Abstract
Since 1996, there have been at least six human norovirus pandemics. All of the pandemic strains are genetically related, segregating in the genogroup II, genotype 4 (GII.4) cluster within the Norovirus genus. Evidence indicates that these strains are closely related but antigenically distinct, supporting immune-driven viral evolution. Thus, norovirus vaccines will likely require periodic reformulation to protect from newly emergent strains. A major obstacle is that the reservoir of emergent strains is unknown. Noroviruses display tight species specificity and there is no evidence supporting zoonotic transmission, so an animal reservoir is considered unlikely. Moreover, available data indicate minimal viral diversity in most natural human infections. In this Gem, we discuss the widely speculated idea that chronically infected immunocompromised individuals are norovirus reservoirs and provide a rationale for the theory that elderly and malnourished hosts may also represent norovirus reservoirs.
INTRODUCTION
Human noroviruses are positive-sense RNA viruses with 7.5-kb genomes divided into three open reading frames (ORFs). ORF1 encodes a large polyprotein that is cleaved into six mature nonstructural proteins, ORF2 encodes a major capsid protein called VP1 that can self-assemble into virus-like particles (VLPs), and ORF3 encodes a minor structural protein called VP2. Human noroviruses are a major cause of gastroenteritis outbreaks across the globe and are the leading cause of both severe childhood diarrhea and food-borne disease outbreaks in the United States. Although norovirus infections typically result in acute and self-limiting gastroenteritis, young children, the elderly, and immunocompromised patients are vulnerable to more severe and prolonged infections. In particular, immunocompromised and transplant patients can be chronically infected with a human norovirus (1). In developing countries, they are likely an even more substantial cause of severe disease, as supported by an estimate that they cause over one million clinic visits and 200,000 deaths in young children annually in this setting (2). Although a recent global enteric multicenter study (GEMS) questioned whether human noroviruses are indeed a significant cause of disease in developing nations (3), it is important to recognize that the viral detection assay used in that study was extremely outdated and much less sensitive than well-established quantitative and broadly reactive reverse transcription-PCR assays in common use today; thus, the GEMS very likely grossly underestimated the norovirus disease burden. This argument is strongly supported by innumerable reports of norovirus detection in ∼20% of children hospitalized with diarrhea from all parts of the world. For example, Ahmed et al. recently documented an 18% norovirus prevalence in 187,336 patients with acute gastroenteritis when analyzing 175 separate and geographically diverse studies (4). Development of effective human norovirus vaccines would thus prevent significant morbidity and mortality worldwide. However, noroviruses are extremely genetically diverse, significantly complicating vaccination strategies. Moreover, new pandemic strains emerge every 2 to 4 years that appear to be antigenically distinct from previously circulating strains. It is thus likely that periodic vaccine reformulations will be required to protect from emergent human norovirus strains, similar to current influenza virus vaccination strategies. Unfortunately, the reservoir(s) of human noroviruses and the mechanisms regulating intra-and interhost norovirus evolutionary patterns remain ill-defined. This information is critical for the design of effective vaccines and to enhance our ability to predict the emergence of new strains on the basis of the efficiency of viral adaptability under different conditions. In this review, we will provide an in-depth analysis of the current view of norovirus reservoirs.
EMERGENCE OF PANDEMIC NOROVIRUS STRAINS
Human norovirus pandemics occurred in 1996-1997, 2002, 2004, 2006, 2009, and 2012. All six pandemics were caused by the emergence and rapid global spread of viral variants segregating within the genogroup II, genotype 4 (GII.4) human norovirus cluster. Most recently, the GII.4 Sydney strain caused a sharp spike in outbreaks globally during the 2012-2013 winter season. On the basis of the epochal pattern of evolution among these six pandemic strains and their high level of genetic relatedness, it is highly likely that population level immunity curbs pandemics after one or two seasons and immune escape mutations result in the emergence of successive pandemic strains. Indeed, available evidence demonstrates that GII.4 pandemic strains are not efficiently recognized by antibodies generated to previously circulating strains (e.g., see reference 5). Specifically, using time-ordered polyclonal and human monoclonal antibodies, time-ordered VLPs, and a surrogate assay for neutralizing antibody detection, multiple studies have shown that contemporary strains evolve mutations that attenuate the ability of ancestral strain immune sera to block binding to histo-blood group antigen (HBGA) carbohydrates. In fact, three highly variable antibody blockade epitopes exist on the surface of the GII.4 virus particle which change during successive waves of pandemic viruses (6). Epitope mapping was performed by using bioinformatic approaches to identify and cluster rapidly evolving amino acid residues on the surface of VLPs and then exchanging these regions between VLPs. By using functional assays, precise mapping of key residues that drive antigenic change in response to human and mouse antibody binding and blockade then provided direct evidence of antigenic variation during GII.4 evolution. Of the epitopes identified, variation in the A epitope domain most closely maps with new strain emergence and pandemic disease outbreaks.
Understanding the evolutionary pressures regulating intra- and interhost norovirus diversity should inform the processes underlying pandemic strain emergence. Viral diversity can enhance the adaptability of a viral swarm by increasing the probability of phenotypically distinct variants. These could include antigenic variants that are not efficiently recognized by immunity to previously circulating strains and variants with increased virulence. Diversity is influenced by viral factors such as the inherent mutational rate of the viral replicative enzyme, the replication rate of the virus, and host factors, with the magnitude and nature of the immune response playing a dominant role. The norovirus RNA-dependent RNA polymerase (RdRp) has a high error rate, with available evidence indicating that the RdRp of GII.4 human norovirus strains has a higher rate of mutation than other genotypes (7). In spite of this, available data—admittedly limited in scope—indicate that the complexity of the norovirus quasispecies in infected individuals with a normal immune system is quite low. For example, Bull et al. observed only one single nucleotide polymorphism (SNP) in the ORF2/partial ORF3 genes over a 9-day period of infection in a healthy subject (8); and Vega et al. identified no nucleotide differences from a published virus sequence among 10 VP1 clones from an immunocompetent subject (9). While future studies using deep sequencing of the entire viral genome from larger cohorts are necessary to more accurately determine in vivo viral evolution rates, early data do support surprisingly limited viral diversity in immunocompetent individuals. Considering this, where do pandemic strains arise? Although noroviruses have been identified in a diverse range of animals, there is no evidence of zoonotic transmission (10), so it is reasonable to presume that variant viruses arise within the human population.
EVIDENCE FOR IMMUNOCOMPROMISED HOSTS AS NOROVIRUS RESERVOIRS
Immunocompromised and transplant patients can develop chronic norovirus infections that last months and even years (1). Because of the extended nature of these infections and the reduced immune pressure restricting viral mutation, many groups have speculated that the immunocompromised host represents a norovirus reservoir. Indeed, multiple studies have investigated longitudinal human norovirus diversity in chronically infected immunocompromised patients, and a high degree of viral sequence heterogeneity was observed in each case (8, 9, 11–15). For example, in the same study detecting one SNP in a healthy host over 9 days of infection, 18 SNPs were detected over 288 days of infection in an immunocompromised subject (8). Furthermore, the quasispecies appeared to be quite complex in the immunocompromised host, in striking contrast to the homogeneous viral population observed in four healthy subjects (8). However, a critical distinction between viral diversity and viral evolution must be recognized—if immunocompromised individuals are unable to mount a robust antiviral antibody response, it is unlikely that viral mutations resulting in immune escape will become dominant in the viral quasispecies. The phylodynamic framework for RNA virus evolution proposed by Grenfell et al., in fact, argues that intermediate levels of immunity should result in maximal intrahost viral adaptation (16) (Fig. 1). This framework is based on the idea that a robust immune response in immunocompetent individuals will fully suppress viral replication, thereby restricting the production of fitter viruses; an absence of immunity in severely immunocompromised hosts will result in a complex quasispecies but no pressure to select for fitter variants; while a weak immune response will fail to completely shut down viral replication while simultaneously providing some selective pressure for escape mutants. There is support for this model in relation to human norovirus evolution; Siebenga et al. revealed that the number of VP1 amino acid changes over time was higher in patients with intermediate immunocompromise than in more severely immunocompromised patients (15). Collective preliminary data thus suggest that human norovirus adaptation is suppressed in healthy people, presumably by a robust immune response; occurs maximally in hosts with intermediate levels of immunity, where selective pressure is applied by the immune response but it is not robust enough to completely clear infection; and is minimal in severely immunocompromised individuals who fail to generate selective pressure.
FIG 1.
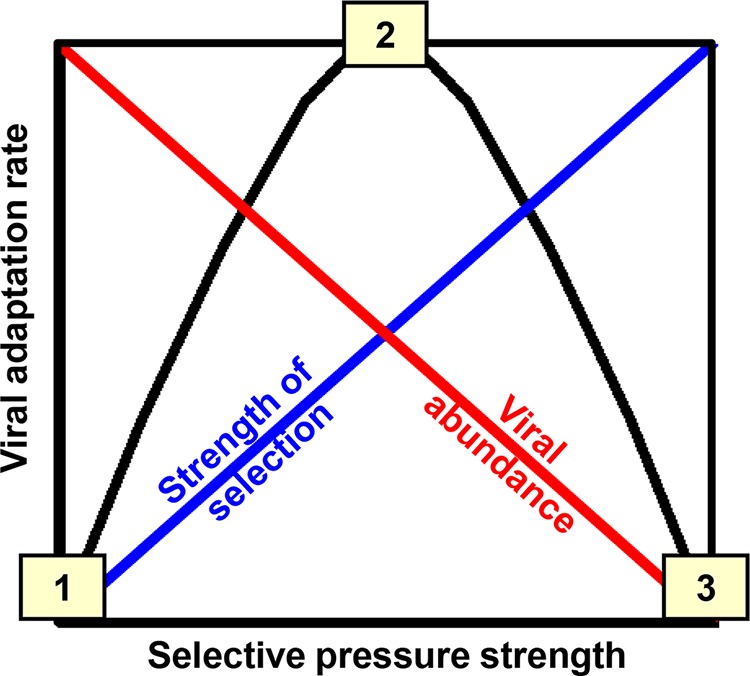
Model of RNA virus evolution. The current framework for the adaptation rate of fast-evolving RNA viruses like noroviruses (black line) is the net result of the complexity of the viral quasispecies resulting from the inherent error rate of the RdRp (red line), the strength of the selective pressures applied to this viral population by the host (blue line), and the replication rate of the virus (not accounted for in this model). In this model, immunocompromised hosts (point 1) have a complex quasispecies because of the high error rate of the RdRp but low adaptation because of reduced selective pressure, healthy hosts (point 3) develop a robust immune response that limits the development of a complex quasispecies, and hosts with intermediate immunity (point 2) display the highest adaptation rate because of the development of weak selective pressure—this pressure is insufficient to completely control viral replication, so a complex quasispecies develops, but it is sufficient to drive the evolution of the virus. Adapted from reference 16.
To directly test whether increased viral quasispecies complexity translates into clinically relevant phenotypic variability, it is necessary to examine whether viral mutations arising in immunocompromised hosts provide fitness benefits (e.g., increased viral replication, antigenic variability, enhanced virulence). However, until very recently human noroviruses could not be cultured in the laboratory and no efficient reverse genetic system was available, precluding direct testing of the effects of viral mutations on antibody binding/neutralization and viral replication efficiency. In the absence of these basic virological tools, researchers have relied on the ability of the norovirus VP1 protein to self-assemble into VLPs, which facilitates genetic testing of mutations arising in this viral protein; and a surrogate neutralization assay whereby antibodies are tested for the ability to prevent VLP binding to an HBGA attachment factor. One recent study by Debbink et al. utilized these tools to probe the critical question of whether increased quasispecies complexity in immunocompromised hosts is indicative of the presence of phenotypically distinct viral variants (17). Specifically, VLPs were synthesized from ORF2 genes containing either the GII.4-2006 pandemic strain or the consensus sequences from two temporal samples of a chronically infected subject 302 days apart. These VLPs displayed differences in carbohydrate binding, antibody reactivity, and antibody blockade activity, providing support for the emergence of phenotypically distinct viral variants in a chronically infected immunocompromised host. Future phenotypic analyses of viral mutations arising in immunocompromised hosts will be greatly facilitated by the recent development of a human norovirus cell culture system (18) and a helper-free human norovirus reverse genetics system (19). Moreover, it will be important to consider whether elderly individuals can act as viral reservoirs since they are immunosuppressed and can have more severe and prolonged norovirus infections than younger adults.
EVIDENCE FOR NUTRITIONAL REGULATION OF NOROVIRUS EVOLUTION
Considering the premise that viral evolution occurs most readily under conditions of weak immunity (16), we have begun to test whether malnourished hosts support higher levels of norovirus evolution than their healthy counterparts and could thus represent a potential viral reservoir. This hypothesis is based on the well-established knowledge that malnourished people mount suboptimal immune responses and are highly susceptible to severe and prolonged viral infections. A murine model of norovirus infection was used to test this hypothesis because it allowed us to examine the impact of malnutrition on viral evolution in the absence of confounding host and environmental variability. Indeed, mice fed a diet low in protein (reflecting a common form of malnutrition observed in children in developing nations referred to as protein energy malnutrition) displayed greater viral diversity, as indicated by a significantly higher number of nonsynonymous substitutions in the VP1 protein, than healthy mice at 50 days postinfection (20). In particular, there was a significantly higher number of virion surface-exposed mutations in the malnourished mice than in their healthy counterparts. This increased viral diversity correlated with increased severity of infection, reduced viral clearance, and a substantially reduced mucosal antiviral antibody response in spite of increased virus titers. Considering the likely scope of norovirus infections in malnourished people each year (2), their potential as reservoirs of phenotypically distinct norovirus strains warrants further study.
Although we have focused our discussion on intrahost norovirus evolution, pandemic strain emergence is also likely shaped by population phylodynamics. For example, neutral viral evolution occurring during widespread outbreaks combined with positive selection of new antigenic variants could explain GII.4 evolutionary patterns. Another factor known to contribute to norovirus evolution is the ability of the viral genome to recombine, with a recombination hot spot identified at the ORF1-ORF2 border.
PERSPECTIVES
Although the epidemiological pattern of human norovirus pandemics is very similar to that of influenza viruses, a major distinction is the apparent absence of animal reservoirs of noroviruses that support viral mixing and zoonotic transmission. Thus, a critical goal in tackling future norovirus vaccine design is identifying the reservoir of emergent pandemic strains. It has been widely speculated that chronically infected immunocompromised subjects may represent a reservoir of emergent strains. This idea stems from the well-supported observation that the viral quasispecies in immunocompromised hosts are complex, in striking contrast to the very stable viral populations in healthy hosts. We propose in this Gem that the degree of immunocompromise in individual patients will play a major role in determining the extent of viral evolution (in contrast to viral diversity) that arises. We also propose that elderly and malnourished hosts represent other likely viral reservoirs because of their suboptimal—but not absent—immune response to viral infections; this idea is supported by recent work with the murine norovirus model system. Unequivocal demonstration of the emergence of novel viral variants requires phenotypic testing of adaptive mutations. Unfortunately, limitations in experimental human norovirus systems have precluded an in-depth analysis of whether viral diversity in the setting of immunocompromise translates to phenotypic variability. One recent study did demonstrate antigenic variability arising in a single immunocompromised host by using existing research tools, warranting continued study of a larger number of subjects with defined but variable levels of immunocompromise and malnutrition. Moreover, recent advances in human norovirus research tools, including a cell culture system and an efficient reverse genetics system, should greatly advance the ability of the field to test for phenotypic consequences associated with viral genetic changes.
REFERENCES
- 1.Bok K, Green KY. 2012. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med 367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel MM. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Ryan M, Vidal R, Del Canto F, Carlos Salazar J, Montero D. 25 February 2015. Vaccines for viral and bacterial pathogens causing acute gastroenteritis: part I: overview, vaccines for enteric viruses and Vibrio cholerae. Hum Vaccin Immunother doi: 10.1080/21645515.2015.1011019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinjé J, Baric RS. 2013. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J Infect Dis 208:1877–1887. doi: 10.1093/infdis/jit370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debbink K, Lindesmith LC, Donaldson EF, Baric RS. 2012. Norovirus immunity and the great escape. PLoS Pathog 8:e1002921. doi: 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull RA, Eden J-S, Rawlinson WD, White PA. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog 6:e1000831. doi: 10.1371/journal.ppat.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull RA, Eden J-S, Luciani F, McElroy K, Rawlinson WD, White PA. 2011. Contribution of intra- and interhost dynamics to norovirus evolution. J Virol 86:3219–3229. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega E, Donaldson E, Huynh J, Barclay L, Lopman B, Baric R, Chen LF, Vinjé J. 2014. RNA populations in immunocompromised patients as reservoirs for novel norovirus variants. J Virol 88:14184–14196. doi: 10.1128/JVI.02494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm B, Waddell L, Greig J, Rajić A, Houde A, McEwen SA. 2015. A scoping review of the evidence for public health risks of three emerging potentially zoonotic viruses: hepatitis E virus, norovirus, and rotavirus. Prev Vet Med 119:61–79. doi: 10.1016/j.prevetmed.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Carlsson B, Lindberg AM, Rodriguez-Díaz J, Hedlund K-O, Persson B, Svensson L. 2009. Quasispecies dynamics and molecular evolution of human norovirus capsid P region during chronic infection. J Gen Virol 90:432–441. doi: 10.1099/vir.0.005082-0. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann D, Hutzenthaler M, Seebach J, Panning M, Umgelter A, Menzel H, Protzer U, Metzler D. 2012. Norovirus GII.4 and GII.7 capsid sequences undergo positive selection in chronically infected patients. Infect Genet Evol 12:461–466. doi: 10.1016/j.meegid.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson M, Hedlund K-O, Thorhagen M, Larson G, Johansen K, Ekspong A, Svensson L. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J Virol 77:13117–13124. doi: 10.1128/JVI.77.24.13117-13124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schorn R, Höhne M, Meerbach A, Bossart W, Wüthrich RP, Schreier E, Müller NJ, Fehr T. 2010. Chronic norovirus infection after kidney transplantation: molecular evidence for immune-driven viral evolution. Clin Infect Dis Off Publ Infect Dis Soc Am 51:307–314. doi: 10.1086/653939. [DOI] [PubMed] [Google Scholar]
- 15.Siebenga JJ, Beersma MFC, Vennema H, van Biezen P, Hartwig NJ, Koopmans M. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J Infect Dis 198:994–1001. doi: 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- 16.Grenfell BT, Pybus OG, Gog JR, Wood JLN, Daly JM, Mumford JA, Holmes EC. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 17.Debbink K, Lindesmith LC, Ferris MT, Swanstrom J, Beltramello M, Corti D, Lanzavecchia A, Baric RS. 2014. Within-host evolution results in antigenically distinct GII.4 noroviruses. J Virol 88:7244–7255. doi: 10.1128/JVI.00203-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and murine norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama K, Murakami K, Sharp TM, Guix S, Oka T, Takai-Todaka R, Nakanishi A, Crawford SE, Atmar RL, Estes MK. 2014. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc Natl Acad Sci U S A 111:E4043–E4052. doi: 10.1073/pnas.1415096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman D, Jones MK, Zhu S, Kirpatrick E, Ostrov DA, Wang X, Ukhanova M, Sun Y, Mai V, Salemi M, Karst SM. 2014. The effect of malnutrition on norovirus infection. mBio 5:e01032–13. doi: 10.1128/mBio.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


