to generate site-specific phospho-Tat antibodies, but our attempts were unsuccessful in several trials. Instead, we used anti-phospho-Thr and anti-phospho-Ser antibodies after immunoprecipitation to assess intracellular Tat phosphorylation by PKR at these Ser or Thr sites. Intracellular Tat phosphorylation at Thr sites (T23 and T40) was markedly impaired by either T23A or T40A substitution (Fig. 4E, upper panel). Among the three PKR Ser target sites (S46, S62, and S68), Tat phosphorylation was significantly attenuated by the S46A substitution (Fig. 4E, lower panel) in PKR-normal cells. However, in PKRKD (sh-PKR) cells, intracellular Tat phosphorylation was barely detected or undetectable by phosphor-Ser/Thr antibodies (Fig. 4E). These data demonstrated the intracellular phosphorylation of Tat at specific Ser/Thr sites by PKR. However, we cannot exclude the possibility that another kinase(s) might be involved in intracellular Tat phosphorylation, because a small amount of Tat phosphorylation was often detected even in the absence of PKR (sh-PKR), as shown in Fig. 4E (upper panel). S46 phosphorylation by PKR was further evidenced by ESI-MS/MS analysis (Fig. 4F). In addition, the shifted Tat bands observed in wt Tat-expressing cells were not detected in 23-40-46A mt Tat-expressing cells (Fig. 4G). These data suggested that (i) PKR phosphorylated Tat predominantly at residues T23, T40, and/or S46 and (ii) T40-S46-phosphorylation may affect subsequent phosphorylation events. This assumption was verified by in vitro kinase assays. The 40-46A mt Tat-72 (with T40A-S46A substitutions) was barely phosphorylated by PKR even though 3 remaining phosphorylation sites were intact, while 40-46D mt (Ser/Thr-to-Asp substitution, pseudophosphorylated form) was significantly phosphorylated by PKR over time, and the intensity of each band was much stronger than that of wt Tat-72 (Fig. 4H). In contrast, there was no difference in Tat phosphorylation between the 62-68A and 62-68D mutants, and band intensity was slightly attenuated compared to that of wt Tat (Fig. 4H). These data suggest that PKR-mediated Tat phosphorylation is a series of allosteric reactions (the first reaction facilitates the following reactions) starting from T40-S46 phosphorylation.
FIG 4.
PKR phosphorylates HIV-1 Tat on five different Ser/Thr residues. (A) wt and 9 mt Tat72 proteins were incubated with preactivated PKR in the presence of [γ-32P]ATP. The reaction products were separated on a 12% SDS-polyacrylamide gel (upper panel) and assessed by autoradiography (lower panel). Phosphor-Tat bands were quantified using TINA 2.0 software (Raytest, Straubenhardt, Germany) and represented by the percentage of the relative intensity of each band. Arrowheads indicate postulated PKR-mediated Tat phosphorylation sites. (B) GST-attached wt and mt (5 Ala substitutions at positions 23, 40, 46, 62, and 68) Tat86 (left) and Tat72 (right) proteins were incubated with preactivated PKR, Tat phosphorylation was assessed by autoradiography following SDS-PAGE. (C) PKR-mediated phosphorylation sites on the functional domain of HIV-1 (HXB2) Tat. (D) Amino acid sequences of the Tat proteins of typical HIV-1 variants in the United States and Europe (see Table S4 in the supplemental material). Five PKR target Ser/Thr sites are indicated by yellow boxes. (E) PKR-normal or PKRKD HEK293 cells were transfected with Flag-tagged wt (86 aa) and single mt Tat plasmids. Cell extracts were immunoprecipitated with anti-Flag antibody and then assessed by Western blotting with phospho-Thr (upper panel)- and phospho-Ser (lower panel)-specific antibodies. (F) ESI-MS/MS of tryptic peptides of Tat. The assigned spectrum matched the given phosphopeptide sequence corresponding to residues 42 to 49 of HIV-1 Tat (ALGIpSYGR, m/z). *, phosphorylation site on the peptide. (G) HEK293T cells were cotransfected with Flag-tagged wt and 23-40-46A triple mt Tat86-expressing plasmids and PKR-expressing (Myc-PKR) plasmids. Samples were assessed by Western blotting with anti-Flag and anti-Myc antibodies. (H) His-tagged wt and double mt (S/T-to-D/A substitutions) Tat72 proteins were incubated with preactivated PKR (GST-PKR) for the indicated periods of time. Reaction products were assessed by autoradiography after SDS-PAGE (left). The relative band intensity of each sample is presented in a histogram (right).
PKR-mediated Tat phosphorylation inhibits Tat activity.
To examine the effects of Tat phosphorylation on Tat activity, we constructed recombinant Tat plasmids encoding single or multiple amino acid substitutions of Asp (D; phospho-mimic amino acid) and Ala (A; non-phospho amino acid) at the five PKR target Ser/Thr sites. Tat activity was significantly attenuated by D substitution at each Ser/Thr site except S62D, while the activities of corresponding A-mt Tats, except T23, were equivalent to or slightly higher than that of wt Tat (Fig. 5A). In addition, Tat activity was slightly attenuated by multiple A mutations (Fig. 5B), while being almost obliterated by multiple D mutations near the arginine-rich motif (ARM) region (46D, 4046D, 404662D, and 40466268D) (Fig. 5C). When the S23D mutation was added to the multiple D mutations around the ARM (234046D, 23404662D, and 2340466268D), Tat activity was completely abrogated (Fig. 5C), suggesting an additional important role for T23 in Tat activity. Tat T23 is highly conserved (over 90%) in HIV-1 strains that are prevalent in the United States and Europe but is uncommon (<4%) in African strains (Fig. 5D), most of which encode the natural T23N substitution (Fig. 5E). Our findings indicate that PKR-mediated Tat phosphorylation, at least within the five Ser/Thr residues of the first exon, suppresses Tat activity, followed by inhibition of HIV-1 replication after infection.
FIG 5.
Effects of PKR-mediated Tat phosphorylation on Tat activity. (A) Single mt (Ser/Thr to non-phospho amino acid Ala [A] or phospho-mimic amino acid Asp [D]) Tat activity in luciferase reporter assay. HeLa cells were cotransfected with each Flag-tagged wt or mt Tat plasmid (100 ng) and pLTR-luc reporter plasmid (200 ng), with pCMV-LacZ (20 ng) as a transfection control. Two days after transfection, luciferase activity was assessed in triplicates, and fold activities normalized with the transfection control are represented as means ± SD (n = 3). *, P < 0.05; **, P < 0.01 (mt versus wt Tat). (B and C) The activities of multiple A mutant (B) and D mutant (C) Tats were assessed by LTR-mediated luciferase reporter assay. (D) The sequence conservation ratio of each PKR target Ser/Thr site on HIV-1 Tat was assessed among 206 HIV-1 strains (101 strains from the United States, 30 from Europe, and 75 from Africa). Information on the 206 HIV-1 strains and their respective Tat amino acid sequences is shown in Table S4 in the supplemental material. (E) Alignment of HIV-1 Tat sequences of 6 African strains. Five PKR target sites, designated by yellow boxes, are mostly mutated with the non-phospho-form amino acids.
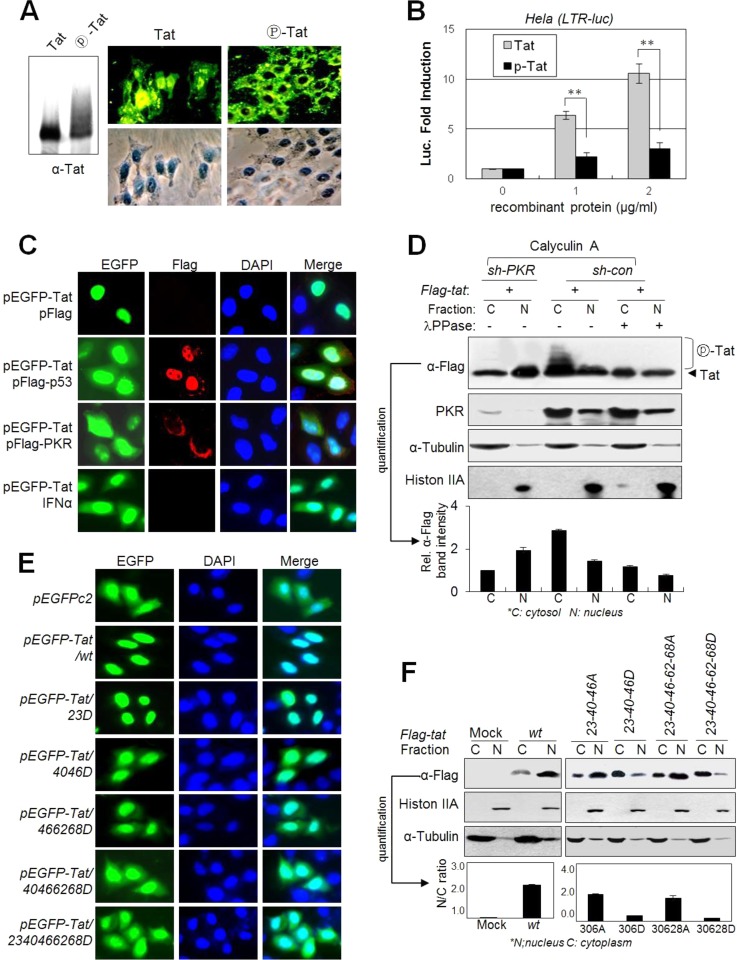
PKR-mediated Tat phosphorylation inhibits Tat nuclear localization.
HIV-1 Tat is a nuclear protein, and the ARM (overlapped with the protein transduction domain [PTD]) is essential for Tat nuclear localization (36, 37) and plasma membrane transduction potential (38, 39). Residues T40, S46, S62, and S68 are adjacent to the ARM in the three-dimensional (3-D) structure of Tat (40). We examined the effects of PKR-mediated Tat phosphorylation on the membrane transduction potential and nuclear localization of Tat. When HeLa cell monolayers were incubated with phospho-free recombinant Tat protein in culture medium, Tat protein was observed mainly in the nucleus, but once fully phosphorylated by PKR in vitro, phospho-Tat protein resided mostly in the cytoplasm after membrane transduction (Fig. 6A). When HeLa(LTR-luc) cells were treated, recombinant phospho-free normal Tat protein effectively induced expression of the luciferase gene in the nucleus in a dose-dependent manner, compared to ineffective induction by fully phosphorylated Tat protein (Fig. 6B). These data suggest that nuclear localization after membrane transduction of Tat protein was markedly impaired by PKR-mediated phosphorylation. Even though intracellular enhanced green fluorescent protein (EGFP) resides both in the cytoplasm and in the nucleus, ectopically expressed EGFP-Tat localized mainly to the nucleus. However, the nuclear localization of EGFP-Tat was inhibited by coexpression of p53 or PKR as shown by the distribution of control EGFP (Fig. 6C). This inhibitory phenomenon was also observed when cells were treated with IFN-α, which is a well-known PKR activator (Fig. 6C). In addition, phospho-shifted Tat bands were detected only in the cytoplasmic fraction of PKR-normal (sh-con) cells, whereas the majority of Tat protein in PKRKD (sh-PKR) cells was detected in the nuclear fraction (Fig. 6D). Analysis of D-mt Tats to assess the effect of phosphorylation on the nuclear localization of Tat revealed that Tat nuclear localization was significantly impaired by D mutations at the PKR target Ser/Thr sites adjacent to the ARM (Fig. 6E). The intracellular distribution patterns of EGFP-Tat40-46-62-68D and EGFP-Tat23-40-46-62-68D were similar to that of control EGFP, while the T23D mutation alone did not affect nuclear localization of Tat (Fig. 6E), suggesting that Tat phosphorylation near the ARM might impair the nuclear localization signal of Tat, probably by disrupting the positive charge of the ARM. These results were further supported by analysis of the nuclear and cytoplasmic fractions. D-mt Tat proteins, particularly all-D-mt Tat, were detected mostly in the cytoplasmic fraction, whereas A-mt Tats were detected mainly in the nuclear fraction (Fig. 6F). These data indicate that the nuclear localization capacity of Tat was strongly impaired by PKR-mediated Tat phosphorylation, particularly adjacent to the ARM domain.
FIG 6.
PKR-mediated Tat phosphorylation blocks Tat nuclear localization. (A) Phospho-free normal Tat and phospho-Tat (fully phosphorylated by PKR for 4 h) recombinant proteins were assessed by Western blotting (left panel) with anti-Tat Ab. HeLa cell monolayers were incubated with 1 μg/ml normal or fully phosphorylated Tat proteins in DMEM. After 1 h of incubation, cells were washed and gently trypsinized (1× trypsin-EDTA) for 2 min at 37°C to remove membrane-bound unpenetrated Tat protein. Intracellular Tat was assessed by immunocytochemistry with anti-Tat and FITC-conjugated secondary antibodies. (B) HeLa cells transfected with pLTR-luc (200 ng) and pCMV-lacZ (20 ng) plasmids were treated with increasing amounts of normal Tat and phospho-Tat proteins (0 to 2 μg/ml). After 24 h, luciferase activity was assessed and is represented as means ± SD (n = 3). *, P < 0.01. (C) pEGFP-tat-transfected HeLa cells were cotransfected with pcDNA3-Flag-p53 or pcDNA3-Flag-PKR or treated with 1,000 U/ml of IFN-α. Twenty-four hours after transfection, cells were photographed under a fluorescence microscope (magnification, ×400). (D) PKR-normal (sh-con) and PKRKD (sh-PKR) HEK293 cells were transfected with pFlag-tat. Two days after transfection, cells were treated with 50 nM calyculin A for 1 h prior to harvest, and then nuclear and cytosolic fractions from the cell extracts were prepared using the NER/PER kit (Pierce) and assessed by Western blotting. Shifted Tat bands are indicated. Relative intensities of Tat bands in the Western blot are represented as means ± SEM (n = 3) after quantification three times using TINA 2.0 software (Raytest, Straubenhardt, Germany). (E) HeLa cells were transfected with GFP-conjugated wt or D-mt Tat plasmids. Two days after transfection, cells were fixed and photographed under a fluorescence microscope (magnification, ×200). (F) HEK293 cells were transfected with Flag-tagged wt or mt Tat plasmids as indicated. Nuclear and cytosolic fractions from the cell extracts were assessed by Western blotting with appropriate antibodies. Relative intensities of the Tat bands in the cytoplasm and the nucleus are represented.
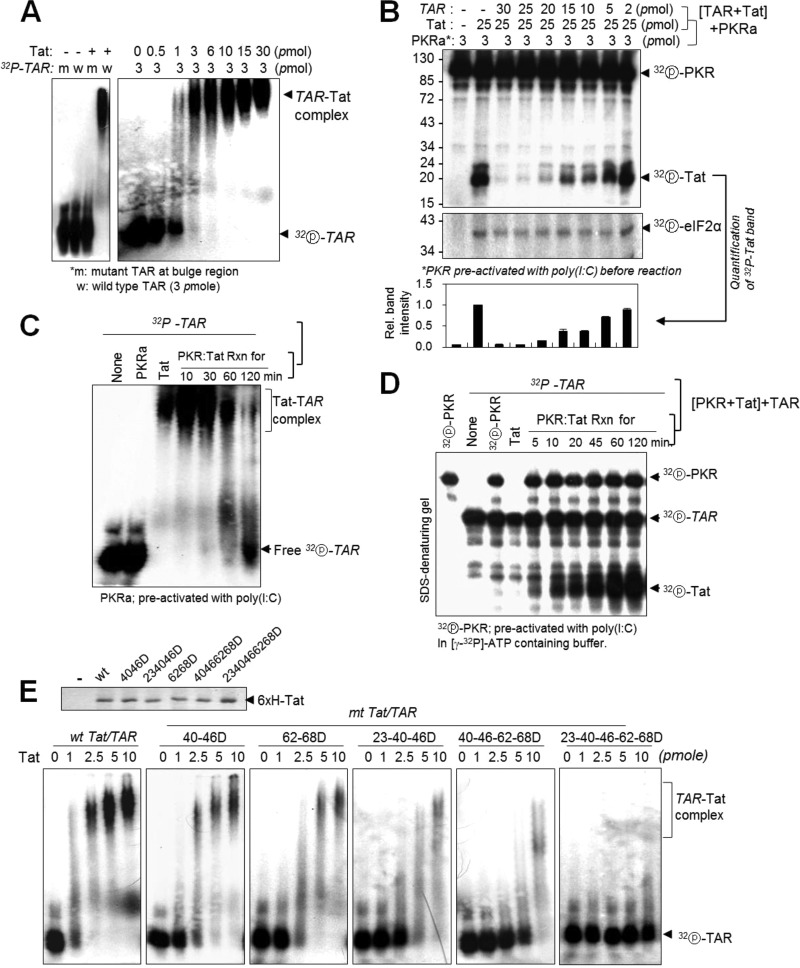
PKR-mediated Tat phosphorylation disrupts Tat binding to TAR RNA.
The Tat-TAR interaction is essential for provirus transcription, and the ARM domain on Tat plays a crucial role in Tat binding to TAR RNA (36, 41). We performed electrophoretic mobility shift assay (EMSAs) with a 32P-TAR probe and recombinant Tat protein at different molar ratios to determine the optimal concentrations for binding. 32P-labeled TAR RNA was fully shifted by Tat at a 1:1 molar ratio for Tat (3 pmol) and TAR (3 pmol), while mutant TAR RNA was not shifted at all (Fig. 7A). When Tat was preincubated with different amounts of TAR RNA and then incubated with active PKR (PKRa) [preactivated with poly(I·C)], TAR-bound Tat was not phosphorylated by PKRa, while a molar excess of Tat unbound to TAR RNA was strongly phosphorylated (Fig. 7B), implying that once bound to TAR, Tat was protected from PKR-mediated phosphorylation. Conversely, when Tat was preincubated with PKRa, it was unable to shift TAR RNA in an EMSA (Fig. 7C). In SDS-denaturing gel experiments, the non-TAR-bound free Tat in Fig. 7C was found to be strongly phosphorylated by PKRa, depending on the preincubation time (Fig. 7D). These data strongly suggested that PKR-mediated Tat phosphorylation impaired Tat binding to TAR RNA, probably due to a conformational change of TAR-binding sites on Tat. These findings were further supported by additional EMSAs with TAR RNA and Tats with D substitutions at PKR target Ser/Thr sites. Tat-TAR interactions were significantly attenuated by accumulation of D mutations on Tat and were almost abolished by combined 23-40-46-62-68 D mutations (Fig. 7E).
FIG 7.
PKR-mediated Tat phosphorylations disrupt Tat binding to TAR RNA. Tat-TAR interaction was assessed by RNA retardation assay as described in Materials and Methods. (A) Left panel, 32P-labeled wild type (w) and mutant (m) (22-UCU-24 to CUC at bulge region) TAR RNAs (3 pmol) were incubated with 3 pmol of Tat protein on ice for 15 min, separated on a 3% native polyacrylamide gel, and autoradiographed. Right panel, increasing amounts of Tat protein were incubated with 3 pmol of 32P-labeled wt TAR RNA and then assessed as described above. (B) Tat protein (25 pmol) was preincubated with different amounts of TAR RNA, and the reaction mixtures were further incubated with PKR preactivated with poly(I·C) in a kinase reaction buffer containing [γ-32P]ATP. The reaction products were separated on a 12% SDS-polyacrylamide gel and assessed by autoradiography. Relative intensities of phosphor-Tat bands are represented as means ± SD (n = 3). (C and D) Tat protein (15 pmol) was incubated with preactivated PKR (3.5 pmol) in the presence of cold ATP (C) or [γ-32P]ATP (D) for the indicated time periods. The reaction mixtures were further incubated with 3 pmol of 32P-TAR RNA on ice for 15 min, separated on 3% native (C) or 12% denaturing (D) polyacrylamide gels, and analyzed by autoradiography. (E) Tat (wt and mt) proteins were fully normalized (upper panel), and then the indicated amounts of wt and mt Tat proteins were incubated with 3 pmol of 32P-labeled TAR RNA, separated on a 3% native polyacrylamide gel, and assessed by autoradiography.
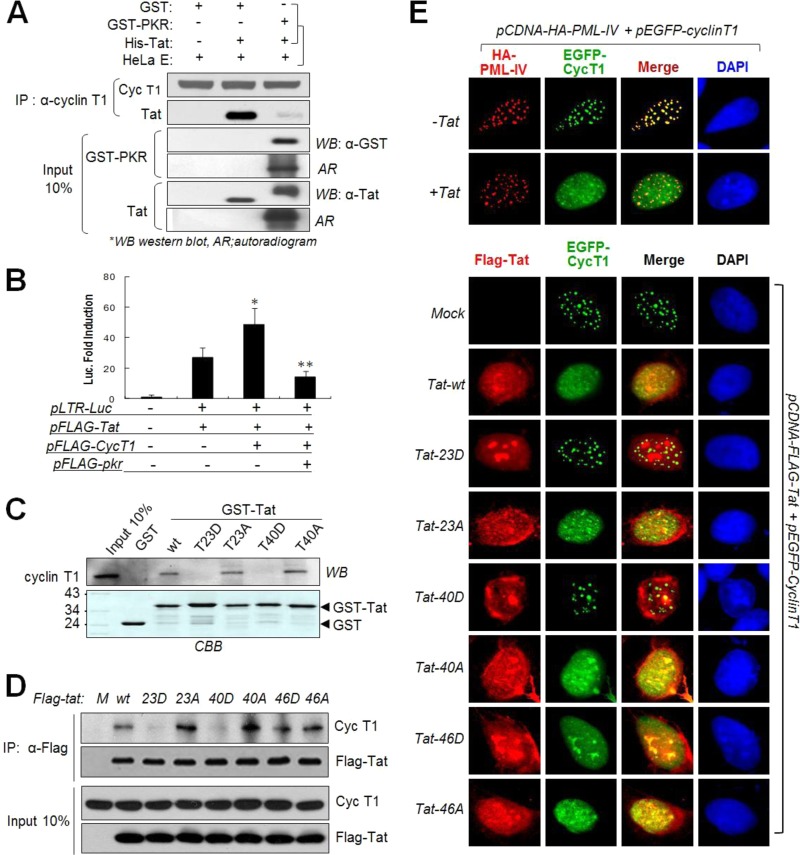
PKR-mediated Tat phosphorylations inhibit Tat-cyclin T1 (P-TEFb) interactions.
The interaction between cyclin T1 (CycT1) and HIV-1 Tat is well established with regard to efficient transcriptional elongation of viral RNAs (42, 43). To determine the effects of Tat phosphorylation on the Tat-CycT1 interaction, we performed co-IP assays. Control recombinant Tat was coimmunoprecipitated with endogenous CycT1, while Tat which had been fully phosphorylated by PKR was not (Fig. 8A). Tat-CycT1-mediated reporter (LTR-luc) expression was significantly downregulated by coexpression of PKR (Fig. 8B). These data suggest that PKR-mediated Tat phosphorylation disrupts Tat-CycT1 interactions. In a GST pulldown assay, T23D- and T40D-mt Tats were unable to pull down endogenous CycT1, while corresponding A mutants were still effective in the pulldown of CycT1 in HeLa cell extracts (Fig. 8C). In in vivo co-IP assays, 23D- and 40D-mt Tats did not immunoprecipitate CycT1, while 23A-, 40A-, and 46D/A mutants were as effective as wt Tat in co-IP with CycT1 (Fig. 8D). These results suggest that T23 and/or T40 is involved in the interaction with CycT1 and that PKR-mediated Tat phosphorylation at one or both of these sites disrupted Tat-CycT1 interactions.
FIG 8.
PKR-mediated Tat phosphorylation disrupts Tat-cyclin T1 interaction. (A) Fully phosphorylated or unphosphorylated Tat (His-Tat) was incubated with HeLa cell extracts at 4°C for 3 h, immunoprecipitated with anti-CycT1 antibody (Santa Cruz), and assessed by Western blotting (WB) and autoradiography (AR). (B) CytT1-mediated enhancement of Tat activity is abrogated by interferon treatment which blocks Tat-CycT1. pFlag-tat, pFlag-pkr, CycT1-expressing plasmid, and pLTR-luc reporter genes were cotransfected together with a pCMV-β-Gal transfection control into HeLa (eIF2αCA) cells. After 48 h, luciferase activities were assessed in triplicates from each sample, normalized with the transfection control, and then represented as means ± SD (n = 3). *, P < 0.01; **, P < 0.05. (C) GST-tagged wt and mt Tat proteins were incubated with HeLa cell nuclear extracts and subjected to GST pulldown assay followed by Western blotting. Input GST-Tat proteins were stained (bottom). (D) Co-IP of CycT1 and wt and A/D-mt Tat in vivo. HEK293 cells were transfected with Flag-tagged wt or D/A-mt Tat plasmids (pcDNA3-Flag-tat). After 48 h, cell extracts were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting assay. (E) Upper panels, pHA-PML-IV(red) and pEGFP-cyclin T1 (green) plasmids were cotransfected with or without the Flag-Tat(wt) plasmid into U2OS cells. Cells were stained with phycoerythrin (PE)-conjugated MAb. Lower panels, U2OS cells were transfected with Flag-tagged wt or mt Tat plasmid (pcDNA-Flag-tat) and pEGFP-cyclin T1, fixed, and stained with PE-conjugated anti-Flag MAb. Images were taken with a confocal microscope.
CycT1 has been reported to physically interact with promyelocytic leukemia (PML) nuclear bodies, and Tat expression redistributes CycT1 out of PML bodies via the Tat-CycT1 interaction (44). Consistent with previous reports, EGFP-CycT1 fluorescence was diffused in the nucleoplasm in the presence of wt Tat (Fig. 8E, upper panels). Similar CycT1 diffusion was observed in cells expressing 23A-mt, 40A-mt, 46A-mt, or 46D-mt Tat but not in cells expressing 23D- or 40D-Tat mutants (Fig. 8E, lower panels). Endogenous CycT1 speckles were also redistributed by wt Tat but were not by 23D- or 40D-Tat mutants (data not shown). These data strongly support our assertion that T23 and/or T40 phosphorylation impedes Tat-CycT1 interaction.
PKR plays a crucial role in the p53-mediated HIV-1 restriction via Tat phosphorylation.
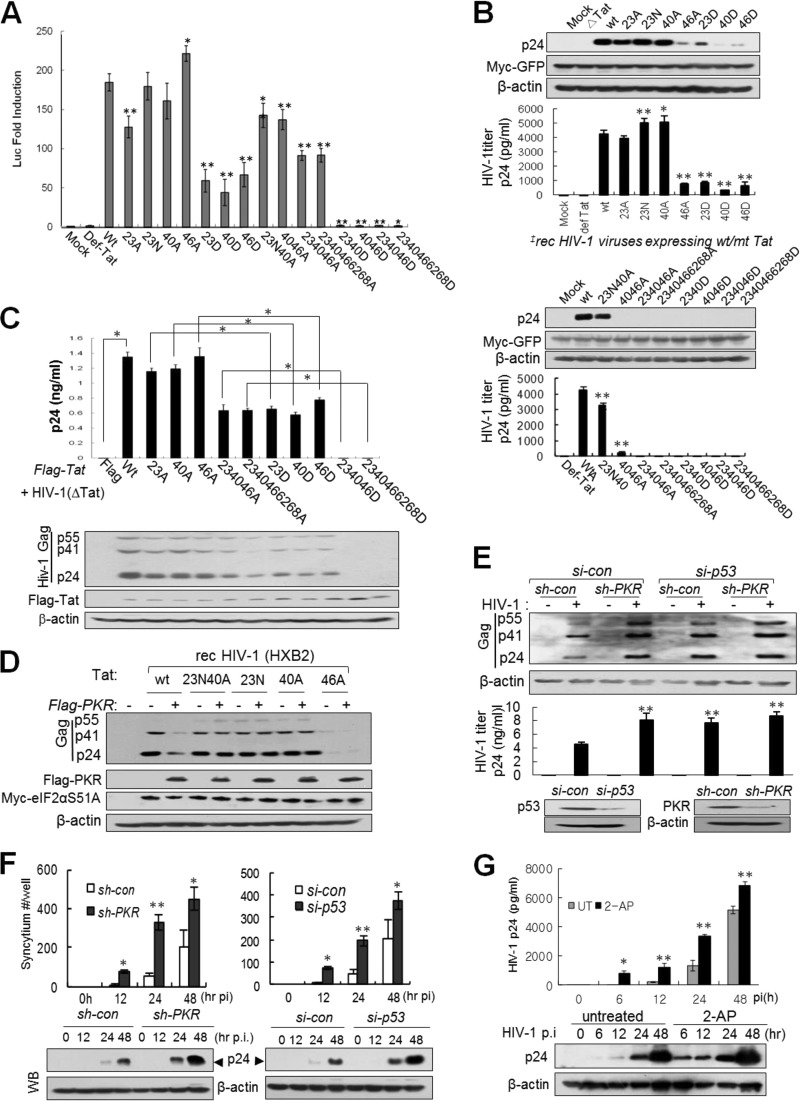
To examine the effects of Tat phosphorylation on HIV-1 replication, we constructed recombinant HIV-1 cDNAs harboring mt Tat by subcloning of cDNA fragment at unique NheI/NcoI sites after PCR-based mutagenesis and generated recombinant viruses by transfection of these cDNAs. In a reporter assay, luciferase activity was significantly reduced or completely abolished when HeLa(LTR-luc) cells were transfected with HIV-1 cDNAs encoding single D-mt or multiple D-mt Tat, respectively, while the luciferase activities of the single A/N-mt Tat-encoding HIV-1 cDNAs (except 23A-Tat) were similar to that of wt-Tat HIV-1 cDNA (Fig. 9A). The natural amino acid substitution of Asp (N) for Thr23 (T23N) on Tat, which is prevalent in African stains, has been reported to increase Tat activity (16). Among the recombinant HIV-1 cDNAs, only those with T23N tat or T40A tat genes produced high titers of progeny viruses after transfection of cDNAs, while the recombinant cDNAs with single or multiple corresponding Tat D mutations produced significantly reduced or undetectable levels of progeny viruses in the same transfection experiments (Fig. 9B). We also performed a complementation assay by the cotransfection of Tat-defective HIV-1 cDNA and mt/wt-Tat-expressing plasmids. Single or multiple A-mt Tats at PKR target sites were much more effective than compatible D-mt Tats for the production of Tat-defective HIV-1 (Fig. 9C). The replication capacity of wt HIV-1 was significantly inhibited by ectopic overexpression of PKR, while that of recombinant HIV-1 progeny viruses possessing Tat with 23N, 40A, or 23N40A substitutions was not as significantly inhibited compared to that of wt HIV-1 under the same conditions, even in the eIF2αCA cells (Fig. 9D). In contrast, HIV-1 (HXB2) replication was markedly enhanced by PKR knockdown and/or p53 knockdown (Fig. 9E). However, the HIV-1 replication in PKRKD cells was not additionally increased by p53/PKR double knockdown (Fig. 9E), suggesting that p53-mediated host restriction of HIV-1 replication is mostly dependent on PKR activity in infected cells. To observe the physiological effects of p53 and PKR on the active replication of HIV-1 in infected T cells, syncytium formation was measured in HIV-1IIIB-infected p53KD or PKRKD C8166 cells. The number of syncytia was also substantially enhanced in PKRKD (sh-PKR) or p53KD (si-p53) cells compared to that in normal (si-con) cells (Fig. 9F). In good agreement with the results shown in Fig. 9F, treatment of HIV-1-infected cells with 2-AP enhanced HIV-1 (HXB2) replication in p24 ELISA (Fig. 9G). All our experimental results strongly suggest that cellular PKR plays a crucial role in the p53-mediated host restriction of HIV-1 replication via Tat phosphorylation and inactivation in three different ways, as summarized in Fig. 10.
FIG 9.
Effects of PKR-mediated Tat phosphorylation on HIV-1 replication (A) HeLa cells were cotransfected with recombinant HIV-1 cDNA (HXB2) containing a wt or mt Tat gene and pLTR-luc, together with pCMV-lacZ as a transfection control. After 48 h, luciferase activity was assessed. Values are means ± SD (n = 4). *, P < 0.05; **, P < 0.01 (wild-type versus mutant viruses). (B) C8166 cells were cotransfected with HIV-1 cDNA (HXB2) containing wt, single (upper panels) or multiple (lower panels) A/D-mt Tat, and pMyc-GFP as a transfection control. After 4 days, HIV-1 titers in the infected cells and the culture supernatants were assessed by Western blotting and p24 ELISA (means ± SD, n = 3). *, P < 0.05; **, P < 0.01 (wild-type versus mutant viruses). (C) Complementation assay of Tat-defective HIV-1 production in Tat-expressing cells. C8166 cells (1 × 106 cells/sample) were cotransfected with 2.5 μg of pcDNA3-Flag-tat86 (wt or mt) and 5 μg of HXB2rTat cDNA (Tat-defective HIV-1) by electroporation. Five days after electroporation, HIV-1rTat titers in the supernatants were assessed by p24 ELISA (upper panel) and intracellular HIV-1 was assessed by Western blotting (lower panels). (D) C8166 cells were cotransfected with pcDNA3-Flag-PKR and pSLX-Myc-eIF2α51A plasmids. After 24 h, cells were infected with wt- or mt-Tat-expressing recombinant HIV-1 for 48 h. Cells were harvested and subjected to Western blot analysis. (E) p53KD (si-p53) and/or PKRKD (sh-PKR) C8166 cells were infected with HIV-1 at an MOI of 1 for 48 h. HIV-1 titers in the infected cells (upper panels) and the culture supernatants (lower panels) were assessed by Western blotting and p24 ELISA (means ± SD, n = 3). **, P < 0.01 (control versus knockdown cells). (F) PKRKD or p53KD C8166 cells were infected with HIV-1IIIB at an MOI of 1. Upper panels, the numbers of syncytia counted in 3 different cultures at 48 h postinfection are represented as means ± SD (upper). *, P < 0.05; **, P < 0.01. Lower panels, cells were harvested and subjected to Western blot analysis with anti-p24 antibody. (G) C8166 cells were infected with HIV-1 (HXB2) at an MOI of 1 for 1 h, and then cultured in the presence or absence of 2 mM 2-AP. The HIV-1 titer in culture supernatants was assessed using a p24 ELISA kit (Perkin-Elmer) and represented as means ± SD (n = 3) (upper panel), and HIV-1 p24 in cell lysates was assessed by Western blotting with anti-p24 antibody (lower panel). *, P < 0.05; **, P < 0.01 (2-AP-treated versus untreated [UT] cells).
FIG 10.
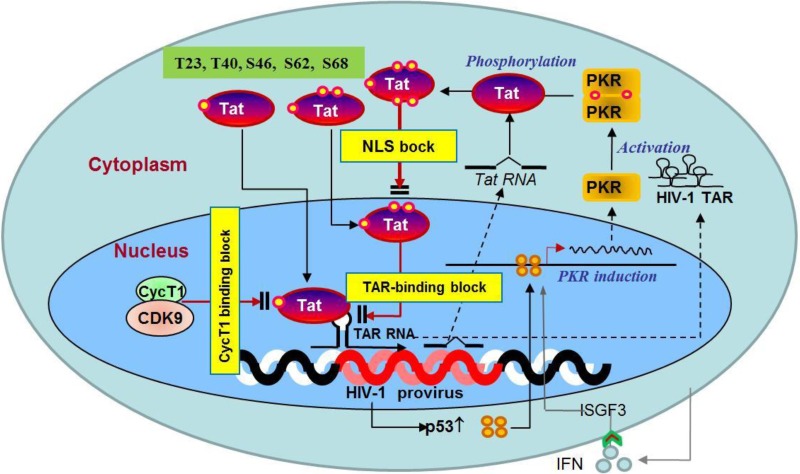
Schematic illustration of p53-derived host restrictions against HIV-1 replication. HIV-1 provirus integration activates p53, followed by the induction of PKR. PKR phosphorylates Tat at least five Ser/Thr residues (T23, T40, S46, S62, and S68), which inhibits Tat function in three different ways: (i) phosphorylation near the ARM inhibits Tat translocation into the nucleus, (ii) Tat phosphorylation abolishes Tat-TAR binding, and (iii) Tat phosphorylation at T23 and/or T40 obliterates the Tat-cyclin T1 interaction.
DISCUSSION
HIV-1 infections are detrimental to host survival and reproduction, and hosts have evolved numerous restriction mechanisms to protect themselves from HIV-1 replication (3, 45). It has been evidenced that p53 may play an important role in HIV-1 restriction. p53 was first suggested to inhibit HIV-1 replication by direct binding to the TFIID and inhibits HIV-1 LTR promoter activity (7). One year later, it was reported that p53 suppresses Tat-mediated LTR transcription (6). Recently, it was found that overexpression of wild-type p53 prevents the phosphorylation of serine 2 in the carboxyl-terminal domain (CTD) of RNA polymerase II on the LTR (8). It has also been reported that HIV-1 infection upregulates p53 expression and activation in primary CD4+ T cells (9, 10) and activates DNA-dependent protein kinase, leading to CD4+ T cell depletion (46). The combined reports indicate that p53 is a strong candidate for HIV-1 restriction under physiological conditions; however, the detailed mechanism underlying p53-mediated restriction of HIV-1 replication is not completely understood.
Here, we found that HIV-1 infection activates p53, followed by induction of PKR, which plays a pivotal role in p53-derived Tat suppression (Fig. 1 and 2) via Tat phosphorylation and inactivation (Fig. 3 to 8), resulting in the inhibition of HIV-1 replication (Fig. 9). The amount of p53 significantly increased at 4 h after HIV-1 infection at a high MOI (data not shown), suggesting that HIV-1 infection induces p53 expression in the early stages of infection, probably due to the previously reported provirus integration-mediated DNA damage response after infection (46). PKR phosphorylates Tat at least five Ser/Thr residues; most of them are highly or relatively conserved among HIV-1 variants prevalent in Europe and the United States (Fig. 4D; see Table S4 in the supplemental material), and these phosphorylations attenuate Tat activity to a greater or lesser extent (Fig. 5) by blocking (i) nuclear localization of Tat (Fig. 6), (ii) Tat binding to TAR (Fig. 7), and (iii) Tat-CycT1 interactions (Fig. 8). Our findings suggest that HIV-1 infection activates the p53-PKR host restriction pathway, which inhibits HIV-1 replication (Fig. 9) via Tat phosphorylation and inactivation in three critical steps, as summarized in Fig. 10. We found that p53/PKR-mediated Tat phosphorylation was restricted to the 5 Ser/Thr sites on the first exon of Tat86. However, our findings do not rule out the possibility of other phosphorylation sites on the second exon of Tat comprising more than 86 amino acids (aa).
PKR has been reported to phosphorylate Tat at residues S62, T64, and S68 (24), and Tat phosphorylation at these three residues enhances Tat binding to TAR RNA (26). However, our repeated experimental results (Fig. 7) demonstrated that Tat-TAR binding is inhibited rather than enhanced by PKR-mediated Tat phosphorylation. It is worth noting that that we identified major PKR target sites on Tat as T23, T40, and S46 rather than residues S62 and S68 (Fig. 3G and 4), and residue T64 was very unlikely to be phosphorylated by PKR (Fig. 4A and B). PKR-mediated Tat phosphorylation at S46 was further substantiated by ESI-MS/MS analysis (Fig. 4F). However, other phosphorylations were not confirmed by MS/MS analysis, presumably due to the many Arg/Lys/Cys residues on Tat. Instead, PKR-mediated Tat phosphorylation in vivo at T23, T40, S46, S62, and S68 was further verified using phospho-Thr/Ser-specific antibodies (Fig. 4E).
Consistent with a previous report (47), higher concentrations of TAR RNA inhibited PKR activation, followed by attenuation of PKR-mediated Tat phosphorylation (data not shown). These data suggest that accumulation of TAR RNA during HIV-1 infection could be another potential mechanism to overcome the PKR-mediated host restriction of HIV-1 replication in the later stages of infection.
Cellular enzymes such as acetyltransferase (20, 21), methyltransferase (48), or E3 ubiquitin ligase (22) control Tat activity by modifying Tat. Acetylation at Lys28 of Tat by p300/CBP-associating factor (PCAF) enhances Tat binding to the CycT1 subunit of the positive transcription elongation factor complex b (P-TEFb), while acetylation at Lys50 by p300 promotes dissociation of Tat and CycT1 from TAR, facilitating transcription elongation of RNA polymerase II (20, 21). In contrast to the positive effect of Tat acetylation on HIV-1 replication, PKR-mediated Tat phosphorylation at T23 and/or T40 disrupts Tat-CycT1 interaction (Fig. 8), leading to the inhibition of HIV-1 replication (Fig. 9). That might be due to the positions of these two amino acids in the Tat structure. The T23 and T40 residues appear to localize to the CycT1 binding pocket and on the gate of the binding pocket, respectively, in the 3-D structure of Tat. Therefore, phospho-mimic substitution of T23D or T40D would be enough to disrupt Tat-CycT1 interaction, as shown in Fig. 8C to E, resulting in the inhibition of HIV-1 replication (Fig. 9B). Our findings are, to a certain extent, consistent with previous reports showing that the C22G and K41A substitutions on Tat disrupt Tat-CycT1 interaction in vitro (49–51). The phenotypes of PKR-mediated Tat phosphorylation and inactivation in transfected cell lines were quite dramatic. However, viral replication was not affected very much as shown in the Tat functional study, likely because of other factors in the infected cells.
Contrary to our expectations, T23A substitution, to avoid PKR-mediated Tat phosphorylation at T23, did not improve Tat activity but rather impaired it (Fig. 5A and 9A), with a reduction in HIV-1 replication (Fig. 9B and C). This is presumably due to the suboptimal conformational changes of Tat by T23A substitution, which may damage normal Tat activity. Instead, the recombinant HIV-1 cDNAs possessing a tat gene encoding the natural T23N substitution, which is dominant among African strains (Fig. 5E), produced a high titer of recombinant progeny viruses in the transfection experiments (Fig. 9B). The HIV-1 cDNA with 23N40A tat did not produce as much progeny virus as control cDNA (Fig. 9B, lower panels), suggesting that double mutation would be detrimental rather than synergistic, even though each single mutation (T23N and T40A) improved Tat activity. The HIV-1 cDNAs possessing a tat gene encoding the S46A substitution produced a significantly reduced level of progeny viruses (Fig. 9B), even though S46A Tat showed strong Tat activity in a reporter assay (Fig. 9A). This might be due to the disturbance of the Kozak sequence of the overlapping gene rev during S46A Tat mutagenesis, as S46 on Tat is located right above the rev initiation AUG codon. Several reports have shown that the strength of upstream AUGs can influence initiation at downstream AUGs in HIV-1 transcription (52–54). A subset of env mRNAs carry the upstream rev and vpu initiation codons, and mutation of a weak upstream HIV-1 rev Kozak sequence to a strong initiating sequence resulted in decreased leaky scanning and poor initiation at the downstream vpu AUG (55).
It has been previously reported that Tat proteins from African strains are much more active than those from Europe and the United States (15), and the natural amino acid substitution T23N on Tat is present in a broad spectrum of African strains that are fast replicating (16). These reports support our findings that PKR targets five Ser/Thr sites on Tat that are highly conserved in HIV-1 subtype B strains (Fig. 4D), such as Bru (56) and Jr (57), which are prevalent in the United States and Europe. In contrast, strains Eli and Mal from Zaire (58) and other clade A, C, D, and G strains demonstrated strong polymorphisms at the corresponding sites, with the exception of S46 (Fig. 5D and E). These data suggest that the replication capacity of HIV-1 strains is, at least in part, associated with the susceptibility to p53/PKR-mediated Tat phosphorylation at five specific Ser/Thr sites. Tat phosphorylation by PKR inhibits Tat activity, leading to restriction of HIV-1 replication after infection. Therefore, amino acid substitutions at PKR target sites on Tat would be an attractive strategy for HIV-1 to escape p53/PKR-mediated host restriction in African strains.
HIV-1 Tat is a potent trans-activator essential for HIV-1 replication. It has been suggested that p53 suppresses Tat functions. Here we report for the first time, to our knowledge, the detailed molecular mechanisms underlying p53-mediated Tat inactivation, followed by inhibition of HIV-1 replication. We found that p53-mediated Tat inactivation could be attributed to PKR-mediated Tat phosphorylation at five specific Ser/Thr sites adjacent to the ARM, which decreased nuclear localization of Tat, Tat's ability to bind TAR RNA and P-TEFb, and Tat's transactivation capacity. Our findings may contribute to further research focusing on the investigation of potential therapeutic targets for HIV-1.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Sodroski for providing JK-Tat, pSV2-tat72, HXB2 virus, and plasmid pHXB2 and B. Vogelstein for isogenic p53−/− HCT116 cells. We are grateful to the AIDS Research and Reference Reagent Program (ARRRP) (NIH, USA) for providing U373MAGI-CXCR4PC3 cells and the HIV-1IIIB strain. We thank J. W. Kim, E. S. Lee, D. S. Lee, J. E. Ha, and J. Kim for their assistance with this project.
This work was supported by a Specific Basement Grant (2009-0084683) and Bio & Medical Technology Development Program Grant (2012M3A9B402826) from the Korea National Research Foundation (NRF) funded by the Korean Ministry of Education Science and Technology.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03087-14.
REFERENCES
- 1.Cullen BR. 1993. Does HIV-1 Tat induce a change in viral initiation rights? Cell 73:417–420. doi: 10.1016/0092-8674(93)90126-B. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson M. 2003. HIV-1 pathogenesis. Nat Med 9:853–860. doi: 10.1038/nm0703-853. [DOI] [PubMed] [Google Scholar]
- 3.Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 4.Choge I, Cilliers T, Walker P, Taylor N, Phoswa M, Meyers T, Viljoen J, Violari A, Gray G, Moore PL, Papathanosopoulos M, Morris L. 2006. Genotypic and phenotypic characterization of viral isolates from HIV-1 subtype C-infected children with slow and rapid disease progression. AIDS Res Hum Retroviruses 22:458–465. doi: 10.1089/aid.2006.22.458. [DOI] [PubMed] [Google Scholar]
- 5.Weinberger LS, Dar RD, Simpson ML. 2008. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet 40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 6.Li CJ, Wang C, Friedman DJ, Pardee AB. 1995. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan L, Ozaki I, Oakes JW, Taylor JP, Khalili K, Pomerantz RJ. 1994. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J Virol 68:4302–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukerjee R, Claudio PP, Chang JR, Del Valle L, Sawaya BE. 2010. Transcriptional regulation of HIV-1 gene expression by p53. Cell Cycle 9:4569–4578. doi: 10.4161/cc.9.22.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imbeault M, Lodge R, Ouellet M, Tremblay MJ. 2009. Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology 393:160–167. doi: 10.1016/j.virol.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Imbeault M, Ouellet M, Tremblay MJ. 2009. Microarray study reveals that HIV-1 induces rapid type-I interferon-dependent p53 mRNA up-regulation in human primary CD4+ T cells. Retrovirology 6:5. doi: 10.1186/1742-4690-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genini D, Sheeter D, Rought S, Zaunders JJ, Susin SA, Kroemer G, Richman DD, Carson DA, Corbeil J, Leoni LM. 2001. HIV induces lymphocyte apoptosis by a p53-initiated, mitochondrial-mediated mechanism. FASEB J 15:5–6. doi: 10.1096/fj.00-0336fje. [DOI] [PubMed] [Google Scholar]
- 12.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon CH, Lee ES, Lim DS, Bae YS. 2009. PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc Natl Acad Sci U S A 106:7852–7857. doi: 10.1073/pnas.0812148106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon CH, Miah MA, Kim KP, Bae YS. 2010. New Cdc2 Tyr 4 phosphorylation by dsRNA-activated protein kinase triggers Cdc2 polyubiquitination and G2 arrest under genotoxic stresses. EMBO Rep 11:393–399. doi: 10.1038/embor.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peloponese JM Jr, Collette Y, Gregoire C, Bailly C, Campese D, Meurs EF, Olive D, Loret EP. 1999. Full peptide synthesis, purification, and characterization of six Tat variants. Differences observed between HIV-1 isolates from Africa and other continents. J Biol Chem 274:11473–11478. [DOI] [PubMed] [Google Scholar]
- 16.Reza SM, Shen LM, Mukhopadhyay R, Rosetti M, Pe'ery T, Mathews MB. 2003. A naturally occurring substitution in human immunodeficiency virus Tat increases expression of the viral genome. J Virol 77:8602–8606. doi: 10.1128/JVI.77.15.8602-8606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Yukl S, Pillai S, Li P, Chang K, Pasutti W, Ahlgren C, Havlir D, Strain M, Gunthard H, Richman D, Rice AP, Daar E, Little S, Wong JK. 2009. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387:98–108. doi: 10.1016/j.virol.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue DA, Kuhl BD, Sloan RD, Wainberg MA. 2012. The viral protein Tat can inhibit the establishment of HIV-1 latency. J Virol 86:3253–3263. doi: 10.1128/JVI.06648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M. 2003. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell 12:167–176. doi: 10.1016/S1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 22.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol 5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 23.Bres V, Gomes N, Pickle L, Jones KA. 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev 19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand SR, Kobayashi R, Mathews MB. 1997. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem 272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 25.Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. 2006. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endo-Munoz L, Warby T, Harrich D, McMillan NA. 2005. Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J 2:17. doi: 10.1186/1743-422X-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DS, Jung KE, Yoon CH, Lim H, Bae YS. 2005. Newly designed six-membered azasugar nucleotide-containing phosphorothioate oligonucleotides as potent human immunodeficiency virus type 1 inhibitors. Antimicrob Agents Chemother 49:4110–4120. doi: 10.1128/AAC.49.10.4110-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung KS, Lee YA, Kim ET, Lee SR, Ahn JH, Choi CY. 2011. Role of the SUMO-interacting motif in HIPK2 targeting to the PML nuclear bodies and regulation of p53. Exp Cell Res 317:1060–1070. doi: 10.1016/j.yexcr.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Poustovoitov M, Serebriiskii I, Adams PD. 2004. A two-step two-hybrid system to identify functionally significant protein-protein interactions. Methods 32:371–380. doi: 10.1016/j.ymeth.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. 1999. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc Natl Acad Sci U S A 96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, Chong BH. 2003. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol 23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson L, Chen CB, Gaynor RP, Sigman DS. 1994. Footprinting RNA-protein complexes following gel retardation assays: application to the R-17-procoat-RNA and tat-TAR interactions. Nucleic Acids Res 22:2255–2263. doi: 10.1093/nar/22.12.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ariumi Y, Kaida A, Hatanaka M, Shimotohno K. 2001. Functional cross-talk of HIV-1 Tat with p53 through its C-terminal domain. Biochem Biophys Res Commun 287:556–561. doi: 10.1006/bbrc.2001.5626. [DOI] [PubMed] [Google Scholar]
- 34.Gabizon R, Mor M, Rosenberg MM, Britan L, Hayouka Z, Kotler M, Shalev DE, Friedler A. 2008. Using peptides to study the interaction between the p53 tetramerization domain and HIV-1 Tat. Biopolymers 90:105–116. doi: 10.1002/bip.20919. [DOI] [PubMed] [Google Scholar]
- 35.Longo F, Marchetti MA, Castagnoli L, Battaglia PA, Gigliani F. 1995. A novel approach to protein-protein interaction: complex formation between the p53 tumor suppressor and the HIV Tat proteins. Biochem Biophys Res Commun 206:326–334. doi: 10.1006/bbrc.1995.1045. [DOI] [PubMed] [Google Scholar]
- 36.Loret EP, Georgel P, Johnson WC Jr, Ho PS. 1992. Circular dichroism and molecular modeling yield a structure for the complex of human immunodeficiency virus type 1 trans-activation response RNA and the binding region of Tat, the trans-acting transcriptional activator. Proc Natl Acad Sci U S A 89:9734–9738. doi: 10.1073/pnas.89.20.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truant R, Cullen BR. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol 19:1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frankel AD, Pabo CO. 1988. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 39.Cardarelli F, Serresi M, Bizzarri R, Giacca M, Beltram F. 2007. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol Ther 15:1313–1322. doi: 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 40.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387>–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southgate CD, Green MR. 1991. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev 5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 42.Cujec TP, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan DO, Peterlin BM. 1997. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev 11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci U S A 96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcello A, Ferrari A, Pellegrini V, Pegoraro G, Lusic M, Beltram F, Giacca M. 2003. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J 22:2156–2166. doi: 10.1093/emboj/cdg205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Melo D, Venkatesh S, Bieniasz PD. 2012. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 37:399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. 2013. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 47.Maitra RK, McMillan NA, Desai S, McSwiggen J, Hovanessian AG, Sen G, Williams BR, Silverman RH. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 48.Van Duyne R, Easley R, Wu W, Berro R, Pedati C, Klase Z, Kehn-Hall K, Flynn EK, Symer DE, Kashanchi F. 2008. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5:40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. 2010. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. doi: 10.1016/S0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 51.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YG, Shaw MH, Warner N, Park JH, Chen F, Ogura Y, Nunez G. 2011. Cutting edge: Crohn's disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. J Immunol 187:2849–2852. doi: 10.4049/jimmunol.1001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Homer CR, Kabi A, Marina-Garcia N, Sreekumar A, Nesvizhskii AI, Nickerson KP, Chinnaiyan AM, Nunez G, McDonald C. 2012. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem 287:25565–25576. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YG, Kamada N, Shaw MH, Warner N, Chen GY, Franchi L, Nunez G. 2011. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity 34:769–780. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson JL, Johnson AT, Howard JL, Purcell DF. 2007. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J Virol 81:4664–4676. doi: 10.1128/JVI.01028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 57.O'Brien WA, Koyanagi Y, Namazie A, Zhao JQ, Diagne A, Idler K, Zack JA, Chen IS. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 58.Shaw MH, Kamada N, Warner N, Kim YG, Nunez G. 2011. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol 32:73–79. doi: 10.1016/j.it.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.