ABSTRACT
Alphaviruses are enveloped positive-sense RNA viruses that exhibit a wide host range consisting of vertebrate and invertebrate species. Previously we have reported that the infectivity of Sindbis virus (SINV), the model alphavirus, was largely a function of the cell line producing the viral particles. Mammalian-cell-derived SINV particles, on average, exhibit a higher particle-to-PFU ratio than mosquito cell-derived SINV particles. Nevertheless, the outcome of nonproductive infection, the molecular traits that determine particle infectivity and the biological importance of noninfectious particles were, prior to this study, unknown. Here, we report that the incoming genomic RNAs of noninfectious SINV particles undergo rapid degradation following infection. Moreover, these studies have led to the identification of the absence of the 5′ cap structure as a primary molecular determinant of particle infectivity. We show that the genomic RNAs of alphaviruses are not universally 5′ capped, with a significant number of noncapped genomic RNA produced early in infection. The production of noncapped viral genomic RNAs is important to the establishment and maintenance of alphaviral infection.
IMPORTANCE This report is of importance to the field of virology for three reasons. First, these studies demonstrate that noncapped Sindbis virus particles are produced as a result of viral RNA synthesis. Second, this report is, to our knowledge, the first instance of the direct measurement of the half-life of an incoming genomic RNA from a positive-sense RNA virus. Third, these studies indicate that alphaviral infection is likely a concerted effort of infectious and noninfectious viral particles.
INTRODUCTION
Alphaviruses are enveloped, positive-sense RNA viruses that are cyclically transmitted between sylvatic vertebrate reservoir hosts and a mosquito vector during the enzootic cycle. The maintenance of this cyclical transmission is vital to viral fitness, as prolonged serial passage within a single host results in attenuation in the alternate host (1–4). Moreover, the ultimate outcome of alphaviral infection differs between vertebrate and invertebrate hosts, as infection of a vertebrate host results in acute cytolytic infection whereas infection of invertebrate hosts often results in persistent infection with minimal cell death (5–11). The widespread geographic distribution of competent vector mosquito species leading to contact with immunologically naive human populations has resulted in several significant outbreaks of alphaviral disease (12–14). Perhaps most notable is the ongoing re-emergence of chikungunya virus, which caused significant morbidity during the height of the 2006 epidemic, with as many as 40,000 new cases per week (13).
Despite the range of diseases and morbidity associated within the genus, the underlying molecular life cycles are highly similar in the two hosts. Since alphaviruses are positive-sense RNA viruses, they function similarly to cellular mRNAs, relying on the translation of the incoming viral genome to initiate viral infection. Translation of the genomic RNA produces the viral RNA synthetic complex that synthesizes progeny genomes through the production and copying of a minus strand RNA replication intermediate. The viral replicase complex consists of the four nonstructural protein products the function of which is regulated by processing of the nonstructural polyprotein. From the minus strand RNA, the viral subgenomic and progeny genomic RNAs are synthesized. Translation of the subgenomic RNAs results in the expression of the viral structural components. The nascent genomic RNAs are then subsequently encapsidated and released as mature viral particles (15).
Viral RNA synthesis is accomplished via the combined activities of the viral nonstructural proteins. Proteolytic processing of the nonstructural polyprotein results in the regulation of viral RNA synthesis at the molecular level. The nsP4 protein is the viral RNA-dependent RNA polymerase. The alphaviral capping reaction is mediated by the viral nsP1 and nsP2 proteins (16–20). The presence of the 5′ cap structure is essential for alphavirus gene expression, as there are no internal ribosome entry site elements present in any of the viral RNAs. During the viral capping reaction, the nsP1 protein methylates GTP, subsequently resulting in the formation of a covalent 7meGMP-nsP1 complex (21). The nascent viral RNA is processed via the RNA triphosphatase activity of nsP2 to a 5′ diphosphate moiety prior to the transfer of the 7meGMP to the viral RNA via the guanylyltransferase activity of nsP1 (22). The result is the formation of the viral type 0 7meGpppA cap, which is found on both the genomic and subgenomic RNAs. The viral type 0 cap structure differs from the cellular type I 7meGppp2meG structure in its lack of a second methylation event.
We have reported previously that the infectivity of a model alphavirus, Sindbis virus (SINV), as measured by the ratio of particles to infectious units, was dependent primarily on the deriving host cell and generally improved with respect to time (23). Additionally, SINV particles produced from mosquito cells were more efficient, in terms of infectivity, than mammalian-cell-derived viral particles. Further characterization of SINV particles produced in mammalian tissue culture cells revealed that SINV particles could be physically separated into two subpopulations on the basis of particle density (24). These viral particles, termed SINVLight and SINVHeavy, were morphologically indistinguishable and hence exhibited differences in particle density due to their mass. Examination of the individual SINV subpopulations revealed that SINVHeavy had host-derived ribosomal components (HDRCs) encapsidated alongside the viral genomic RNA. The SINVLight and SINVHeavy subpopulations exhibited significant differences at the level of particle infectivity. The SINVLight subpopulation was, on average, less infectious than the SINVHeavy subpopulation, exhibiting particle-to-PFU ratios of ∼100:1 and 10:1, respectively. This indicates that a large number of the viral particles in the SINVLight subpopulation were noninfectious and incapable of initiating viral infection. The observed difference in infectivity was not a function of differential viral entry. The SINVHeavy subpopulation exhibited enhanced translation of the viral genomic RNAs early during the infection of mammalian cells relative to SINVLight. A consequence of enhanced translation of the viral genomic RNA was increased viral RNA synthesis and accumulation early during infection. Additionally, infections initiated with SINVHeavy produced less type I interferon (IFN) than those of SINVLight, indicating that the presence of the HDRCs, perhaps via enhanced viral functionality early during infection, is a component of innate immune evasion.
While the production of noninfectious viral particles, defined above as viral particles incapable of initiating viral infection, is well established, the role(s) of the noninfectious particles is unclear. A primary goal of the study reported here was to identify the potential fate and function of the genomic RNA of noninfectious particles, as well as identify the molecular determinants that led to the viral particle being noninfectious. In this work, we found that the genomic RNAs of noninfectious particles are unstable and undergo RNA decay more rapidly that RNAs from infectious particles. We determined that the genomic RNAs of alphavirus particles are not universally 5′ 7meGpppA capped and the absence of the viral 5′ cap is a major determinant of particle infectivity. We report that the presence of noncapped viral particles correlates with the activation of host innate immunity, presumably because of the presence of activating 5′ RNA termini, as measured by the production of soluble type I IFN (IFN-α/β).
MATERIALS AND METHODS
Tissue culture cells.
BHK-21, 293HEK, L929, and C6/36 cells were cultured in minimal essential medium (MEM; Cellgro) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 1× antibiotic-antimycotic solution (Cellgro), 1× nonessential amino acids (Cellgro), and l-glutamine (Cellgro). All of the mammalian cell lines used in this study were maintained at 37°C in the presence of 5% CO2. The Aedes albopictus C6/36 cell line was maintained at 28°C in the presence of 5% CO2.
Preparation and purification of SINV.
Wild-type SINV Toto1101, SINVLM, and SINV p389, a Toto1101-derived strain containing green fluorescent protein (GFP) in frame with nsP3, Ross River virus (RRV) strain T48, and RRVnsp1S79C:L224I were prepared by electroporation as previously described (24). Briefly, 10 μg of in vitro-transcribed RNA was electroporated into BHK-21 cells via a single pulse from a Gene Pulser Xcell system (Bio-Rad) at 1.5 kV, 25 mA, and 200 Ω. Twenty-four hours later, the tissue culture supernatant were collected and clarified via centrifugation at 8,000 × g for 10 min. The titers of the resulting P(0) stocks were determined on BHK-21 cells, and the stocks were either used immediately or stored at −80°C for later use.
SINV particles were purified via the two-step process described below. Typically 2 × 108 BHK-21 cells were infected with SINV at a multiplicity of infection (MOI) of 3 PFU/cell. After aspiration of the inoculum, whole medium was added and the cells were incubated for 18 h prior to the collection of the supernatant as described above. The viral particles were then concentrated by pelleting through a 27% sucrose cushion prepared in HNE buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM EDTA) by centrifugation at 185,000 × g for 1.5 h in a 60Ti rotor. The pelleted virions were then suspended in HNE buffer (supplemented to 40 mM EDTA) prior to being applied to a linear gradient of 15 to 45% (mass/vol) sucrose in HNE. The linear gradients used in this study were prepared with a Gradient Master Apparatus (BioComp Instruments) by using the appropriate preprogrammed settings. SINV particles were banded by centrifugation at 250,000 × g for 2.5 h in an SW41 rotor. The SINV subpopulations were collected via needle aspiration and stored either at 4°C for short-term use or at −80°C in small-volume aliquots.
SINVLM was cultured under either normal conditions, as described above, or in Dulbecco's MEM lacking the amino acid methionine or cysteine for 16 h prior to harvesting, as previously described (25). Viral titer determination was performed by using standard plaque assays on BHK-21 cells. Quantitative assessment of the particle numbers of viral samples was performed as previously described (23, 26). Briefly, viral samples (100 μl) were treated with 10 μg of RNase A for 15 min at 37°C prior to extraction with TRIzol reagent. The extracted RNAs were subsequently used as the template for the synthesis of cDNA via reverse transcription (RT) with oligonucleotide primer SINV nsP1 (5′-AACATGAACTGGGTGGTG-3′). The resulting cDNAs were quantitatively assessed by quantitative RT (qRT)-PCR as previously described, with the following primer set: SINV nsP1F, 5′-AAGGATCTCCGGACCGTA-3′; SINV nsP1R, 5′-AACATGAACTGGGTGGTGTCGAAG-3′. The CT values detected were then compared to internal-standard curves to determine the number of genome equivalents (GE) present in a given sample. The determination of viral infectivity is described by the ratio of total particles to infectious units.
SINV incoming genomic RNA half-life assay.
293HEK cells, cultured in medium supplemented with 50 μM 4-thiouridine (4SU; Sigma), were infected at 4°C with the indicated SINVs at an MOI of 5 PFU per cell. Infections were conducted at 4°C to block viral entry, effectively synchronizing viral infection. The infected monolayers were washed with cold 1× phosphate-buffered saline (PBS) to remove unbound viral particles and supplemented with prewarmed medium supplemented with 50 μM 4SU. At the postinfection times indicated, the tissue culture supernatant was removed and the cell monolayers were washed three times with PBS prior to TRIzol (Invitrogen) extraction. A total of 5 μg of total RNA was biotinylated with HPDP-Biotin (Pierce). The biotinylated RNAs were phenol extracted prior to being bound to Ultralink streptavidin resin (Pierce) to remove newly transcribed viral RNAs. The unbound RNAs were then phenol extracted and ethanol precipitated prior to use as a template for cDNA synthesis via RT with Random Hexamer. The resulting cDNAs were assayed via qRT-PCR to determine the relative abundances of the incoming SINV genomic RNAs normalized to the cellular 18S rRNA as previously described (27, 28). RNA half-lives were calculated via nonlinear regression.
5′ end characterization.
Quantitative assessment of the 5′ end of the SINV genomic RNAs consisted of an enzymatic process followed by qRT-PCR. The input RNA consisted of either genomic RNAs from viral particles (∼108 PFU) or cellular RNA (1 μg) with TRIzol as previously described (24). Input RNA samples were phosphatase treated with Antarctic Phosphatase (NEB) for 20 min at 37°C. The reaction mixtures was heat inactivated and split into two equal aliquots to assay the noncapped and 7meGpppA-capped genomic RNAs in parallel (referred to here as noncapped and capped, respectively). The noncapped reaction mixtures were then treated with T4 PNK (NEB) to produce a pool of 5′-monophosphate and 7meGpppA-capped genomic RNAs. The capped aliquot was treated with the Nudix domain containing RppH (NEB) to decap 7meGpppA-capped genomic RNAs to generate a pool of nonphosphorylated and 5′-monophosphate genomic RNAs (29). The noncapped and capped reaction mixtures were then used in parallel as substrates for a ligation between a 5′ blocked RNA oligonucleotide (5′-12 C spacer-GUUCAGAGUUCUACAGUCCGACCCAUC-3′) and the 5′-monophosphate genomic RNAs via T4 RNA ligase (NEB). Alternatively, the RNA samples were treated with various combinations of the above treatments to focus on individual 5′-terminal structures such as 5′-monophosphates and/or 5′-polyphosphates. After heat inactivation, the samples were used as substrates for RT to generate cDNA specific to the viral genomic RNA (24).
Quantitative assessment of the individual RNA species was performed as follows. The relative amounts of 5′-capped and noncapped viral genomic RNAs was determined by comparing the relative quantities of the linker-containing amplicon species with the total amount of viral genomic RNA present by the ΔΔCT method to obtain the ratios of 5′-capped and noncapped RNAs, depending on the sample treatment, as described above. The following oligonucleotides were used to detect the linker-containing amplicon representing the 5′ end for SINV and RRV samples (5′ Linker, 5′-GTTCAGAGTTCTACAGTCCGACCCATC-3′; SINV genomic Reverse, 5′-CGTCTACGTTTACTACTGGCTTCTCC-3′; RRV genomic Reverse, 5′-CAGCCTCAACATCTACAGTGACC-3′). The total amount of SINV and RRV RNA present in a sample was determined via qRT-PCR with the following primer sets (SINV nsP1F, 5′-AAGGATCTCCGGACCGTA-3′; SINV nsP1R, 5′-AACATGAACTGGGTGGTGTCGAAG-3′; RRV nsP1F, 5′-GAAGGTCACTGTAGATGTGGCT-3′; RRV nsP1R, 5′-GCTCTGGCATTAGCATGGTCATTGG-3′). The determination of specific quantities was accomplished by using internal standard curves and subtractive analyses, as previously described (24).
Immunoprecipitation.
Extracted total RNA from infected cells or UV cross-linked SINV particles were diluted in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.6],150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and bound to either anti-7meG (System Synaptics) or anti-RPS14 (Santa Cruz) antibodies according to the manufacturer's instructions, respectively. After incubation while gently mixing at 4°C for a minimum of 4 h, the lysate was clarified via centrifugation prior to being moved to a fresh tube containing protein G Sepharose beads that had been pretreated with 10 μg of both yeast tRNA and bovine serum albumin. After incubation at 4°C for 1 h, the beads were collected via centrifugation and washed six times prior to being phenol extracted and ethanol precipitated. The resulting immunoprecipitate was used as the template for cDNA synthesis and qRT-PCR detection as previously described (24, 27).
Quantification of type I IFN.
The production of type I IFN was assayed as previously described (24). The IFN-competent L929 cell line was infected with either infectious or UV-inactivated SINV or RRV at an MOI of 10 PFU/cell for 1 h at room temperature. Prior to the addition of whole medium, the inoculum was removed and the cell monolayers were washed at least twice with 1× PBS. Twenty-four hours later, the supernatants were collected and clarified via centrifugation. Infectious viral particles present in the supernatants were inactivated via acidification and UV irradiation. The inactivation of the supernatants was confirmed via standard plaque assay. The inactivated supernatants were then diluted 1:10 in whole medium and then further serially diluted onto fresh L929 cells plated in 96-well plates at a ratio of 1:3. Twenty-four hours later, the treated cells were infected with a fluorescent RRV mKate strain at an MOI of 10 PFU/cell. Viral gene expression, detected as a function of mKate expression, was detected 24 h postchallenge via a Typhoon 9200 phosphorimager and analyzed via densitometry. After 72 h postinfection (hpi), the cells were fixed with formaldehyde and examined for indications of a cytopathic effect, namely, cell death, which was highly consistent with the aforementioned fluorescent gene expression. Relative IFN production was calculated as a function of the dilution required to attain a 50% reduction in viral gene expression.
Statistical analyses.
Unless stated otherwise, the numerical data reported here are the mean values of a minimum of three independent biological replicates. Error bars indicate the standard deviation of the mean. Where indicated, the P values associated with the individual data sets are the result of the Student t test for the corresponding data.
RESULTS
Incoming SINV genomic RNAs exhibit short half-lives.
Host cell mRNAs are highly regulated posttranscriptionally, leading to dynamic gene expression. Cellular transcripts that are aberrant or undesirable are rapidly degraded by way of the host RNA decay machinery. The genomic RNAs of infectious SINV particles are presumed to be functionally similar to mRNAs, despite lacking hallmarks of cellular mRNAs such as exon-junction complexes, as the viral genomic RNA is known to be translated. Nevertheless, what happens to the genomic RNAs of noninfectious particles, which fail to undergo translation, is unclear. We hypothesized that the SINV genomic RNAs from noninfectious viral particles would be more rapidly degraded than those of infectious viral particles early during infection. To this end, we characterized the RNA half-lives of the SINV genomic RNAs from populations of SINV particles with low or high particle-to-infectious-virus ratios.
Previous characterizations of viral RNA half-lives have used either temperature-sensitive mutants or chemical inhibitors of transcription (27, 28). Either of these options is likely to have effects on the host cell environment that may prove detrimental to the determination of the viral RNA half-life. We opted to use a method developed for the global analysis of cellular RNA half-lives via metabolic labeling (30). In this method, cotranscriptional incorporation of 4SU may be used to selectively monitor the abundance of RNAs via affinity purification. We adapted this method to use 4SU incorporation as a means by which the separation of incoming and newly transcribed viral genomic RNAs could be attained. By performing synchronous SINV infections in the presence of 4SU, we were able to separate the incoming viral genomes from those produced during active viral replication/RNA synthesis via biotinylation of the 4SU residues, followed by streptavidin purification. This effectively removes newly synthesized RNA from the sample, leaving the incoming viral genomes and the preexisting unlabeled cellular RNAs in the unbound fraction. The unbound fraction was then subsequently used to determine the half-lives of the incoming viral RNAs. The 293HEK cell line was used for the assessment of viral RNA decay as it had been used in previously published studies for the same purpose (28). Use of the same cell line ensured that any observed differences in RNA half-life were indeed genuine and not a functional difference between cell lines.
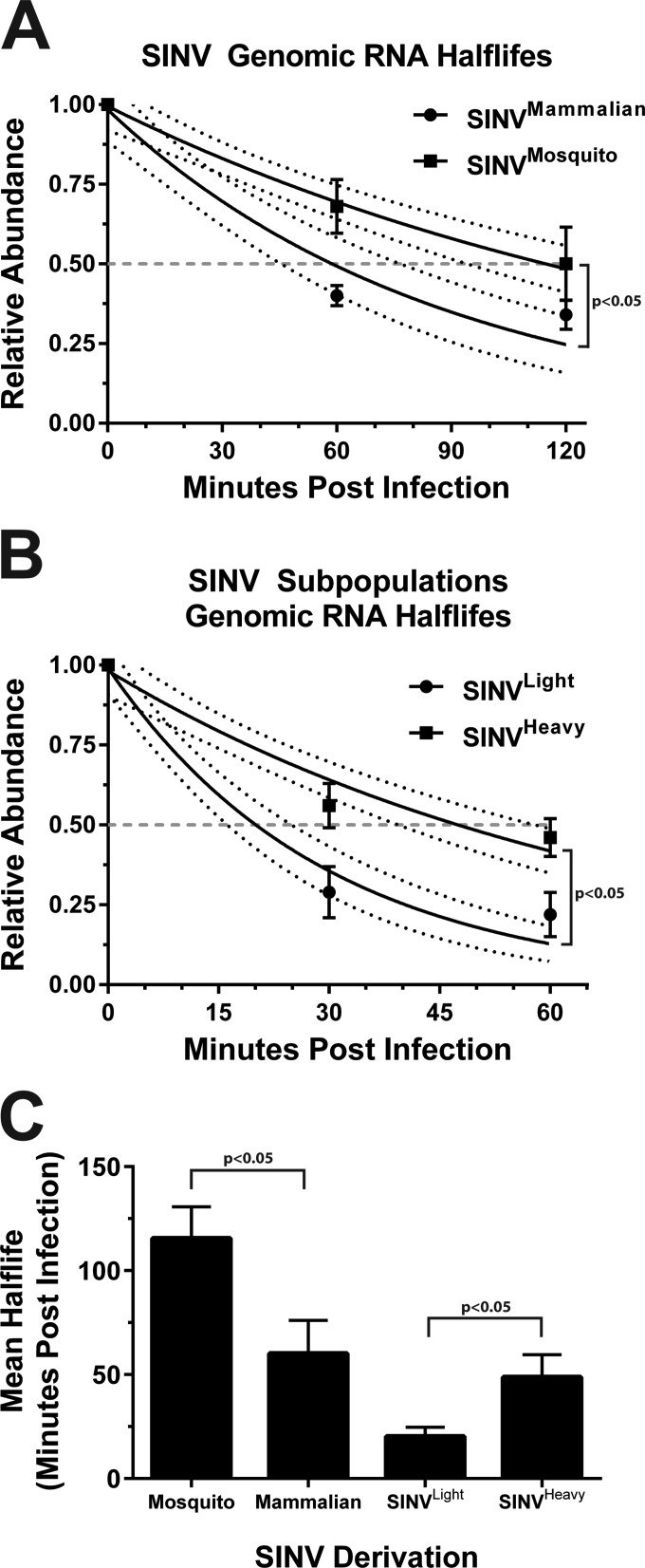
Quantitative analysis of the input SINV genomic RNA levels with respect to time indicated that the SINV genomic RNAs from mammalian-cell-derived particles (which exhibit an average particle-to-PFU ratio of >100:1) exhibited a half-life of 60.2% ± 15.9 min, as calculated by the method described by Dölken et al. (30) (as shown in Fig. 1A). The SINV genomic RNAs from mosquito cell-derived particles (which exhibit an average particle-to-PFU ratio of ∼5:1) exhibited a half-live, on average, of 115.6% ± 15.06 min. Analysis of the tissue culture supernatants indicated that genome loss due to particle desorption was negligible (data not shown), indicating that the viral genomic RNAs were remaining internalized throughout the course of the experiment.
FIG 1.
Comparative analysis of the half-lives of the incoming SINV genomic RNAs. (A) Quantitative assessment of the RNA half-lives of the incoming genomic RNAs of mammalian and mosquito-derived particles. 293HEK cells cultured in the presence of 4SU were infected with SINV derived from either mammalian (BHK-21) or mosquito (C6/36) tissue culture cells at an MOI of 5 PFU/cell. Viral adsorption was conducted at 4°C to synchronize viral infection. At the postinfection times indicated, the total RNA was extracted, biotinylated, and fractionated into the incoming and newly transcribed viral RNAs. The unbound RNAs, consisting of the incoming viral genomic RNAs and unlabeled cellular transcripts, were used to synthesize cDNAs, which were assayed via qRT-PCR to determine the relative abundance of the incoming SINV genomic RNA with respect to time. (B) Essentially identical to that described for panel A, with the exception that the inoculum consisted of the mammalian-cell-derived SINVLight or SINVHeavy subpopulation. (C) Graphic representation of the mean half-lives calculated as described by Dölken et al. (30). All graphical data shown are the mean values of a minimum of three independent biological replicates. The error bars represent the standard deviation of the mean. The upper and lower limits of range of the nonlinear regressions shown in panels A and B are represented by the dashed horizontal lines.
We have previously demonstrated that SINV derived from mammalian tissue culture cells consists of a heterogeneous population of viral particles consisting of at least two subpopulations, SINVLight and SINVHeavy, which differ biologically at the level of infectivity and translational activity (24). As previously reported, SINVLight and SINVHeavy exhibit particle-to-PFU ratios of ∼60:1 and ∼20:1, respectively. As shown in Fig. 1B, assessment of the half-lives of the genomic RNAs of SINVLight and SINVHeavy exhibit mean half-lives of 20.32% ± 4.4 min and 48.75% ± 10.9 min, respectively. A graph summarizing the calculated half-lives of the particle species used in these studies is shown in Fig. 1C.
Taken together, these data strongly suggest that the incoming viral genomic RNAs experience turnover following infection and that the SINV genomes of mosquito cell-derived particles are inherently more stable than those of mammalian-cell-derived particles. Moreover, the rate of decay associated with SINVLight, relative to either the SINVHeavy subpopulation or the parental heterogeneous mammalian population, indicates that the genomic RNAs of SINVLight particles are exceptionally unstable early during infection. The half-lives of mammalian-cell-derived SINV and SINVHeavy are remarkably similar. This is interesting because the SINVLight subpopulation is the predominant particle species present in mammalian-cell-derived SINV particles. While the precise reasons for this are unclear, a direct comparison of the two may be inappropriate. The individual SINV subpopulations may exhibit different behavior when apart from one another. For instance, perhaps the SINVHeavy component of the mixed population is capable of preventing the active degradation of the accompanying SINVLight particles. Regardless, we observe distinct differences between the RNA half-lives of mammalian-cell- and mosquito cell-derived SINV genomic RNAs, and SINVLight and SINVHeavy genomic RNAs, indicating that the stability of the incoming genomic RNAs is derivation and particle specific.
Noncapped SINV genomic RNAs are encapsidated into particles.
The relatively short observed half-lives of the entering SINV genomic RNAs, in particular, the genomic RNAs of SINVLight particles, suggested that the RNA substrates were not stabilized or protected from degradation. Previous studies have indicated that SINV RNAs are highly resistant to deadenylation in vitro and in vivo (27, 28). The observed instability of the incoming viral genomic RNAs combined with the previously reported SINV inhibition of the 3′-to-5′ cellular RNA decay pathway (via the association of the host HuR protein) led to the hypothesis that the 5′ cap structure may be absent from many of the incoming viral genomic RNAs. Hence, we focused our attention on the 5′ end of the SINV genome as a cause of the RNA instability, as a transcript lacking a 5′ cap is often a substrate for rapid turnover.
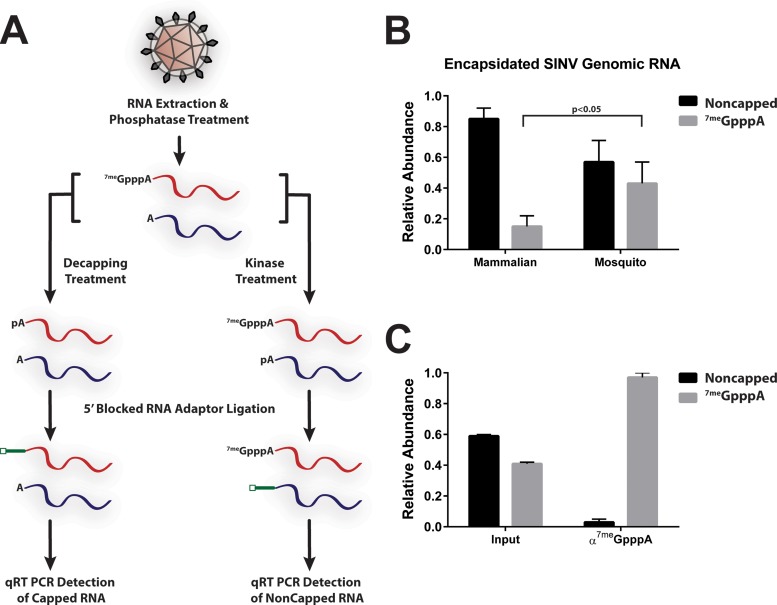
To quantitatively determine whether the 5′ cap was present on or absent from the SINV genome, we developed an enzymatic assay capable of differentiating 5′-capped and noncapped RNAs from encapsidated SINV genomes (Fig. 2A). Prior to treatment, the SINV samples were incubated with RNase A to degrade any extraparticle RNAs. The detection of viral noncapped and capped genomic RNAs consisted of two parallel reaction mixtures intended to determine the noncapped and capped RNAs individually. After extraction but prior to splitting of the sample into two aliquots, the RNAs were phosphatase treated to remove any 5′-phosphates. Following inactivation of the enzyme, the reaction mixtures were treated with polynucleotide kinase to generate a mixture of 5′-monophosphate and 7meGpppA-capped RNAs. These RNAs were then used as substrates for an RNA ligation reaction to a 5′-blocked adaptor RNA oligonucleotide. Detection of 5′ 7meGpppA-capped viral genomic RNAs consisted of an approach similar to that described above. The primary difference between the detection schemes was the use of a decapping enzyme, instead of treatment by polynucleotide kinase, to generate the 5′-monophosphate RNAs for subsequent ligation. Regardless, the result was the specific modification of either noncapped or 7meGpppA-capped viral RNAs with a 5′ adaptor sequence. The modified RNAs were then used as templates for the synthesis of cDNA via RT. The abundances of noncapped and capped viral genomic RNAs were determined via quantitative PCR. The ΔΔCT method was used for quantitative analysis of the abundance of an amplicon specific to the 5′ adaptor sequence and the 5′ end of the genomic RNA normalized to an amplicon internal to the genomic RNA.
FIG 2.
Quantitative assessment of the 5′ terminus of the SINV genomic RNA. (A) Schematic diagram of the quantitative assessment of the viral 5′ terminus. (B) Quantitative assessment of SINV particles produced from either mammalian (BHK-21) or mosquito (C6/36) tissue culture cells. The viral samples were collected 18 hpi and treated as described in Materials and Methods. (C) Quantitative assessment of input and anti-7meG immunoprecipitated intracellular viral RNAs from infected 293HEK cells at 12 hpi. The values shown are the means of a minimum of three independent replicates. Error bars represent the standard deviation of the mean.
As shown in Fig. 2B, noncapped SINV genomic RNAs were detected in mammalian-cell- and mosquito cell-derived particles. The overall ratio of noncapped to capped genomic RNAs differed between SINV particles derived from a mammalian host cell and those derived from a mosquito host cell. Only 15% ± 7% of mammalian-cell-derived particles, in contrast to 43% ± 14% of mosquito cell-derived SINV particles, contained a 7meGpppA-capped genomic RNA (Fig. 2B). Similar levels of capping were observed in both the Toto1101 and AR86 strains of SINV (data not shown). This approach was validated by using an immunoprecipitation approach to select for 5′-capped RNAs. SINV genomic RNAs extracted from tissue culture cells were immunoprecipitated with anti-7meG antibodies prior to processing and quantitative analysis as described above. As shown in Fig. 2C, anti-7meG immunoprecipitation of the intracellular RNAs present at 12 hpi resulted in a population that was 97% ± 3% 5′ 7meGpppA capped. This result indicates that the quantitative assay described above is highly selective and specific.
The genomic RNAs of SINVLight are predominantly noncapped.
As shown in Fig. 1, the genomic RNAs of the SINVLight subpopulation, which consists primarily of noninfectious particles, exhibited a half-life shorter than that of the genomic RNAs of the SINVHeavy subpopulation. Additionally, as shown in Fig. 2, a significant number of SINV particles derived from mammalian tissue culture cells lack the viral 5′ cap structure. It was also notable that the percentage of 5′-capped particles was similar to the overall number of SINVHeavy particles within the mixed population. Collectively, these observations led us to hypothesize that the genomic RNAs of the SINVLight subpopulation were predominantly noncapped, thereby resulting in the short RNA half-lives and inability to cause infection associated with SINVLight. To this end, we assayed the genomic RNAs of SINVLight and SINVHeavy with the enzymatic assay described above.
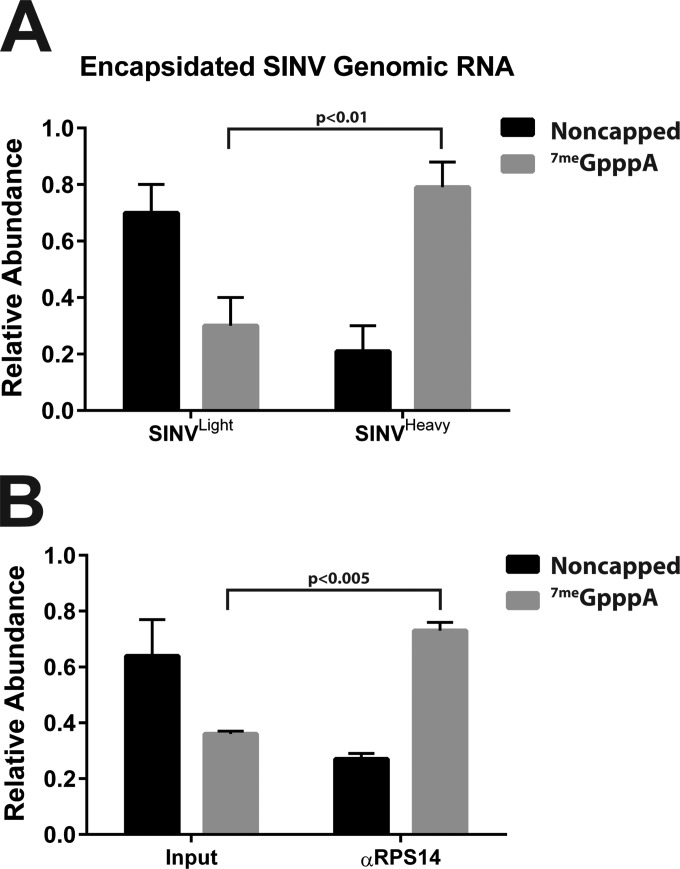
Analysis of the purified mammalian-cell-derived SINVLight and SINVHeavy subpopulations with the enzymatic assay described above revealed that the individual subpopulations differed from one another on the basis of the magnitude of noncapped to 7meGpppA-capped viral genomic RNAs. As shown in Fig. 3A, on average, 30% ± 10% of the genomic RNAs of the SINVLight subpopulation were 7meGpppA capped. In contrast, 79% ± 9% of the genomic RNAs of the SINVHeavy subpopulation were 7meGpppA capped. These results indicated a correlation between the presence of noncapped genomic RNAs and noninfectious particles. Specifically, SINVLight had significantly more noncapped genomic RNAs than SINVHeavy, which consisted of predominantly 5′-capped genomic RNAs. Interestingly, the individual ratios of noncapped and capped viral genomic RNAs appear to be incongruous with that observed for the unpurified mixture of particles produced during mammalian infection (as shown in Fig. 2B). We can readily detect, albeit at relatively low levels, genomic RNA outside the banding zones for SINVLight and SINVHeavy. These genomic RNAs are not associated with infectious particles but are a component of the parental mixture and therefore likely contribute to the abundance of the noncapped component in that mixture.
FIG 3.
The viral 5′ cap structure correlates with HDRC encapsidation. (A) Quantitative assessment of the 5′ terminus of the mammalian-cell-derived SINVLight and SINVHeavy subpopulations purified as described in Materials and Methods. (B) Quantitative assessment of input and anti-RPS14 antibody-immunoprecipitated, UV cross-linked mammalian-cell-derived, SINV particles. The values shown are the means of a minimum of three independent replicates. Error bars represent the standard deviation of the mean.
As previously reported, SINVHeavy and SINV derived from mosquito cells contained encapsidated HDRCs (24). Nevertheless, the determinants leading to encapsidation of the HDRCs into alphaviral particles are unknown, but the observation that a significant proportion of SINVLight, which lacks HDRCs, were noncapped led us to hypothesize that the viral 5′ cap structure may be a determinant of HDRC incorporation. Thus, we used immunoprecipitation of a component of the HDRCs, RPS14 from a mammalian-cell-derived mixed viral population, to determine the relationship of the viral 7meGpppA cap with the encapsidated HDRCs.
Mammalian-cell-derived SINV particles were cross-linked via UV irradiation and, following detergent-mediated disruption, immunoprecipitated with antibodies specific to RPS14. The immunoprecipitated RNAs were then used as substrates for the enzymatic 5′ characterization described above. Analysis of the RPS14 immunoprecipitate indicated that, on average, 73% ± 3% of the RPS14:SINV genomic RNA complexes were 7meGpppA capped (Fig. 3B). Interestingly, this value closely resembled what was observed for the HDRC-containing SINVHeavy subpopulation, which was 79% 7meGpppA capped. Taken together, these data suggest that the encapsidation of HDRCs correlates with the presence of the 7meGpppA cap structure.
Noncapped SINV particles are synthesized and packaged early during infection.
The data reported in Fig. 2 and 3 indicate that a significant number of noncapped genomic RNAs are packaged into alphaviral particles during the infection of mammalian tissue culture cells. Nonetheless, when during viral replication the synthesis of noncapped viral genomic RNAs occurs and to what levels they accumulate were unclear. We have previously reported that the infectivity of SINV, as measured by the ratio of particles to infectious units, improves with time in tissue culture systems (23). Since the ratios of noncapped to 7meGpppA-capped SINV genomic RNAs were highly similar to the ratios of particles to infectious units observed for mammalian-cell- and mosquito cell-derived SINV particles, we hypothesized that the number of noncapped particles (viral particles that contain a noncapped genomic RNA) would decrease, while the number of 7meGpppA-capped particles would increase, with time.
Initially, we sought to determine when the noncapped viral genomic RNAs were being produced during infection. To this end, we infected 293HEK cells with SINV at an MOI of 5 PFU/cell and, at the postinfection times indicated, pulsed the cells with the 4SU nucleoside analogue to specifically label the viral RNAs actively being transcribed. After 3-h pulses of 4SU labeling, the total RNA was extracted from the infected tissue culture cells, biotinylated, and then fractionated on the basis of 4SU incorporation to separate the nascent viral transcripts. The bound fraction was then analyzed as described above to quantitatively assess the 5′ end of the viral genomic RNAs. As shown in Fig. 4A, noncapped viral genomic RNAs are predominantly synthesized early during viral infection. At 9 and 12 hpi, capped and noncapped genomic RNAs are produced in roughly equivalent amounts. Quantitative examination of the entire population of viral RNAs present in the extracted total RNA pool, as shown in Fig. 4B, revealed a similar profile of noncapped RNAs relative to both extracellular particles and ongoing viral RNA synthesis.
FIG 4.
Temporal analysis of the 5′ terminus of the SINV genomic RNA. (A) Quantitative assessment of the rate of noncapped and capped viral genomic RNA synthesis. (B) Quantitative assessment of the intracellular accumulation of noncapped and capped SINV genomic RNAs during the aforementioned infections. (C) Quantitative assessment of the encapsidated SINV genomic RNAs from particles produced in the mammalian 293HEK tissue culture cell line. (D) Determination of the infectivity with respect to time, reported as the ratio of particles to infectious units, of 293HEK-derived SINV. Analysis was performed as previously reported (Fig. 2). Samples were paired with those analyzed as described in panel C. The values shown are the means of a minimum of three independent replicates. Error bars represent the standard deviation of the mean.
To determine if the ratio of noncapped to capped viral particles varied with respect to time, we collected the supernatant of infected tissue culture cells at regular intervals and assayed for viral infectivity and the presence of capped genomic RNA as described above. Temporal quantitative examination of the 5′ terminus of the encapsidated viral genomes indicated that the majority of particles produced early during infection contained noncapped genomic RNAs. Specifically, at 3 and 6 hpi, less than a quarter of the SINV genomic RNAs were 7meGpppA capped. However, by 9 and 12 hpi, the proportion of 7meGpppA-capped genomic RNAs increased to nearly a 1:1 ratio (Fig. 4C). Notably, the profile of noncapped RNA synthesis was highly similar to that observed in released particles at the times assayed during infection. Moreover, these data closely matched the basal level of infectivity of the sample. As shown in Fig. 4D, the infectivity of SINV particles derived from mammalian cells improved as infection progressed. Importantly, the improvement of infectivity (as measured by the particle-to-infectious-unit ratio) was due primarily to an increase in the production of infectious particles rather than a net decrease in particle production (data not shown).
Together, these observations indicate that noncapped genomic RNAs are produced predominantly early during infection; yet, by an unknown mechanism, at later stages of viral infection, the production of 5′ 7meGpppA-capped genomic RNAs increases. This observation is puzzling because alphaviruses express their own viral capping machinery, consisting of the RNA triphosphatase activities of nsP2 and the methyltransferase and guanylyltransferase activities of nsP1. Interestingly, there is no apparent preference to packaging of either 5′-capped or noncapped viral genomes throughout the infection of mammalian cells, as the synthesis and intracellular accumulations (Fig. 4A and B, respectively) of noncapped viral genomes closely mirror what is observed in extracellular particles (Fig. 4C).
The noncapped RNAs are predominantly 5′-monophosphate RNAs.
The data described above indicated that a significant number of encapsidated viral genomic RNAs lacked the canonical type 0 viral 7meGpppA cap structure, but the precise nature of the 5′ terminus of the noncapped viral RNAs was unknown. The 5′ end of RNAs is a known target for host surveillance, as several notable pathogen-associated molecular patterns (PAMPs) may be present. Since the 5′ end of the noncapped viral RNAs were permissive to ligation following phosphatase and kinase treatment we hypothesized that the 5′ end was likely a phosphate moiety. This is significant because 5′ tri- and diphosphate moieties are recognized by cellular innate immune receptors such as RIG-I and the RIG-I-like receptors (RLRs) (31, 32). Recognition of these PAMPs leads to the activation of a robust innate immune response, such as the production and release of IFN. Given our success with an enzymatic approach, as demonstrated above, we restructured our assays to determine the molecular nature of the 5′ end of the noncapped viral genomic RNAs.
Following RNA extraction, the viral RNAs from purified viral particles were split into a series of parallel reaction mixtures and processed enzymatically, as described below, to determine the overall composition of the 5′ moiety. The abundance of viral capped and noncapped genomic RNA extracted from mammalian viral particles was determined as previously described. The direct detection of a 5′-monophosphate RNA species was the simplest, in terms of enzymatic treatment and analysis, as it involved the direct use of the extracted RNAs as a substrate for adaptor ligation. RNAs lacking a 5′-monophosphate, for instance, those without a phosphate or with a di- or triphosphate, cannot function as substrates for T4 RNA ligase and are not modified via addition of the 5′ adaptor instilling a high degree of selectivity to the assay. The abundance of nonphosphate RNA was determined via kinase treatment to generate a 5′-monophosphate and subsequent adaptor ligation. It is of note that the RNA populations detected during the above-described approach consist of signals from both the enzymatically generated and native 5′-monophosphate RNAs originally present in the sample; however, via subtractive analysis, the portion pertaining to the nonphosphate RNAs can be determined. Similarly, the presence of a 5′ diphosphate cannot be detected directly but rather is inferred from the relative abundances of the other 5′ moieties.
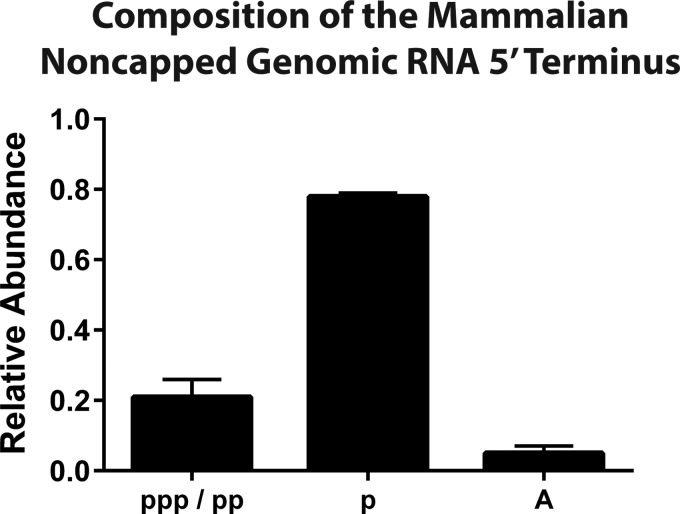
Unexpectedly, as shown in Fig. 5, the noncapped viral genomic RNAs of mammalian-cell-derived particles were predominantly monophosphate RNAs. Nonetheless, enzymatic analysis of the 5′ end of mammalian-cell-derived SINV particles revealed that a significant proportion of the noncapped viral genomic RNAs had tri- and diphosphate species. Non-phosphate-containing RNAs were the minor species of the encapsidated SINV particles derived from either host. The mechanism by which each RNA species is formed, in particular, the 5′-monophosphate, remains unclear.
FIG 5.
Composition of the 5′ terminus of the noncapped viral genomic RNA. Quantitative assessment of the 5′ terminus of mammalian-cell-derived SINV particles to determine the presence of 5′ tri-, di-, and monophosphates as described in Materials and Methods. The values shown are representative of two independent biological replicates. Error bars represent the variation between multiple technical replicates.
Mutation of nsP1 modulates the production of noncapped viral particles.
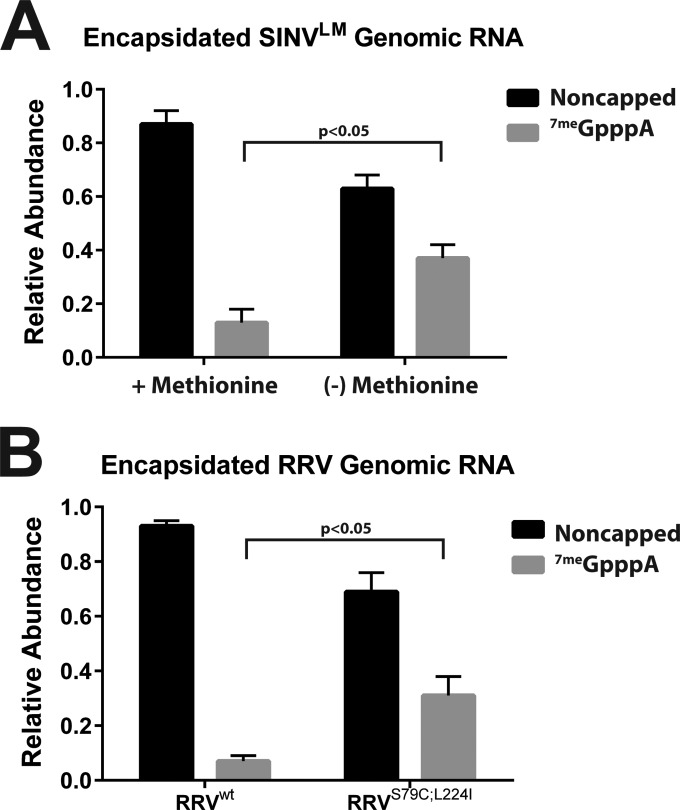
Since the viral nsP1 protein is the alphaviral capping protein, we focused our attention on nsP1 mutants with the intention of identifying alphaviruses that exhibited different capping activities. Previous studies have identified a number of alphaviral nsP1 capping mutants of SINV, RRV, and Semliki Forest virus (16, 18, 25, 33–35). Of particular interest were a set of nsP1 mutants, the SINV nsP1LM mutant that has been previously reported to be resistant to methionine deprivation and the similar RRVS79C;L224I mutant (25, 34). We hypothesized that the nsP1LM mutant SINV (for simplicity, SINVLM) would exhibit increased levels of 5′ 7meGpppA-capped viral genomic RNAs relative to wild-type SINV due to the function of methionine as a component of the methylation reaction. To our surprise, SINVLM produced noncapped viral genomic RNA containing particles at levels similar to those of wild-type SINV when cultured in the presence of methionine. In contrast, SINVLM cultured in the absence of methionine produced, on average, 4-fold more 5′ 7meGpppA-capped viral particles than those cultured in the presence of methionine (Fig. 6A). Similarly, the RRV nsP1 mutant RRVS79C;L224I exhibited similar behavior in terms of viral capping. As shown in Fig. 6B, RRVS79C;L224I produced, on average, 4.5-fold more capped viral particles than wild-type RRV. Taken together, these data indicated that production of noncapped viral RNAs could be modulated via the mutation of nsP1.
FIG 6.
Alphavirus nsP1 mutants modulate the production of noncapped viral RNAs. (A) Quantitative assessment of SINVLM particles produced from mammalian tissue culture cells under high- or low-methionine conditions. (B) Quantitative analysis of RRVS79C;L224I particles produced from mammalian cells. For both panels, the viral samples were collected at 18 hpi and treated as described in Materials and Methods. The values shown are the means of several biological replicates. Error bars represent the standard deviation of the mean.
SINV particles containing noncapped viral genomic RNAs activate the production of type I IFN.
Since the noncapped viral RNAs were produced at high levels during the infection of mammalian cells, we next sought to determine if there were biological consequences to their presence. While the majority of the noncapped viral genomes had a 5′-monophosphate, as shown in Fig. 5, approximately 20% had either 5′ tri- and diphosphate moieties. Cellular RNA sensors, in particular, RIG-I and the RLRs, are capable of recognizing tri- and diphosphate RNAs, thereby activating an antiviral host response (31, 32). It was unclear if the presence of the immune system-activating 5′-phosphate groups were sufficient to activate an antiviral response. Therefore, we next sought to determine if the level of noncapped viral particles correlated with the activation of a host innate immune response known to be activated by RIG-I, specifically, the secretion of soluble type I IFN. We hypothesized that alphavirus populations that contain fewer viral particles with noncapped genome, such as SINVHeavy, SINVLM, and the RRVS79C;L224I mutants, would induce less of an IFN response during infection. To this end, we examined these viruses for their induction of a type I IFN response in vitro.
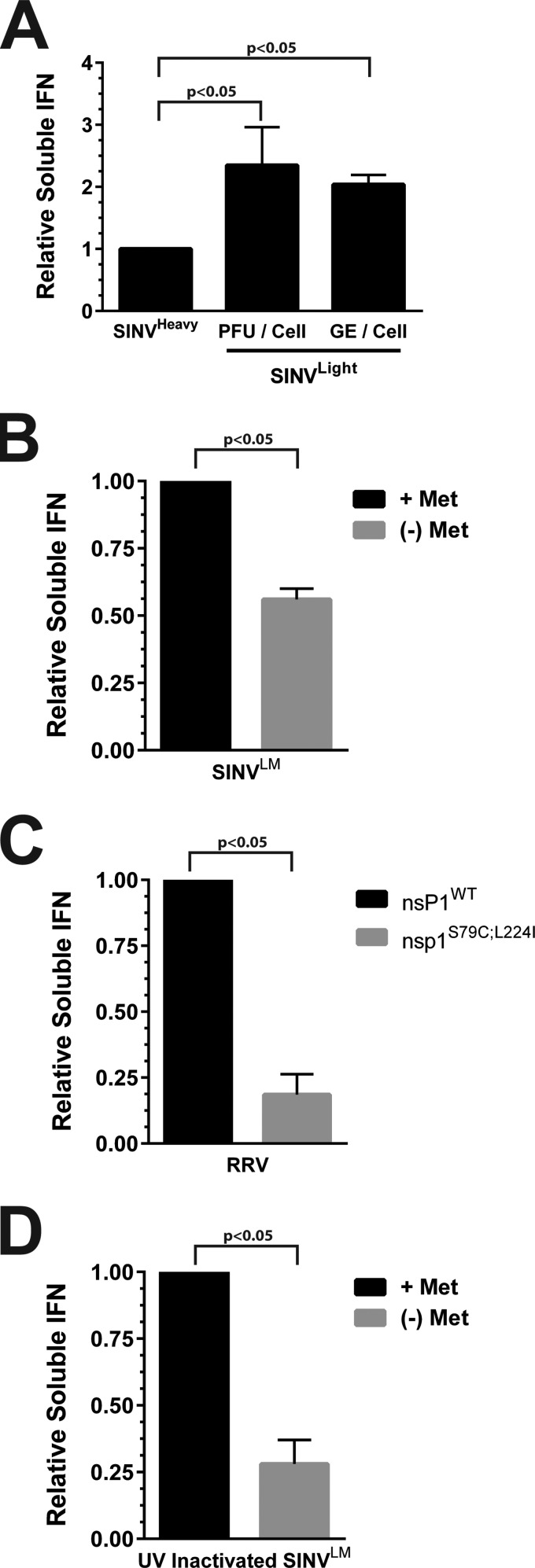
Confluent monolayers of L929 cells were infected with either SINVLight or SINVHeavy. After overnight incubation, the supernatants from the infected cells were harvested and any progeny viruses were inactivated as described in Materials and Methods. The resulting samples were then serially diluted onto fresh L929 cells and 24 h later challenged with a fluorescent RRV. The presence of IFN was detected via viral gene expression, as a function of fluorescence, or cell death. As shown in Fig. 7A, infection with SINVLight, at either equivalent numbers of PFU per cell or equal numbers of GE per cell, resulted in the production of ∼2-fold more soluble type I IFN than infection with SINVHeavy. Together, these data strongly suggest that particle quality, not the sheer number of particles, is responsible for the induction of a type I IFN response.
FIG 7.
Noncapped viral particles elicit an innate immune response. (A) The relative production of soluble type I IFNs during the infection of L929 cells with SINVHeavy or SINVLight at equivalent numbers of PFU per cell or total particles (as a function of the number of GE) per cell relative to that of SINVHeavy, as indicated on the x axis. (B) The relative production of soluble type I IFNs during the infection of L929 cells with infectious SINVLM cultured under either high- or low-methionine conditions, as indicated on the right. (C) Relative production of soluble type I IFN during the infection of L929 cells with either wild-type RRV or RRVS79C;L224I, as indicated on the x axis. (D) The relative production of soluble type I IFNs during the infection of L929 cells with UV-inactivated SINVLM cultured under either high- or low-methionine conditions, as indicated on the right. The values shown are the means of a minimum of three biological replicates. Error bars represent the standard deviation of the mean.
We next sought to determine whether the observed difference in the induction of soluble type I IFN was an intrinsic difference between SINVLight and SINVHeavy or due to the presence of increased numbers of noncapped viral genomic RNAs. As shown in Fig. 7B, SINVLM particles produced under high-methionine conditions, which contain predominantly noncapped genome, led to the production of approximately 2-fold more soluble IFN relative to SINVLM cultured under low-methionine conditions. Additionally, as shown in Fig. 7C, the RRVS79C;L224I mutant induced, on average, 4.5-fold less IFN than wild-type RRV in our in vitro model system. Taken together, these data indicate that the presence of noncapped genome in SINV and RRV viral particles correlates with the induction of a type I IFN response.
Despite having similar noncapped-to-capped genomic RNA ratios, low-methionine SINVLM exhibited a smaller reduction in the amount of IFN than that observed with RRVS79C;L224 (2- and 4.5-fold, respectively). A plausible explanation for this is that nascent RNA synthesis was diminishing the relative effects of the incoming viral particles. As shown in Fig. 5C, a significant amount of noncapped viral RNAs is produced during the early stages of alphaviral infection. Furthermore, as shown in Fig. 6A, SINVLM, when cultured under high-methionine conditions, produces wild-type levels of noncapped viral RNAs. Therefore, the production of increased levels of IFN observed may be causally linked to the synthesis of new noncapped viral RNAs rather than solely the incoming genomes. To determine if nascent synthesis of noncapped SINV RNAs was, in fact, confounding the above analyses, we used UV inactivation to prevent viral RNA synthesis. As shown in Fig. 7D, UV-inactivated SINVLM cultured under high-methionine conditions activated the production of soluble IFN approximately 4-fold more than SINVLM cultured under low-methionine conditions. It should be noted that the inactivation of the viral particles did, in fact, result in a net decrease in soluble-IFN levels (data not shown).
Taken together, these data indicate that the 5′ modification of the incoming genomic RNA influences the activation of host innate immunity. Specifically, viral populations that contain high proportions of noncapped, presumably 5′ tri- and diphosphate genomic RNAs activate the production of type I IFN in the absence of viral RNA synthesis. Moreover, these data indicate that the induction of IFN is additionally exacerbated by early viral RNA synthesis.
DISCUSSION
The data presented here indicate that the genomic RNAs packaged into SINV particles are inherently unstable early during infection and are not, as previously believed, universally 5′ capped with the viral type 0 7meGpppA cap structure. The presence of noncapped genomic RNA containing SINV particles inversely correlates with the basal infectivity of a sample, indicating that the 5′ cap is a primary determinant of viral infectivity. Furthermore, the noncapped SINV genomic RNA species correlated with the activation of an innate immune response, suggesting that the level of noncapped genomic RNAs may potentially influence viral transmission. Taken together, these observations represent a novel contribution to the field and expand our fundamental understanding of alphaviral infection at both the cellular and molecular levels, further underscoring the complexity of alphaviral infection.
Incoming SINV genomic RNAs are unstable and exhibit biphasic decay.
The data shown in Fig. 1 indicate that the half-lives of the incoming SINV genomic RNAs are, relative to most cellular RNAs, comparatively short. This ultimately may not be surprising, as the viral RNA following disassembly of the nucleocapsid core lacks the host factors that regulate the function of the viral transcript and thus the transcript likely appears foreign to the host cell. The half-lives associated with the incoming viral genomic RNA are relatively short and are reminiscent of those of mRNAs involved in immune responses or cytokine production, which require fine-tuned posttranscriptional control to provide adaptability to the cellular environment in response to stress or other stimuli (44). Interestingly, the decay of the incoming viral genomic RNA species appears to be biphasic regardless of derivation, starting with a period of rapid decay for the first 30 min of infection, followed by a prolonged period of relative stability. This phenomenon might be indicative of a period where the viral RNAs are initially unprotected prior to the formation of the functional viral messenger ribonucleoprotein (mRNP) complexes, resulting in decay. Characterization of the root causes of this observed biphasic decay pattern is an ongoing research focus.
At first glance, these findings appear to be contradictory to the RNA half-lives previously reported for the alphavirus RNA species; however, a fundamental difference between the two studies exists (23, 28). This study evaluated the half-lives of the incoming viral genomic RNA immediately after viral infection, whereas the previous studies evaluated viral RNA stability later during infection. It can be safely presumed that the cellular environment has been altered as a result of viral infection, leading to a change in RNA stability. For this reason, we assert that the differences in half-life reported between the two studies are not contradictory. Moreover, the studies reported here for the first time, to our knowledge, directly measured the half-life of an incoming viral genomic RNA.
SINV particles contain noncapped and SINV genomic RNAs.
Further characterization of the genomic RNAs themselves with the aim of identifying why the RNAs were unstable led to an unexpected realization. The observation that noncapped viral genomic RNAs are encapsidated and released in mature viral particles is novel to the field, as prior reports have indicated that the viral genomic RNA is universally capped. The data described in Fig. 2 through 5 indicate that a significant number of viral particles lack the type 0 7meGpppA cap structure. In our experience, noncapped in vitro-transcribed viral genomic RNAs are incapable of initiating viral infection via either electroporation or lipid transfection methods. Additionally, nsP1 mutants that lack guanylyltransferase activity are noninfectious (data not shown). Nonetheless, noncapped viral RNAs have been shown to be infectious, albeit at a significantly lower level than capped viral RNAs (36). Therefore, the viral particles containing noncapped genomic RNAs are likely noninfectious and incapable of initiating viral infection; however, their underlying function during viral infection remains to be seen. The presence of noncapped viral genomic RNA was likely obscured during the early studies of the alphavirus genomic 5′ terminus, which relied on the identification of radiolabeled methyl residues (37–39). Comparison of mammalian-cell- and mosquito cell-derived particles indicated that the overall composition of the viral particle populations differed between the two host species. Mammalian-cell-derived particles consist of a greater number of noninfectious particles than mosquito cell-derived particles, which consisted of roughly equal amounts of noncapped and capped viral particles. The data shown in Fig. 5 indicate that a determinant of the basal level of infectivity of a viral particle is the 5′ cap structure. Moreover, the rate of synthesis and accumulation of intracellular noncapped viral genomic RNAs in mammalian cells indicates that there is no preferential packaging of either 5′-capped or noncapped genomic RNA during infection.
Precisely why there are differences in the ratios of noncapped to capped genomic RNAs released in particles between the host species is unclear. It is well known that the outcome of infection is fundamentally different between the two hosts, as mammalian infection is acute whereas arthropod infection creates a persistent infection (5, 6, 8–10, 40–42). Perhaps these differences between the host species, for instance, the shutoff of host macromolecular synthesis, are a determinant of noncapped viral RNA production. Another possibility is that the formation of the noncapped alphaviral genomic RNAs is linked to the processing of the nonstructural polyprotein during infection. Alternatively, perhaps the greater proportion of capped viral particles observed with mosquito cell-derived particles is due to viral RNA synthesis and packaging being slower in mosquito cells, providing more time for capping to occur. The impetus behind the formation of noncapped viral particles and the apparent differences in production between vertebrate and invertebrate cells are ongoing research focuses.
The precise biochemical events leading to the formation of noncapped viral genomic RNAs are similarly uncertain. As stated in the introduction, the alphaviral capping reaction is a complex process mediated by the viral nsP1 and nsP2 proteins (16, 21, 22). The detection of a significant amount of 5′-monophosphate RNA is puzzling, as it should not be formed as a result of viral replication. Perhaps rapid RNA triphosphatase activity, as a result of differences in the kinetic rates of 5′ processing, results in the formation of 5′-monophosphate viral RNAs which are incapable of acting as substrates for the guanylyltransferase activity of nsP1 during viral RNA synthesis. Interestingly, as shown in Fig. 6, several mutant forms of nsP1, in at least two biologically distinct alphaviruses, SINV and RRV, are capable of altering the ratio of noncapped to capped RNAs. This indicates that the production of noncapped viral genomic RNAs may be modulated, at least in part, by sequence polymorphisms in nsP1.
The encapsidation of HDRCs is not dependent on the 5′ cap structure.
Initially, we had hypothesized that, in addition to being a basal determinant of viral infectivity and genomic RNA stability, the 5′ viral cap structure was a key determinant leading to the encapsidation of HDRCs into viral particles. The data presented in Fig. 3 indicate that the inclusion of HDRCs correlates with the presence of the viral 5′ cap. It is perhaps unsurprising that the inclusion of host factors involved in translation would correlate with the intrinsic ability to be translated. Nonetheless, we have also observed noncapped viral genomic RNAs in association with HDRC-containing particles. Whether the noncapped viral RNAs observed as described above are an inherent contaminant in our assays is unclear. It is possible, and indeed likely, that SINV particles lacking HDRCs are present in the SINVHeavy subpopulation as a consequence of cross contamination during purification. Despite this possibility, the observation of a similar profile of RNAs by an alternative approach, immunoprecipitation of RPS14, indicates that the presence of noncapped viral RNAs in contact with HDRCs is indeed genuine, but at a significantly lower level than for capped viral RNA. Therefore, these observations are insufficient to conclude that the 7meGpppA cap is a necessary or sufficient determinant of HDRC incorporation, but a correlation between the two exists.
Noncapped viral particles activate the cellular innate immune response.
Regardless of how the viral noncapped RNAs are synthesized, they influence viral infection, as the presence of noncapped genomic RNAs in noninfectious particles correlates with the expression of type I IFN. As shown in Fig. 7, the presence of noncapped viral particles correlated with the activation of a type I IFN response in vitro. Moreover, the ongoing synthesis of noncapped genomic RNAs likely exacerbates the innate immune response early during infection. Collectively, these results provide a potential molecular mechanism responsible for the differences in IFN elicitation by the infection of mosquito cell-derived versus mammalian-cell-derived particles (43). In addition, these data suggest that during alphaviral infection a significant number of noninfectious innate-immunity-enhancing viral particles are released prior to the exponential growth of infectious particles. The precise effects of this phenomenon on alphaviral biology are unclear; however, the RRVS79C;L224I mutant has been previously shown to exhibit attenuation in terms of viral titer and tissue clearance and increased sensitivity to IFN in mouse models of alphaviral infection (34). This is highly interesting, as it links viral RNA capping efficiency to viral fitness within a mammalian host. Furthermore, this is highly suggestive that the production of noncapped viral RNAs, and the release of noncapped viral particles, is functionally important to the replication and maintenance of viral infection within the vertebrate host, leading to pathology.
At first glance, it appears contrary to conventional wisdom that the production of noncapped viral RNAs, and hence the induction of an increased type I IFN response, would be beneficial to viral infection. At the organismal level, the production of noncapped viral RNAs, and hence viral particles, correlates with increased severity of disease (34). The cause of this phenomenon is unknown and represents an active area of interest; perhaps the activation of a local immune response is important for dissemination within the host. Another possibility is that the noncapped viral RNAs function to regulate the host cellular and local environment via an unknown molecular mechanism to an end that enhances viral infection. Nonetheless, the data presented here show that the production of noncapped viral RNAs is common to two evolutionarily divergent alphaviruses, observed in both invertebrate and vertebrate hosts, temporally regulated during infection, and biologically significant to the establishment and outcome of alphaviral infection.
A new molecular model of alphaviral infection.
Genuine alphaviral infection consists, at its basis, of the exposure of a permissive host cell to a mixture of infectious and noninfectious viral particles. Upon entry into the host cell cytoplasm, the genomic RNAs of infectious and noninfectious viral particles exhibit different biological activities. The genomic RNAs of noninfectious particles, which are noncapped, undergo rapid turnover following nucleocapsid disassembly and genome release. Nevertheless, a fraction of the noninfectious viral genomic RNAs attain stability via an unknown mechanism, perhaps as a result of host cell factors associating with the viral RNA. Given that the noncapped and capped viral RNAs exhibit different functions, at the very least on the basis of translation, during infection, in all likelihood, the mRNP complexes of the incoming noncapped and capped viral genomic RNAs are different. Regardless, the incoming noncapped viral genomic RNAs, as well as those produced early during infection, potentiate an innate immune response. Infectious particles with translationally capable genomic RNAs also exhibit RNA decay, albeit at a significantly lower rate than those of noninfectious particles. We postulate that this may be due to the time necessary to assemble the proper viral mRNP complex leading to viral gene expression. Viral particles that contain HDRCs may be able to more rapidly assemble the functional mRNP complex, as shown by the enhanced rate of translation exhibited by this subclass of particles. Collectively, the establishment of a productive alphaviral infection is the sum of all of the above processes.
ACKNOWLEDGMENTS
We thank the members of the Hardy and Mukhopadhyay labs for reading and editing the manuscript.
The work reported here was supported by grant R01 AI090077 from the NIH/NIAID to R.W.H., by grant F32AI104217 from the NIH/NIAID to K.J.S., and by grant R01 AI08725 from the NIH/NIAID to T.E.M.
REFERENCES
- 1.Coffey LL, Vignuzzi M. 2011. Host alternation of chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J Virol 85:1025–1035. doi: 10.1128/JVI.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. 2008. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A 105:6970–6975. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. 2005. Effect of alternating passage on adaptation of Sindbis virus to vertebrate and invertebrate cells. J Virol 79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, Brault AC, Kang W, Holland JJ. 1999. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol 73:4316–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garmashova N, Atasheva S, Kang W, Weaver SC, Frolova E, Frolov I. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J Virol 81:13552–13565. doi: 10.1128/JVI.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorchakov R, Frolova E, Frolov I. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol 79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpf AR, Blake JM, Brown DT. 1997. Characterization of the infection of Aedes albopictus cell clones by Sindbis virus. Virus Res 50:1–13. doi: 10.1016/S0168-1702(97)01461-5. [DOI] [PubMed] [Google Scholar]
- 9.Stollar V, Thomas VL. 1975. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology 64:367–377. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]
- 10.Stollar V, Shenk TE, Koo R, Igarashi A, Schlesinger RW. 1975. Observations of Aedes albopictus cell cultures persistently infected with Sindbis virus. Ann N Y Acad Sci 266:214–231. doi: 10.1111/j.1749-6632.1975.tb35103.x. [DOI] [PubMed] [Google Scholar]
- 11.Tooker P, Kennedy SI. 1981. Semliki Forest virus multiplication in clones of Aedes albopictus cells. J Virol 37:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson MA, Powers AM, Pesik N, Cohen NJ, Staples JE. 2014. Nowcasting the spread of chikungunya virus in the Americas. PLoS One 9:e104915. doi: 10.1371/journal.pone.0104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis 49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 14.Farnon EC, Sejvar JJ, Staples JE. 2008. Severe disease manifestations associated with acute chikungunya virus infection. Crit Care Med 36:2682–2683. doi: 10.1097/CCM.0b013e3181843d94. [DOI] [PubMed] [Google Scholar]
- 15.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahola T, Laakkonen P, Vihinen H, Kaariainen L. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol 71:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HL, Stollar V. 1999. Synthesis and methyltransferase activity of nonstructural protein nsP1 in Sindbis virus-infected Aedes albopictus cells. J Microbiol Immunol Infect 32:90–98. [PubMed] [Google Scholar]
- 18.Wang HL, O'Rear J, Stollar V. 1996. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology 217:527–531. doi: 10.1006/viro.1996.0147. [DOI] [PubMed] [Google Scholar]
- 19.Mi S, Stollar V. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184:423–427. doi: 10.1016/0042-6822(91)90862-6. [DOI] [PubMed] [Google Scholar]
- 20.Mi S, Durbin R, Huang HV, Rice CM, Stollar V. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170:385–391. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 21.Ahola T, Kaariainen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A 92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasiljeva L, Merits A, Auvinen P, Kaariainen L. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J Biol Chem 275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 23.Sokoloski KJ, Hayes CA, Dunn MP, Balke JL, Hardy RW, Mukhopadhyay S. 2012. Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res 167:26–33. doi: 10.1016/j.virusres.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokoloski KJ, Snyder AJ, Liu NH, Hayes CA, Mukhopadhyay S, Hardy RW. 2013. Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J Virol 87:12216–12226. doi: 10.1128/JVI.02437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi S, Stollar V. 1990. Both amino acid changes in nsP1 of Sindbis virusLM21 contribute to and are required for efficient expression of the mutant phenotype. Virology 178:429–434. doi: 10.1016/0042-6822(90)90340-W. [DOI] [PubMed] [Google Scholar]
- 26.Snyder AJ, Sokoloski KJ, Mukhopadhyay S. 2012. Mutating conserved cysteines in the alphavirus e2 glycoprotein causes virus-specific assembly defects. J Virol 86:3100–3111. doi: 10.1128/JVI.06615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. 2008. The 3′ untranslated region of Sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J Virol 82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song MG, Bail S, Kiledjian M. 2013. Multiple Nudix family proteins possess mRNA decapping activity. RNA 19:390–399. doi: 10.1261/rna.037309.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dölken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski UH. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblum CI, Scheidel LM, Stollar V. 1994. Mutations in the nsP1 coding sequence of Sindbis virus which restrict viral replication in secondary cultures of chick embryo fibroblasts prepared from aged primary cultures. Virology 198:100–108. doi: 10.1006/viro.1994.1012. [DOI] [PubMed] [Google Scholar]
- 34.Stoermer Burrack KA, Hawman DW, Jupille HJ, Oko L, Minor M, Shives KD, Gunn BM, Long KM, Morrison TE. 2014. Attenuating mutations in nsP1 reveal tissue-specific mechanisms for control of Ross River virus infection. J Virol 88:3719–3732. doi: 10.1128/JVI.02609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz CC, Suthar MS, Montgomery SA, Shabman R, Simmons J, Johnston RE, Morrison TE, Heise MT. 2010. Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein. Virology 399:1–10. doi: 10.1016/j.virol.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice CM, Levis R, Strauss JH, Huang HV. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol 61:3809–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubin DT, Stollar V. 1975. Methylation of Sindbis virus “26S” messenger RNA. Biochem Biophys Res Commun 66:1373–1379. doi: 10.1016/0006-291X(75)90511-2. [DOI] [PubMed] [Google Scholar]
- 38.Hefti E, Bishop DH, Dubin DT, Stollar V. 1975. 5′ nucleotide sequence of Sindbis viral RNA. J Virol 17:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersson RF, Soderlund H, Kaariainen L. 1980. The nucleotide sequences of the 5′-terminal T1 oligonucleotides of Semliki-Forest-virus 42-S and 26-S RNAs are different. Eur J Biochem 105:435–443. doi: 10.1111/j.1432-1033.1980.tb04518.x. [DOI] [PubMed] [Google Scholar]
- 40.Raghow RS, Grace TD, Filshie BK, Bartley W, Dalgarno L. 1973. Ross River virus replication in cultured mosquito and mammalian cells: virus growth and correlated ultrastructural changes. J Gen Virol 21:109–122. doi: 10.1099/0022-1317-21-1-109. [DOI] [PubMed] [Google Scholar]
- 41.Raghow RS, Davey MW, Dalgarno L. 1973. The growth of Semliki Forest virus in cultured mosquito cells: ultrastructural observations. Arch Gesamte Virusforsch 43:165–168. doi: 10.1007/BF01249360. [DOI] [PubMed] [Google Scholar]
- 42.Mims CA, Day MF, Marshall ID. 1966. Cytopathic effect of Semliki Forest virus in the mosquito Aedes aegypti. Am J Trop Med Hyg 15:775–784. [DOI] [PubMed] [Google Scholar]
- 43.Shabman RS, Morrison TE, Moore C, White L, Suthar MS, Hueston L, Rulli N, Lidbury B, Ting JP, Mahalingam S, Heise MT. 2007. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J Virol 81:237–247. doi: 10.1128/JVI.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. 2005. Stability regulation of mRNA and the control of gene expression. Ann N Y Acad Sci 1058:196–204. doi: 10.1196/annals.1359.026. [DOI] [PubMed] [Google Scholar]