ABSTRACT
The nucleoprotein (NP) is a major component of the viral ribonucleoprotein (vRNP) complex. During the replication of influenza virus, the vRNP complex undergoes nuclear-cytoplasmic shuttling, during which NP serves as one of the determinants. To date, many phosphorylation sites on NP have been identified, but the biological functions of many of these phosphorylation sites remain unknown. In the present study, the functions of the phosphorylation sites S9, Y10, and Y296 were characterized. These residues are highly conserved, and their phosphorylation was essential for virus growth in cell culture and in a mouse model by regulating the activity of the viral polymerase and the nuclear-cytoplasmic shuttling of NP. The phosphorylation and dephosphorylation of S9 and Y10 controlled nuclear import of NP by affecting the binding affinity between NP and different isoforms of importin-α. In addition, the phosphorylation of Y296 caused nuclear retention of NP by reducing the interaction between NP and CRM1. Furthermore, tyrosine phosphorylation of NP during the early stage of virus infection was ablated when Y296 was mutated to F. However, at later stages of infection, it was weakened by the Y10F mutation. Taken together, the present data indicate that the phosphorylation and dephosphorylation of NP control the shuttling of NP between the nucleus and the cytoplasm during virus replication.
IMPORTANCE It is well known that phosphorylation regulates the functions of viral proteins and the life cycle of influenza A virus. As NP is the most abundant protein in the vRNP complex of influenza A virus, several phosphorylation sites on this protein have been identified. However, the functions of these phosphorylation sites were unknown. The present study demonstrates that the phosphorylation status of these sites on NP can mediate its nuclear-cytoplasmic shuttling, which drives the trafficking of vRNP complexes in infected cells. The present data suggest that the phosphorylated residues of NP are multistep controllers of the virus life cycle and new targets for the design of anti-influenza drugs.
INTRODUCTION
Influenza A viruses cause major respiratory infectious diseases in birds and mammals and are a global burden to public health, as exemplified by the swine-origin influenza H1N1 viruses from 2009 and the avian influenza H7N9 viruses from 2013. The influenza A virus genome is composed of eight negative-sense, single-stranded RNA segments (viral RNAs [vRNAs]) (1, 2). Each vRNA segment is encapsulated by multiple copies of the nucleoprotein (NP) (3). During the early stage of influenza virus infection, the viral ribonucleoprotein (vRNP) complex utilizes the nuclear localization signals (NLSs) on NP for nuclear import (4). During the late stage of infection, the nuclear export protein (NEP) and matrix protein 1 (M1) of influenza A virus guide the newly assembled vRNP complexes from the nucleus into the cytoplasm by interacting with NP, and NP itself also plays a role in the export of vRNP complexes (5–7).
Two NLSs (NLS1 [amino acids {aa} 3 to 13] and NLS2 [aa 198 to 216]) and one nuclear accumulation signal (NAS [aa 327 to 345]) control nuclear import and accumulation of NP, while three nuclear export signals (NES1 [aa 24 to 49], NES2 [aa 183 to 197], and NES3 [aa 248 to 274]) transport NP into the cytoplasm (6–11). The nuclear import of NP relies on the importin-α/β transport system, and importin-α binds NP by recognition of its NLSs (12). During vRNP complex export, NP also plays an important role by binding to the CRM1 (chromosome region maintenance 1) cellular export receptor (7, 13).
Posttranslational modifications of influenza A virus proteins, such as phosphorylation, regulate the viral life cycle (14–18). The functionality of NLS1 is also regulated by phosphorylation of NP serine 3 in influenza virus A/Puerto Rico/8/1934 (H1N1) (19). Treatment with a stimulator (tetradecanoyl phorbol acetate [TPA]) or inhibitor (H7) of protein kinase C (PKC) changes the cellular localization of NP (20). Indeed, many aspects of the viral life cycle, including vRNA synthesis, shuttling of viral proteins between the nucleus and the cytoplasm, protein synthesis, and the release of virus particles, can be affected by kinase inhibitors (21–24).
Recently, S165 phosphorylation of NP was shown to have a negative effect on the viral polymerase and the dissociation of the NP oligomers (14, 25, 26). Other phosphorylated NP residues, including S9, Y10, and Y296, have also been reported (14). However, the functions of these phosphorylation sites have not been elucidated, i.e., whether and how these phosphorylation sites affect NP trafficking have not yet been determined. In the present study, the S9, Y10, and Y296 phosphorylation sites were found to be essential for viral growth and the regulation of polymerase activities. By performing immunofluorescence assays (IFAs) and coimmunoprecipitation (co-IP) assays, we demonstrated that S9 and Y10 control the nuclear import of NP. In addition, we found that Y296 plays an important role in the nuclear export of NP by regulating the binding affinity of NP for CRM1 via phosphorylation and dephosphorylation.
MATERIALS AND METHODS
Virus and cells.
Influenza A virus A/WSN/33 (H1N1) was rescued from cDNAs and propagated in the allantoic cavities of 10-day-old specific-pathogen-free embryonic chicken eggs. Human embryonic kidney (293T) cells, human alveolar epithelial (A549) cells, and Madin-Darby canine kidney (MDCK) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (both from Invitrogen) at 37°C and 5% CO2.
Antibodies and reagents.
A rabbit polyclonal antibody against NP and a mouse monoclonal antibody against M1 were generated as previously described (27). Mouse anti-β-actin polyclonal antibody, anti-HSP70 (3A3) antibody, anti-lamin B1 (A-11) antibody, anti-p-Tyr (sc-508) antibody, and anti-c-MYC (9E10) antibody were purchased from Santa Cruz Biotechnology, Inc. Mouse anti-FLAG antibody and rabbit anti-c-MYC antibody were purchased from Sigma-Aldrich. All secondary antibodies were obtained from Bai Hui Zhong Yuan Biotechnology. An RNase inhibitor was purchased from Promega, and protease inhibitor cocktail and phosphatase inhibitor phosSTOP were purchased from Roche.
Plasmid construction.
The full-length NP gene obtained from the A/WSN/33 virus was cloned into the pCDNA4/TO vector for prokaryotic expression. For co-IP assays, IFAs, and luciferase assays, the NP gene was cloned into the pcDNA3-Flag, pCMV-MYC, and pCDNA4/TO vectors. Mutations in full-length NP in pHH21, pcDNA4/TO, pcDNA3-Flag, and pCMV-MYC were generated using a site-directed mutagenesis kit (Newpep, China). The full-length CRM1, importin-α1, importin-α3, importin-α5, and importin-α7 genes were cloned into pcDNA3-Flag.
Generation of recombinant influenza A viruses.
The wild-type (WT) influenza A/WSN/1933 (H1N1) virus and its NP mutants were generated using a 12-plasmid reverse genetic system (28). Briefly, 293T cells grown to 90% confluence in 60-mm dishes were transfected with 1 μg each of the 12 plasmids of the virus rescue system. Six hours later, the medium was replaced with DMEM containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin. The cells were further cultured for 72 h at 37°C in 5% CO2, and the supernatant containing the recombinant viruses was harvested and centrifuged at 5,000 × g for 5 min to remove cell debris.
Plaque assays.
Plaque assays were performed as previously described (27). Briefly, MDCK cell monolayers (5 × 106 cells at 100% confluence in a 12-well plate) were washed with phosphate-buffered saline (PBS) and infected with different dilutions of virus for 1 h at 37°C. The virus inoculums were removed by washing with PBS. Cell monolayers were then overlaid with agar overlay medium (DMEM supplemented with 1% low-melting-point agarose and 1 μg/ml TPCK-treated trypsin) and incubated at 37°C. Visible plaques were counted at 3 days postinfection, and the virus titers were determined. All data are expressed as the means of values from three independent experiments.
IP and Western blot analyses.
293T cells were lysed in lysis buffer containing 1% Triton X-100, 150 mM NaCl, 20 mM HEPES, 10% glycerol, and 1 mM EDTA (pH 7.4) supplemented with Complete protease inhibitor cocktail and a phosphatase inhibitor (5 mM Na3VO4). After an incubation period of 40 min at 4°C, insoluble components were removed by centrifugation at 12,000 × g for 15 min. Lysates were incubated with anti-FLAG M2 affinity gel for ≥8 h. Following five washes in wash buffer (1% Triton X-100, 300 mM NaCl, 20 mM HEPES, 10% glycerol, and 1 mM EDTA [pH 7.4]), the precipitated proteins were separated by SDS-PAGE and then transferred to Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Billerica, MA). The membranes were blocked for 2 h at 4°C in blocking solution (5% skim milk powder and 0.5% Tween 20 in PBS), and proteins were detected using appropriate antibodies, followed by the addition of anti-rabbit or anti-mouse secondary antibody coupled to horseradish peroxidase. Proteins were visualized by use of chemiluminescence detection reagents.
Pathogenicity in mice.
Seventy-six 6- to 8-week-old (∼17 g) female BALB/c mice (Vital River Laboratory, Beijing, China) were anesthetized with tiletamine-zolazepam (Zoletil; Virbac) (25 μg/g of body weight) and inoculated intranasally with 50 μl of virus diluent in PBS. Mock-infected control animals were inoculated with 50 μl PBS. Three mice from each group were euthanized at 3, 5, and 7 days postinfection (p.i.), and their lungs were collected, weighed, and homogenized using a Qiagen TissueLyser II machine (30 cycles/s; 4 min) in 1 ml of cold PBS under sterile conditions. The solid debris was pelleted by centrifugation at 5,000 × g for 10 min, and the homogenates were used for virus titrations in MDCK cells. The other mice were monitored daily for clinical signs, including body weight and mortality, for 14 days. All animal research was approved by the Chinese Academy of Sciences of Research Ethics Committee and complied with the Beijing Laboratory Animal Welfare and Ethical Guidelines of the Beijing Administration Committee of Laboratory Animals.
Luciferase assay of influenza virus polymerase activity.
The plasmids for expression of the PA, PB1, PB2, and NP (WT or mutant) proteins were simultaneously transfected into 293T cells by use of luciferase reporter plasmids (pHH21-cNS-Luc and pcDNA-β-gal) as described by Li et al. (29), with some modifications. As a negative control, 293T cells were transfected with the same plasmids, excluding the NP expression plasmid. After transfection, the cells were incubated at 37°C for 36 h, and then the amount of luciferase activity in the transfected cells was measured and normalized to the amount of β-galactosidase activity, as measured by use of standard kits (Promega, Madison, WI).
RNA extraction, cDNA synthesis, and real-time quantitative PCR.
Total RNA was extracted from 293T cells by use of TRIzol (Invitrogen) according to the manufacturer's instructions. Samples were digested with DNase I and subjected to reverse transcription-PCR (RT-PCR). RNA was reverse transcribed using the following PCR primers: mRNA primer, oligo(dT); cRNA primer, 5′-AGTAGAAACAAGG-3′; and vRNA primer, 5′-AGCGAAAGCAGG-3′. A mock reaction was performed with no reverse transcriptase added to the reaction mixture. The analysis of relative M1 gene expression was performed using an ABI 7300 machine and the following PCR primers: M1 forward (5′-TCTGATCCTCTCGTCATTGCAGCAA-3′) and reverse (5′-AATGACCATCGTCAACATCCACAGC-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control, using the PCR primers GAPDH forward (5′-GGTGGTCTCCTCTGACTTCAACA-3′) and GAPDH reverse (5′-GTTGCTGTAGCCAAATTCGTTGT-3′). The cycling conditions comprised an initial denaturation step of 30 s at 95°C, followed by 40 two-step cycles (95°C for 5 s and 60°C for 31 s). Dissociation curve analysis was performed after each assay to ensure specific target detection.
IFAs.
IFAs were performed with an Olympus FV500 and Leica SP8 confocal laser scanning microscope. Coverslips carrying 293T cells were washed with PBST (PBS plus 1% Triton X-100) and then fixed with 4% paraformaldehyde. Cells were then blocked with 4% bovine serum albumin (BSA) dissolved in PBST and stained with anti-NP and anti-MYC antibodies. The secondary antibodies were fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG and tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-rabbit IgG.
Phosphate-affinity SDS-PAGE and preparation for nano-LC-MS/MS analysis.
Influenza A virus-infected 293T cells were lysed in lysis buffer (20 mM HEPES [pH 7.4], 1% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM EDTA) supplemented with Complete protease inhibitor cocktail (Roche Diagnostics) and a phosphatase inhibitor (5 mM Na3VO4; Sigma). The NP was purified with protein G agarose beads prebound to a mouse anti-NP polyclonal antibody for 3 h at 4°C. Proteins were separated by 15% Mn2+-Phos-tag SDS-PAGE as described previously (15). Briefly, normal polyacrylamide gel electrophoresis was conducted according to the TaKaRa protocol, with an acrylamide-pendant phosphate-tagged (Phos-tag) ligand (50 μM) and 0.1 mM MnCl2 (Sigma) added to the separating gel before polymerization. The gel was silver stained, and the separated bands were subjected to nano-liquid chromatography–tandem mass spectrometry (nano-LC-MS/MS) identification at the Technological Platform of the Institute of Zoology, Chinese Academy of Sciences (LCQ Deca XP Plus; Thermo).
Computer modeling and statistical analyses.
The three-dimensional crystal structure of NP was manipulated with the program PyMOL (Schrödinger). Statistical analyses were performed using MegAlign software (DNAstar Software, San Diego, CA).
RESULTS
Effects of S9, Y10, and Y296 phosphorylation on virus replication in cell culture and in a mouse model.
Sequence data indicated that residues S9, Y10, and Y296 are highly conserved among different subtypes of influenza A viruses (Table 1). The sequence alignment was performed with viruses of the H1N1, H2N2, H3N2, H5N1, H7N7, H9N2, and recent avian H7N9 subtypes (400 different isolates of the H1N1, H3N2, and H5N1 subtypes, 110 isolates of the H2N2 subtype, 60 isolates of the H7N7 subtype, and 347 isolates of the H7N9 subtype were analyzed by MegAlign). Residues Y10 and Y296 were found to be highly conserved in all of the analyzed isolates. Residue S9 was conserved in >90% of isolates.
TABLE 1.
Conservation of S9, Y10, and Y296 among NPs of different subtypes of influenza A virus
| Residue | Kinase predictiona | Residue conservation (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| H1N1 | H2N2 | H3N2 | H5N1 | H7N7 | H7N9 | H9N2 | ||
| S9 | PKG | 100.0 | 100.0 | 100.0 | 99.0 | 90.7 | 100.0 | 100.0 |
| Y10 | INSR | 100.0 | 99.1 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| Y296 | INSR | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
For the WSN virus sequence. Kinase prediction was performed by use of KinasePhos (http://kinasephos.mbc.nctu.edu.tw/). PKG, protein kinase G; INSR, insulin receptor.
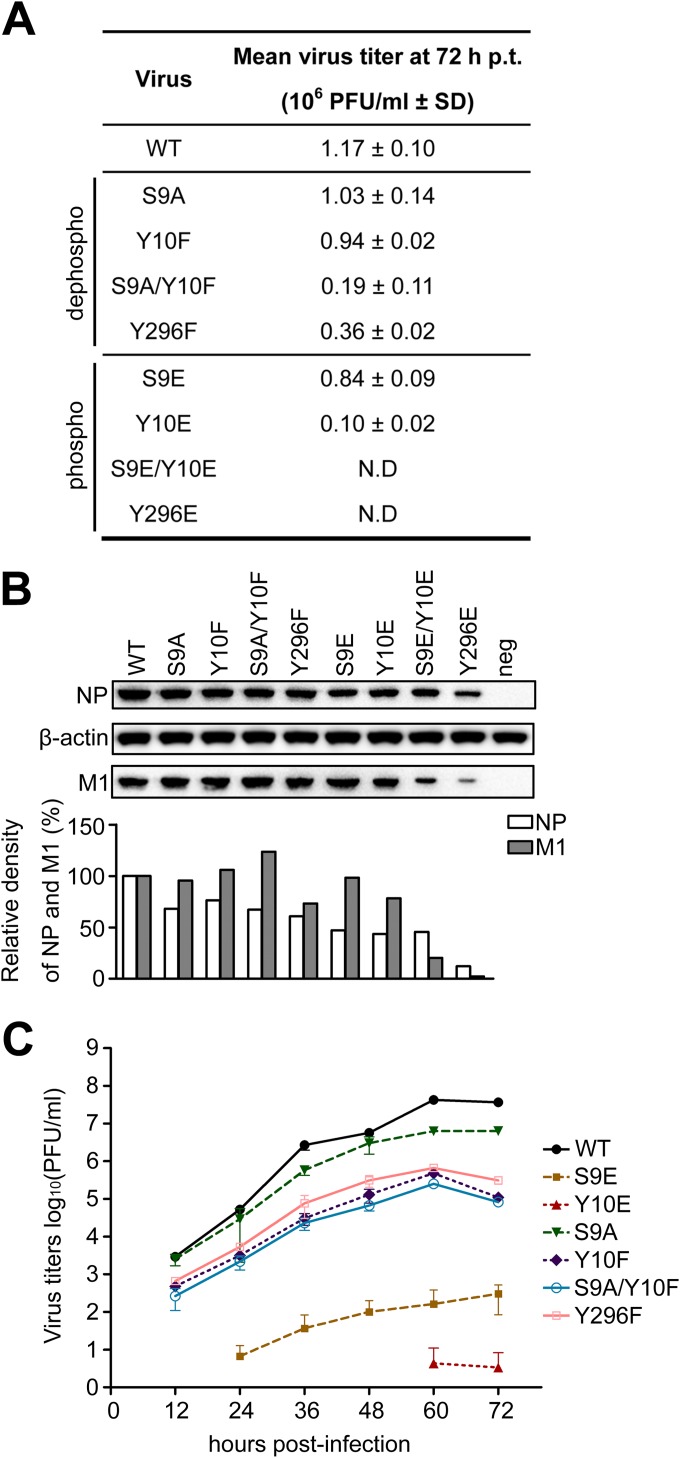
To elucidate the effects of these three phosphorylation sites on influenza A virus rescue, they were mutated in both the pCDNA-NP and pPolI-NP plasmids of the 12-plasmid reverse genetic system, as follows: we introduced an alanine (A) substitution (at S) or a phenylalanine (F) substitution (at Y) to mimic the dephosphorylated residues, while acidic amino acid substitutions (E) mimicked the phosphorylated residues. These plasmids were then transfected into 293T cells as part of the reverse genetic system. At 72 h posttransfection (p.t.), the rescued virus in the supernatant was titrated on MDCK cells. We tried to generate the mutant viruses in five independent attempts, and in each assay, the WT virus was successfully generated. The experiments were divided into two groups: the dephospho-mimetic group and the phospho-mimetic group. The data indicated that the titers of the S9A and Y10F mutants were similar to that of the WT in the dephospho-mimetic group, but the titer of the S9A/Y10F double mutant was 5-fold lower than that of the WT. Notably, the S9E and Y10E phospho-mimetic mutants had virus titers of approximately 80% and 10% that of the WT, respectively, while the double S9E/Y10E mutant failed to generate recombinant viruses. In the case of the Y296 residue, the Y296F mutant reduced the virus titers to approximately 35% of the WT level, while the Y296E mutant did not yield any infectious viruses (Fig. 1A). The expression levels of NP and M1 in the above-mentioned transfected cells were also examined by Western blotting, and they were found to be equal for most mutants and the WT, with the exception of the S9E/Y10E and Y296E mutants, which showed reduced NP and M1 protein expression levels (Fig. 1B). The NP and M1 signals from the S9E/Y10E and Y296E mutants were much weaker than those from the WT. These results provide evidence that the phosphorylation of S9, Y10, and Y296 may play an important role in progeny virus generation.
FIG 1.
Effects of S9, Y10, and Y296 phosphorylation on WSN virus growth and replication. (A) A 12-plasmid reverse genetic system was used to rescue recombinant viruses with WT and mutant NPs. At 72 h p.t., the culture supernatants were harvested and subjected to plaque assays on MDCK cells. N.D, failed rescue in five independent virus rescue assays; SD, standard deviation. (B) At 72 h p.t., the transfected cells were lysed for Western analysis. NP and M1 were detected with their respective antibodies. β-Actin was used as a loading control. Quantification of NP and M1 was performed by determining the ratio of NP (or M1) to actin. (C) Recombinant viruses with mutations (S9A, S9E, Y10F, Y10E, S9A/Y10F, and Y296F) were attenuated during multiple-cycle replication in A549 cells.
To determine if the WSN virus could maintain a similar virus replication efficiency without phosphorylation of S9, Y10, and Y296, we measured the multiple-cycle growth of WSN WT and mutant viruses. A549 cells were inoculated with the recombinant WT virus and the mutants (S9A, S9E, Y10F, Y10E, S9A/Y10F, and Y296F) at a multiplicity of infection (MOI) of 0.001. We found that the S9A mutation reduced virus replication 10-fold and that the Y10F mutation reduced it nearly 1,000-fold. These data are consistent with a report by Hutchinson et al. in which the S9A mutation reduced viral titers 10-fold and the Y10A mutation reduced them 100-fold in MDBK cells (14). The growth rates of the S9E and Y10E mutant viruses were greatly attenuated, and the titer of the WT WSN virus at 72 h postinfection (p.i.) was approximately 1,000-fold and 100-fold higher than those of the S9A/Y10F and Y296F mutants, respectively (Fig. 1C). These results indicate that dephospho-mimetic mutations of these three sites attenuated virus replication.
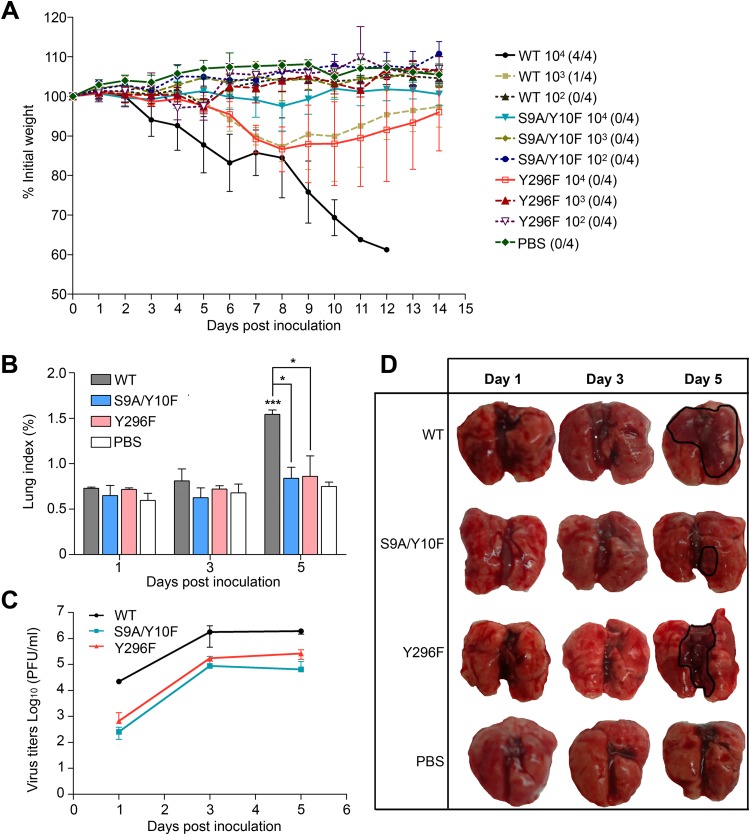
To further assess the effects of phosphorylation on the replication and pathogenicity of the WSN virus in vivo, we inoculated mice with WSN WT and mutant (S9A/Y10F and Y296F) viruses. One hundred percent survival was observed in all mutant-infected groups, compared with 100% lethality in the 104 PFU WT-infected group and 25% lethality in the 103 PFU WT-infected group. For mice in the PBS control, 102 PFU WT-infected, 102 PFU and 103 PFU S9A/Y10F mutant-infected, and 102 PFU and 103 PFU Y296 mutant-infected groups, no loss of body weight was observed. In contrast, mice in the 104 and 103 PFU WT-infected groups rapidly lost weight and succumbed to infection, whereas animals in the 104 PFU Y296 mutant-infected group displayed weight losses similar to those of the 103 PFU WT-infected group. Furthermore, the 104 PFU S9A/Y10F mutant-infected group displayed only a slight peak of weight loss, at 7 to 9 days p.i. (Fig. 2A). We also examined lung samples from infected mice at different days p.i. to determine the lung indexes, virus titers in the lung, and presence of lung lesions, because the WSN virus mainly affects the lungs of infected animals. The lung index of the 104 PFU WT-infected group was ∼2-fold higher than those of the S9A/Y10F and Y296F mutant-infected groups for the same PFU at 5 days p.i. (Fig. 2B). The virus titers in the lungs reached a plateau at 3 days p.i. As expected, virus titers in the WT-infected lungs were 10-fold higher than those with the Y296F mutant and 100-fold higher than those with the S9A/Y10F mutant at 5 days p.i. (Fig. 2C). Furthermore, necropsies indicated significant gross pneumonia lesions at 5 days p.i. in mice infected with each virus, especially the WT WSN virus (Fig. 2D). Together, these data provide evidence that a lack of the possibility of phosphorylation of residues S9, Y10, and Y296 reduces WSN virus replication efficiency and pathogenicity in vivo, which is consistent with our cell culture data.
FIG 2.
Growth of WSN WT and recombinant mutant (S9A/Y10F and Y296F) viruses in a mouse model. (A) Weight curves for specific-pathogen-free mice (n = 4 per group) intranasally infected with 104, 103, or 102 PFU of the indicated viruses. Numbers in parentheses indicate the numbers of mice that succumbed to infection/number of mice per group. The data are the mean body weights for five mice, and the error bars represent standard errors of the means. (B to D) Three mice from each 104 PFU-infected group were euthanized on days 1, 3, and 5 postinfection to determine the lung index and lung virus titer and for necropsy examination. The lung index was calculated as the lung wet weight/body weight × 100. Statistically significant differences between groups were determined using the Student t test. *, P < 0.05; ***, P < 0.001.
Effects of S9, Y10, and Y296 phosphorylation on viral polymerase activity.
Because NP is the major component of the vRNP complex, mutations of several conserved residues in NP result in a loss of polymerase activity (29). We found that the S9E/Y10E and Y296E mutations decreased the expression levels of NP and M1 (Fig. 1B). Therefore, we used a minireplicon system to assess the effects of the S9, Y10, and Y296 phospho-status on the activity of the vRNP complex (30). The luciferase activities from the groups with the dephospho-mimetic S9A, Y10F, and S9A/Y10F mutants were similar to those from groups with the WT virus, whereas the single phospho-mimetic S9E and Y10E mutants displayed slightly reduced levels of luciferase activity. Moreover, the double S9E/Y10E mutant caused a 50% decrease in luciferase activity (Fig. 3A). Remarkably, the Y296E mutant caused a drastic decrease in luciferase activity, while the Y296F mutant displayed only a slight reduction (Fig. 3B). These data demonstrate that phospho-mimetic mutations of S9, Y10, and Y296 affect the polymerase activity.
FIG 3.
Phosphorylation of S9, Y10, and Y296 affects the replication and transcriptional activity of the viral polymerase. (A and B) Activities of vRNPs containing WT NP and NP mutants (S9A/E, Y10F/E, S9A/Y10F, S9E/Y10E, and Y296F/E) in luciferase assays. The effects of WT NP and NP mutants on polymerase activity were determined. Luciferase activity was measured 30 h after the transfection of viral proteins (PB1, PB2, and PA) and the vNS-luc-expressing plasmids. The luciferase activities of mutant NPs were compared with that generated by the WT NP. (C) Production of mRNA, cRNA, and vRNA with WT NP and NP mutants (S9E/Y10E, S9A/Y10F, and Y296E/F) by real-time PCR. All data are means and standard deviations and were determined for three independent experiments. Differences from the WT were tested using one-sample t tests. *, P < 0.05; ***, P < 0.001.
To further determine which steps of viral replication were affected by the mutations of these sites, real-time quantitative PCR was performed to monitor the changes of vRNA, cRNA, and mRNA of the virus. We transfected 293T cells with a 12-plasmid reverse genetic system expressing WT NP or the Y296E, Y296F, S9E/Y10E, or S9A/Y10F mutant. RNAs were extracted from transfected cells at 48 h p.t. and subjected to RT-PCR with specific primers for reverse transcription of M1 mRNA, cRNA, and vRNA. The relative quantities of the three types of RNAs of the M1 gene were assessed by real-time PCR with M1-specific primers. The Y296E mutant displayed remarkably reduced expression levels of all M1 RNA species (Fig. 3C). The above data indicated that both the replication and transcriptional activity of the polymerase were impaired when Y296 was phosphorylated, and consequently, the expression levels of viral proteins were highly decreased (Fig. 1B). The S9E/Y10E mutant reduced the transcription level of viral polymerase but enhanced its replication level (Fig. 3C), which may explain the 50% reduction of luciferase activity in the minireplicon system (Fig. 3A). Nevertheless, the dephospho-mimetic mutations of these three residues contributed to viral replication but had little or no effect on transcriptional activity (Fig. 3C).
Both S9 and Y10 control the nuclear import of NP.
Exogenously expressed NP can shuttle between the cytoplasm and the nucleus, which plays a crucial role in vRNP import and the assembly of virus particles, thus affecting viral growth and viral protein synthesis (4, 31). NP bears two NLSs, which help its transport into the nucleus during the early stage of both viral infection and plasmid transfection. The N-terminal NLS1 plays a dominant role in NP import, and mutations of several residues within this NLS, such as R8A and S3E mutations, cause cytoplasmic accumulation of NP (19, 32). Therefore, we hypothesized that S9 and Y10 would also regulate the nuclear import of NP. To test this hypothesis, the intracellular localization of a series of NP mutants (S9E, Y10E, S9A, Y10F, S9A/Y10F, and S9E/Y10E) was examined by IFAs. All of the NP mutants were expressed in 293T cells, and the cells were fixed at 10 h p.t., a time when WT NP is detected exclusively in the nucleus during transfection (7). Imaging revealed that the cellular localization of the S9A, Y10F, and S9A/Y10F mutants was the same as that of WT NP, but the nuclear (N) fraction of these three mutants was significantly increased relative to that of the WT as determined by cell counting (Fig. 4A). Both the S9E and Y10E mutants displayed cytoplasmic and nuclear localization, but there was more of the Y10E mutant than the S9E mutant in the cytoplasm. As we expected, the S9E/Y10E mutant was completely located in the cytoplasm (Fig. 4A). Furthermore, we extended the sampling time to confirm the effect of phosphorylation of these two residues on the intracellular transport of NP from 4 to 24 h p.t. The S9E/Y10E mutant displayed only cytoplasmic localization at all time points (data not shown). In addition, the results of nuclear and cytoplasmic fractionation experiments also strongly demonstrated that phospho-mimetic mutations of S9 and Y10 reduced the nuclear distribution of NP (Fig. 4B). These results support our hypothesis that constitutive phosphorylations of both S9 and Y10 together prevent nuclear import of NP.
FIG 4.
S9 and Y10 phosphorylation prevents nuclear import of NP. (A) Effects of S9 and Y10 on nuclear import of NP. WT NP and its mutants (S9A/E, Y10E/F, S9A/Y10F, and S9E/Y10E) were expressed in 293T cells. At 10 h p.t., cells were washed and fixed, and the subcellular distribution of the NPs (red) was analyzed by IFAs. At least 600 cells in each group for three independent assays were scored as predominantly nuclear (N), nuclear and cytoplasmic (N+C), or predominantly cytoplasmic (C). The nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole; blue). Differences between each group and the WT were tested using t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.0005. (B) 293T cells were transfected with a plasmid expressing WT NP or the S9E/Y10E mutant and harvested at 20 h p.t. The cells were separated into the nuclear fraction (N) and the cytoplasmic fraction (C). Each fraction was examined by Western blotting, and an anti-NP antibody was used for protein detection. The difference between the N fractions of the WT and the S9E/Y10E mutant from three independent assays was tested using the one-sample t test. **, P < 0.01.
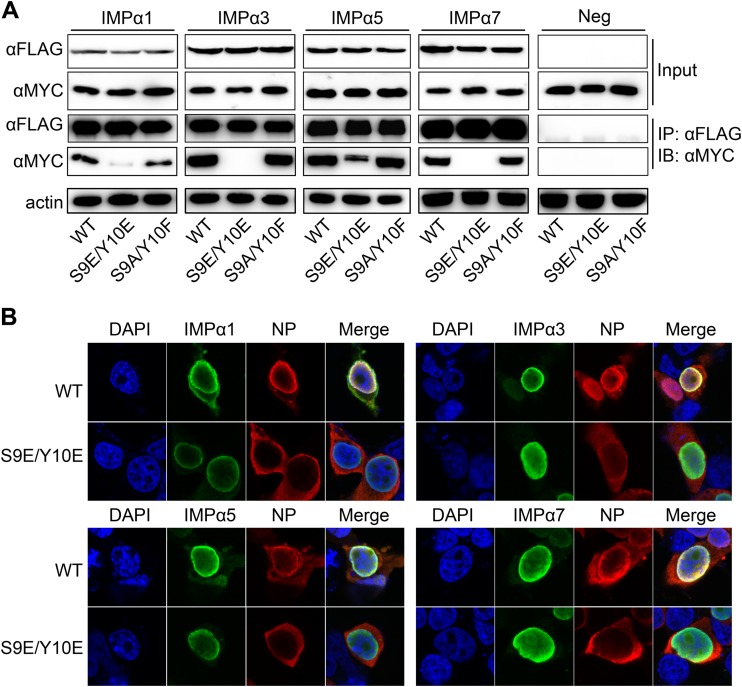
Given that nuclear import of NP is mediated by the cellular import receptors of the importin-α/β family, the S9E/Y10E mutations may inhibit nuclear import by impairing the interaction of NP with importin-α. To investigate the role of the phospho-status of S9 and Y10 in the binding of NP to importin-α and to identify which isoform of importin-α is involved in the NP import regulated by S9 and Y10, we transfected plasmids encoding WT NP or the S9A/Y10F or S9E/Y10E mutant and one of four isoforms of importin-α (α1, α3, α5, and α7) into 293T cells for co-IP assays. WT NP could bind to all four isoforms of importin-α, and the S9A/Y10F mutant displayed the same binding affinity for all importin-α isoforms. Remarkably, the S9E/Y10E mutant displayed weakened binding to all four importin-α receptors (Fig. 5A), indicating that phospho-mimetic mutations of S9 and Y10 inhibit NP import by reducing the association between NP and four importin-α receptors. In contrast, dephospho-mimetic mutation of these two sites recovered the interaction and nuclear import. Moreover, it is noteworthy that small amounts of importin-α1 and -α5 could still bind to the S9E/Y10E mutant. IFAs were performed to visualize the effect of the dissociation of NP from importin-α on the location of NP. 293T cells transfected with WT NP or the S9A/Y10F mutant and different FLAG-importin-α isoforms were fixed at 24 h p.t. The four importin-α receptors were localized mainly in the nucleus and only minimally diffused into the cytoplasm. WT NP displayed colocalization with the four importin-α isoforms in the nuclear membrane. In contrast, the S9E/Y10E mutant displayed only a cytoplasmic localization, even when it was coexpressed with the four importin-α isoforms. Thus, it was obvious that the S9E/Y10E mutant–importin-α association in the nucleus was disrupted (Fig. 5B).
FIG 5.
S9 and Y10 phosphorylation weakens binding of NP to importin-α. (A) Interaction of WT NP and its mutants with importin-α family members. Plasmids encoding WT NP or its mutants appended with a MYC tag were cotransfected with pCDNA3-Flag-importin-α (1/3/5/7) into 293T cells. FLAG-importin-α (1/3/5/7) was immunoprecipitated (IP) with anti-FLAG agarose, and the associated NP was detected by immunoblotting (IB) with the anti-MYC antibody. (B) Effects of phosphorylation of S9 and Y10 on the association of NP and importin-α, as assessed by IFAs. NP and FLAG-importin-α were coexpressed in 293T cells, which were stained with anti-NP (red) and anti-FLAG (green) antibodies. The nucleus was stained with DAPI (blue).
Y296 is crucial for nuclear export of NP.
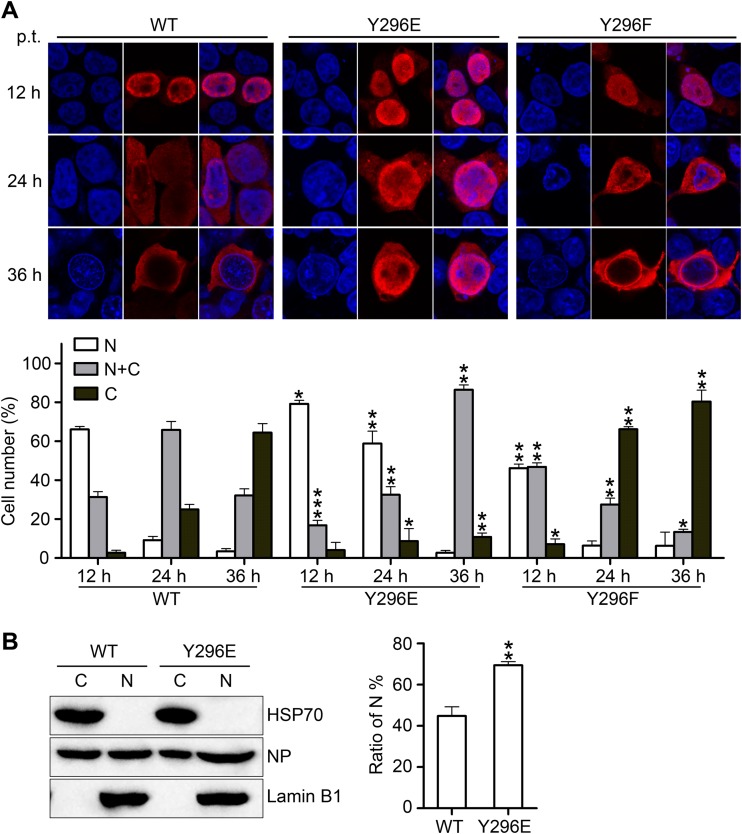
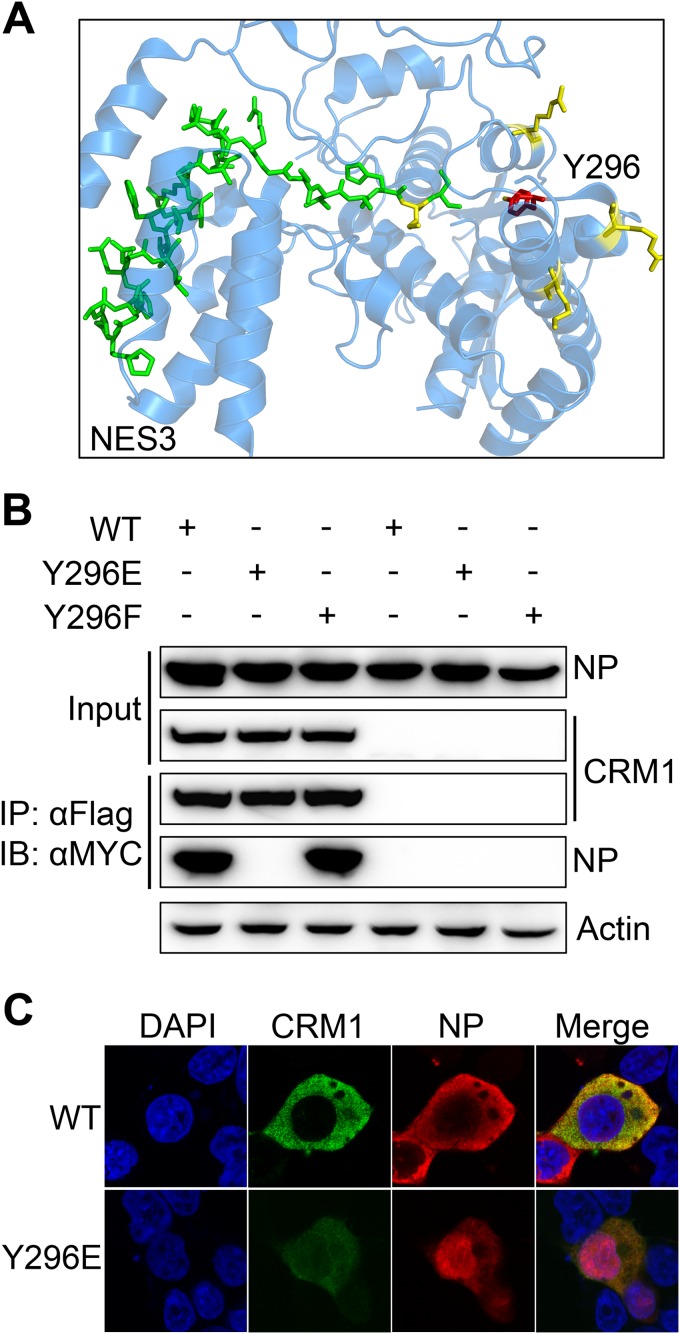
To observe the effect of the phospho-status of Y296 on the intracellular localization of NP, plasmids encoding WT NP or the Y296E or Y296F mutant were transfected into 293T cells. IFAs were then performed to examine the cellular localization of NPs, and the transfected cells were fixed at three time points (12, 24, and 36 h p.t.). WT NP exhibited nuclear localization at 12 h p.t. and a preferentially cytoplasmic localization at 36 h p.t. Notably, we found that the Y296E mutant was located in the nucleus at all time points. In contrast, the Y296F mutant displayed WT-like nuclear shuttling (Fig. 6A). Furthermore, cell counting data clearly demonstrated the nuclear retention caused by the Y296E mutation and the accelerated nuclear export caused by the Y296F mutation. Fractionation experiments were also performed to confirm the cellular localization of NP. Cells transfected with plasmids expressing WT NP or the Y296E mutant were fractionated into nuclear and cytoplasmic groups. The cytoplasmic accumulation of the Y296E NP mutant was significantly decreased compared to that of WT NP (Fig. 6B). These results indicate that phospho-mimetic mutation of Y296 is a key mediator of the nuclear export of NP.
FIG 6.
Phosphorylation of Y296 inhibits the nuclear export of NP. (A) The localization of WT NP and its mutants (Y296E/F) was determined using IFAs. 293T cells were transfected with plasmids expressing mutant NPs or WT NP. At 12, 24, and 36 h p.t., at least 600 cells in each group for three independent assays were scored as predominantly nuclear (N), nuclear and cytoplasmic (N+C), or predominantly cytoplasmic (C). The transfected cells were fixed and stained with anti-NP antibody (red). The nucleus was stained with DAPI (blue). (B) Cells transfected with WT NP and the Y296E mutant were separated into N and C fractions. Each fraction was analyzed by Western blotting using anti-NP antibody, anti-lamin B1 antibody, and anti-HSP70 antibody. Differences between each group and the WT were tested by t tests. *, P < 0.05; **, P < 0.01.
In the NP structure (PDB ID 2IQH) (Fig. 7A), Y296 is located in close proximity to NES3, which functions in the CRM1 transport pathway for nuclear export (9), so we hypothesized that phosphorylation of Y296 might affect the interaction between NP and CRM1 to result in nuclear retention. To test this hypothesis, we transfected both FLAG-CRM1 and MYC-NP (WT, Y296E, or Y296F) expression plasmids into 293T cells and performed co-IP assays. WT NP and the Y296F mutant displayed similar binding affinities for CRM1, whereas the Y296E mutation greatly impaired the interaction between NP and CRM1 (Fig. 7B). These results support the hypothesis that phosphorylation of Y296 blocks nuclear export of NP by inhibiting the binding of NP and CRM1. In line with this, dephosphorylation recovered the interaction. IFAs were also utilized to validate the effect of phosphorylation of Y296 on the colocalization of NP and CRM1. The cells expressing CRM1 and the NPs were fixed and observed at 24 h p.t. We found that WT NP displayed a cytoplasmic distribution when coexpressed with CRM1, unlike the results described above and shown in Fig. 6A. This phenomenon, which was also observed by Elton et al. (13), is likely due to the overexpression of CRM1, which may alter the equilibrium of the nuclear shuttling of NP. In all fields observed, FLAG-CRM1 displayed an obvious cytoplasmic distribution and colocalized with WT NP in the cytoplasm. In the case of the Y296E mutant, the overexpressed CRM1 did not promote the nuclear translocation of the mutant or colocalization with the Y296E mutant. These data indicate that the disassociation of NP and CRM1 caused by phospho-mimetic mutation of Y296 leads to nuclear retention of NP.
FIG 7.
Phosphorylation of Y296 affects the binding of NP to CRM1. (A) Position of Y296 in the NP crystal structure of WSN (PDB ID 2IQH). NES3 is indicated in stick representation (green). Y296 is indicated in red. The basic amino acids around Y296 are also shown, in yellow. (B) Co-IP assays of WT NP and its mutants with CRM1. Plasmids expressing WT NP or NP mutants (Y296E and Y296F) with a MYC tag were cotransfected with plasmid pcDNA3-Flag-CRM1 into 293T cells. FLAG-CRM1 was immunoprecipitated with anti-FLAG agarose, and the associated NP was detected using the anti-MYC antibody. (C) The effect of Y296 phosphorylation on the association of NP and CRM1 was assessed by IFAs. NP and FLAG-CRM1 were coexpressed in 293T cells and stained with anti-NP (red) and anti-FLAG (green) antibodies. The nucleus was stained with DAPI (blue).
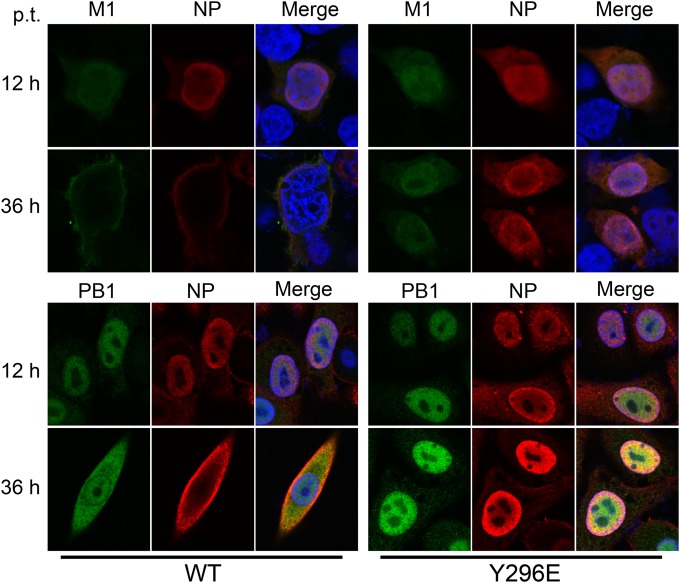
The nuclear export of newly synthesized vRNP complexes is mediated by the CRM1-NEP-M1-vRNP nuclear export complex, and NP also plays a role in mediating this process (7, 13). Because a recombinant virus with the Y296E NP mutant could not be generated, we transfected 293T cells with the 12-plasmid reverse genetic system for WT NP or the Y296E mutant to examine the effect of Y296 phosphorylation on nuclear export of vRNP complexes. IFAs were performed to examine the cellular localization of NP and M1 (PB1), which was representative of vRNP as one of the components of the complex. At all time points, the Y296E mutant was located in the nucleus, in contrast to WT NP (Fig. 8); in the meantime, M1 (PB1) was colocalized with NP. The data indicated that phosphorylation of Y296 retarded the nuclear export of vRNP complexes, consequently reducing the assembly of virus particles and polymerase activity.
FIG 8.
Effect of Y296 phosphorylation on nuclear export of the vRNP complex. 293T cells were transfected with a 12-plasmid reverse genetic system expressing WT NP or the Y296E mutant. At 12 and 36 h p.t., the transfected cells were fixed and stained with anti-NP antibody (red) and anti-M1 (PB1) antibody (green). The nucleus was stained with DAPI (blue).
The phosphorylation states of Y10 and Y296 differ in the early and later stages of virus infection.
We attempted to determine the mechanism by which phosphorylation of S9, Y10, and Y296 occurs during actual virus infection; thus, we performed a previously established Phos-tag SDS-PAGE protocol (15) to enrich the phosphorylated NP, and LC-MS/MS was utilized to detect the phosphorylation sites on NP. S9 was detected to be phosphorylated at 12 h p.i. (Fig. 9). In addition, we performed a pY (phosphorylated tyrosine) detection experiment to examine the phosphorylation state of Y10 and Y296 during infection. We infected A549 cells with WT, S9A/Y10F, or Y296F WSN virus at an MOI of 1. Initially, we observed the cellular localization of the NPs from the WSN WT, S9A/Y10F, and Y296F viruses at 6 and 12 h p.i. (representing the early and later infectious stages, respectively), confirming the nuclear location of NPs at 6 h p.i. and the cytoplasmic location of NPs at 12 h p.i. (Fig. 10A and C). We then treated the infected cells with phosSTOP (a phosphatase inhibitor) for 1 h to accumulate phospho-NPs at 6 and 12 h p.i. The cell lysate was incubated with NP antibody for 1 h and with protein G-agarose for 6 h to enrich NPs, and then the anti-p-Tyr antibody was used to detect the phosphorylated tyrosines of NP. The pY band of the Y296F groups was barely detected, in contrast to those of the WT and S9A/Y10F groups, at 6 h p.i.; at 12 h p.i., the S9A/Y10F signal was weaker than the WT and Y296F signals (Fig. 10B and D). These data suggest that during the early stage of infection, NP Y296 is phosphorylated and Y10 is not. In contrast, during the later stage of infection, Y10 is phosphorylated, and Y296 is dephosphorylated.
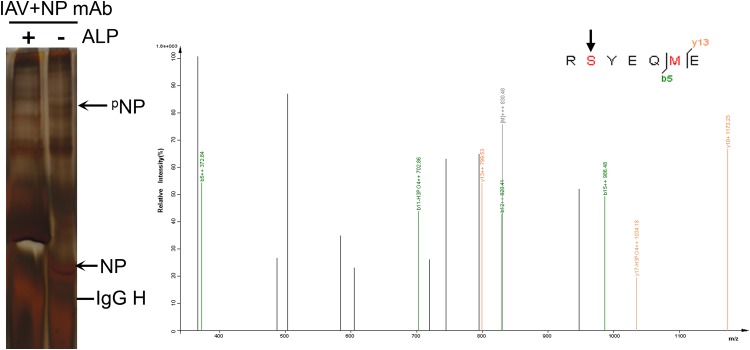
FIG 9.
LC-MS/MS detection of phospho-S9 of NP in the WSN virus. The NP of WSN virus was purified using an anti-NP antibody according to previously described procedures (15). Briefly, lysates of 293T cells infected with WSN virus at an MOI of 3 at 12 h p.i. were incubated with anti-NP antibody and protein G-Sepharose. The immunoprecipitated NP was treated with or without alkaline phosphatase (ALP). An ALP-sensitive band in the Phos-tag SDS-PAGE gel was presumed to be phosphorylated NP and subjected to LC-MS/MS, which showed that the band was indeed NP and revealed that S9 was one of the phosphorylated sites on NP. The identified phosphorylation sites are labeled in red.
FIG 10.
Phosphorylation state of WSN NP (WT and S9A/Y10F and Y296F mutants) at different times postinfection. A549 cells were infected with WT WSN or the S9A/Y10F or Y296F mutant at an MOI of 1. (A and C) Cellular localization of NP at 6 and 12 h p.i. for each group, as visualized by IFAs. (B and D) Phosphorylation sites of WSN NP were different at 6 and 12 h p.i. Lysates of A549 cells infected with viruses at 6 and 12 h p.i. were incubated with NP antibody and protein G-Sepharose. The immunoprecipitated NPs of the WSN WT, S9A/Y10F, and Y296F viruses carrying phosphorylated tyrosine were detected using an anti-p-Tyr antibody. Loss of phosphorylation of Y296 reduced the total tyrosine phosphorylation of NP at 6 h p.i., while loss of phosphorylation of Y10 reduced the total tyrosine phosphorylation of NP at 12 h p.i.
DISCUSSION
In the present study, we observed the effects of phosphorylation and dephosphorylation at residues S9, Y10, and Y296 on the nuclear-cytoplasmic shuttling of NP and the vRNP complex, the polymerase activity of the vRNP complex, and the production of influenza virus particles. We suggest that phosphorylation affects NP trafficking by interfering with the interactions between NP and its nuclear import and export partners.
NP NLS1 has a predominant role in the nuclear import of NP. Mutations within this motif, such as K4/K7R and R8A mutations, cause a loss of NP import ability (6, 33). In addition to phosphorylation, SUMOylation also regulates the cellular trafficking of NP (34). In the present study, phosphorylation of S9 and Y10 reduced the interaction of NP with different isoforms of importin-α. During all phases of transfection, the phospho-mimetic S9E/Y10E mutant was distributed only in the cytoplasm, with a minimal amount imported into the nucleus, which may have been caused by NP NLS2 and the attenuated interaction between NLS1 and importin-α1 and -α5. Moreover, the contribution of NLS1 to RNA replication and transcription is smaller than that of NLS2 (33), perhaps accounting for the polymerase activity being reduced only 50% when the interaction between NLS1 and importin-α was blocked. This result is consistent with that of Ozawa et al. (33), who showed that mutation of NP K7 and R8 to alanine led to cytoplasmic retention and a 50% reduction of RNA polymerase activity of the vRNP complex.
The structure of the N-terminal 13 residues of NP, containing the unconventional NLS1 motif, is still unsolved. Therefore, we do not have structural information regarding these phosphorylation sites on NP and can only hypothesize on the effects of phospho-mimetic or dephospho-mimetic substitutions on NLS1. However, it is known that NP is transported into the nucleus by the classical importin-α/β pathway, which recognizes the typical basic residue-containing NLSs (NP residues K4, K7, and R8 in the case of influenza virus A/WSN/1933) (7, 32). We hypothesize that phosphorylation of S9 and Y10 may change the charge environment of the NLS1 domain and therefore inhibit importin-α recognition. This situation may be similar to that for the phosphorylation of NP S3 (19). Our results also suggest that dephosphorylation of S9 and Y10 is required for nuclear localization and viral amplification.
We also hypothesized a mechanism underlying the phosphorylation of residue Y296 decreasing the binding of NP and CRM1. In our previous work, three NESs were identified as responsible for the nuclear export of NP (9). Among these NESs, NES3 is important for virus growth, because mutation of NES3 results in a failure to rescue mutant virus (35). The position of Y296 in the NP crystal structure (36) is close to that of NES3, suggesting that phosphorylation of Y296 may affect the function of the CRM1-dependent NES3. Phosphorylation of Y296 may attract several positively charged residues to the surrounding area (K31, K273, R293, and R305) (Fig. 6A), changing the conformation of NES3 and disrupting the interaction between NP and CRM1. Therefore, the Y296E mutant cannot yield recombinant virus. However, we know that NES3 is not the only NES that plays a role in NP export, because even though NP accumulates in the nucleus in the Y296E mutant, a large quantity of NP still exists in the cytoplasm.
Here we found that phosphorylation mutations of NP at S9 and Y10 (S9A/Y10A) lead to decreased RNA polymerase activity of the vRNP complex. This is most easily explained by disrupted nuclear import of the NP mutants and the corresponding RNP complex. However, we do not know whether there is any causal relationship between the decreased RNA polymerase activity of the vRNP complex and its nuclear retention in Y296 phosphorylation mutants of NP (Y296E). It seems that nuclear retention gives the vRNP complex more time to synthesize vRNA and mRNA. One possible explanation may be that phosphorylation of NP Y296 inhibits the polymerase complex (PA-PB1-PB2) by direct interaction.
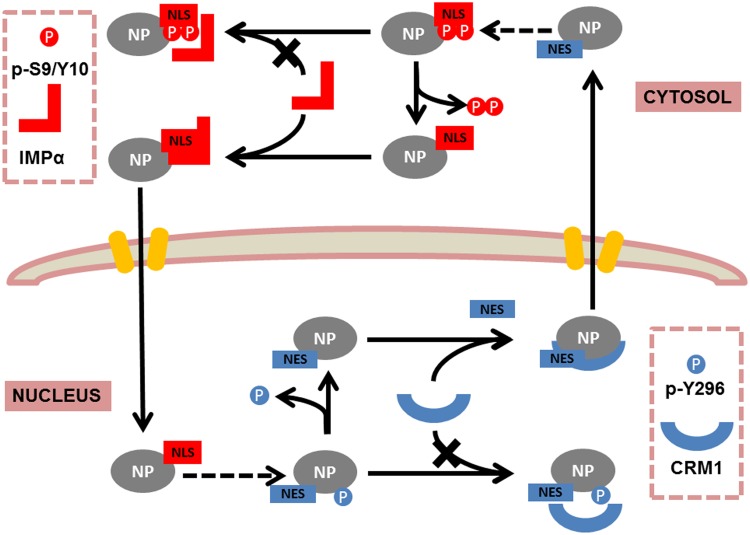
The S9A/Y10F and Y296F mutants displayed a cellular localization similar to that of the WSN WT virus in the immunofluorescence images, but the replication efficiencies of the mutant viruses were attenuated in both cell culture and the animal model. Indeed, their pathogenicity and lethality were especially reduced in mice. We also found that the WT virus displayed an overwhelming advantage when A549 cells were coinfected with these two mutants (data not shown). Until now, there was no obvious evidence to explain this phenomenon. However, increased nuclear retention of the S9A/Y10F mutant and increased cytoplasmic localization of the Y296F mutant compared to the WT were observed in our cell counting data. Thus, we hypothesize that a loss of phosphorylation of these three residues perturbs the well-balanced virus replication process. NP undergoes subtle phosphorylation and dephosphorylation steps during the infectious cycle (37), implying that different phosphorylation sites play various roles during different stages of virus multireplication. Based on our observations, we propose the following model of nuclear-cytoplasmic shuttling of NP mediated by phosphorylation and dephosphorylation: phosphorylation of sites S9, Y10, and Y296 can block (or weaken, in the case of Y296) the import and export of NP, and only dephosphorylation of the three sites can restore transport (Fig. 11). We hypothesize that during the early stage of infection, Y296 is phosphorylated to inhibit nuclear export, and S9 and Y10 are dephosphorylated so that importin-α and importin-β can transport NP into the nucleus. In the later stages of infection, S9 and Y10 are phosphorylated, and the phosphate group on Y296 is removed. These changes lead to decreased nuclear import and increased nuclear export. This model presents a new framework for studying NP trafficking mechanisms in influenza virus infection and identifies targets for anti-influenza agent design. Furthermore, a recently published paper revealed the mechanism for how phosphorylation of S165 controls the self-oligomerization of NP (26), suggesting that phosphorylation may play a role in both NP nuclear trafficking and self-oligomerization, as well as in other aspects of NP function. Multiple sites controlling one function (such as S9 and Y10) or single sites controlling multiple functions (e.g., phosphorylated Y296) may affect polymerase activity by different mechanisms. Such sites in viral proteins and during the viral life cycle will be researched further in our future work.
FIG 11.
Proposed model for nuclear-cytoplasmic shuttling of NP mediated by phosphorylation and dephosphorylation of S9, Y10, and Y296. Phosphorylation of S9 and Y10 inhibits the binding of NP to importin-α, while dephosphorylation of S9 and Y10 restores the binding and thereby increases the nuclear import of NP. Phosphorylation of Y296 reduces the affinity of NP for CRM1, while dephosphorylation of Y296 promotes the interaction and hence facilitates nuclear export.
ACKNOWLEDGMENTS
We thank Shuang Zhang, Xi Liang, and Yun Li for technical support and Joal Haywood for English editing. We also thank Xiaolan Zhang for assistance with confocal microscopy analysis.
This work was supported by grants from the National Key Technologies Research and Development Program of China (grant 2013ZX10004-610), China Ministry of Science and Technology (MOST) Project 973 (grants 2012CB955501 and 2011CB504705), the Key Research Program of the Chinese Academy of Sciences (grant KSZD-EW-Z-005-001), and the National Natural Science Foundation of China (NSFC) (grant 31402216) and by IDRC-APEIR Project funding. G.F.G. and W.L. are principal investigators of the NSFC Innovative Research Group (grant 81321063).
REFERENCES
- 1.Samji T. 2009. Influenza A: understanding the viral life cycle. Yale J Biol Med 82:153–159. [PMC free article] [PubMed] [Google Scholar]
- 2.Portela A, Digard P. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 83:723–734. [DOI] [PubMed] [Google Scholar]
- 3.Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. 2009. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog 5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson EC, Fodor E. 2012. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 30:7353–7358. doi: 10.1016/j.vaccine.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 5.Whittaker G, Bui M, Helenius A. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol 70:2743–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Palese P, O'Neill RE. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol 71:1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J Virol 71:9690–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey J, Dimmock NJ, Colman A. 1985. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell 40:667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- 9.Yu M, Liu X, Cao S, Zhao Z, Zhang K, Xie Q, Chen C, Gao S, Bi Y, Sun L, Ye X, Gao GF, Liu W. 2012. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J Virol 86:4970–4980. doi: 10.1128/JVI.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Digard P, Elton D, Bishop K, Medcalf E, Weeds A, Pope B. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J Virol 73:2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber F, Kochs G, Gruber S, Haller O. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 12.O'Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. 1995. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem 270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 13.Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol 75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson EC, Denham EM, Thomas B, Trudgian DC, Hester SS, Ridlova G, York A, Turrell L, Fodor E. 2012. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog 8:e1002993. doi: 10.1371/journal.ppat.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Zhao Z, Bi Y, Sun L, Liu X, Liu W. 2013. Tyrosine 132 phosphorylation of influenza A virus M1 protein is crucial for virus replication by controlling the nuclear import of M1. J Virol 87:6182–6191. doi: 10.1128/JVI.03024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiang TY, Zhou L, Krug RM. 2012. Roles of the phosphorylation of specific serines and threonines in the NS1 protein of human influenza A viruses. J Virol 86:10370–10376. doi: 10.1128/JVI.00732-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrese M, Portela A. 1996. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J Virol 70:3385–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitzner D, Dudek SE, Studtrucker N, Anhlan D, Mazur I, Wissing J, Jansch L, Wixler L, Bruns K, Sharma A, Wray V, Henklein P, Ludwig S, Schubert U. 2009. Phosphorylation of the influenza A virus protein PB1-F2 by PKC is crucial for apoptosis promoting functions in monocytes. Cell Microbiol 11:1502–1516. doi: 10.1111/j.1462-5822.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 19.Bullido R, Gomez-Puertas P, Albo C, Portela A. 2000. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J Gen Virol 81:135–142. [DOI] [PubMed] [Google Scholar]
- 20.Bui M, Myers JE, Whittaker GR. 2002. Nucleo-cytoplasmic localization of influenza virus nucleoprotein depends on cell density and phosphorylation. Virus Res 84:37–44. doi: 10.1016/S0168-1702(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 21.Kumar N, Liang Y, Parslow TG, Liang Y. 2011. Receptor tyrosine kinase inhibitors block multiple steps of influenza A virus replication. J Virol 85:2818–2827. doi: 10.1128/JVI.01969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol 3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa M, Ochiai H, Nakajima K, Niwayama S. 1990. Inhibitory effect of protein kinase C inhibitor on the replication of influenza type A virus. J Gen Virol 71:2149–2155. doi: 10.1099/0022-1317-71-9-2149. [DOI] [PubMed] [Google Scholar]
- 24.Root CN, Wills EG, McNair LL, Whittaker GR. 2000. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J Gen Virol 81:2697–2705. [DOI] [PubMed] [Google Scholar]
- 25.Chenavas S, Estrozi LF, Slama-Schwok A, Delmas B, Di Primo C, Baudin F, Li X, Crepin T, Ruigrok RW. 2013. Monomeric nucleoprotein of influenza A virus. PLoS Pathog 9:e1003275. doi: 10.1371/journal.ppat.1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turrell L, Hutchinson EC, Vreede FT, Fodor E. 2015. Regulation of influenza A virus nucleoprotein oligomerization by phosphorylation. J Virol 89:1452–1455. doi: 10.1128/JVI.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Sun L, Yu M, Wang Z, Xu C, Xue Q, Zhang K, Ye X, Kitamura Y, Liu W. 2009. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol 11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 28.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Watanabe T, Hatta M, Watanabe S, Nanbo A, Ozawa M, Kakugawa S, Shimojima M, Yamada S, Neumann G, Kawaoka Y. 2009. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J Virol 83:4153–4162. doi: 10.1128/JVI.02642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Liu X, Zhao Z, Xu C, Zhang K, Chen C, Sun L, Gao GF, Ye X, Liu W. 2011. Cyclophilin E functions as a negative regulator to influenza virus replication by impairing the formation of the viral ribonucleoprotein complex. PLoS One 6:e22625. doi: 10.1371/journal.pone.0022625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulo S, Akarsu H, Ruigrok RW, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res 124:12–21. doi: 10.1016/j.virusres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki Y, Hagiwara K, Kakisaka M, Yamada K, Murakami T, Aida Y. 2013. Importin alpha3/Qip1 is involved in multiplication of mutant influenza virus with alanine mutation at amino acid 9 independently of nuclear transport function. PLoS One 8:e55765. doi: 10.1371/journal.pone.0055765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Q, Chang C, Li L, Klenk C, Cheng J, Chen Y, Xia N, Shu Y, Chen Z, Gabriel G, Sun B, Xu K. 2014. Sumoylation of influenza A virus nucleoprotein is essential for intracellular trafficking and virus growth. J Virol 88:9379–9390. doi: 10.1128/JVI.00509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chutiwitoonchai N, Kakisaka M, Yamada K, Aida Y. 2014. Comparative analysis of seven viral nuclear export signals (NESs) reveals the crucial role of nuclear export mediated by the third NES consensus sequence of nucleoprotein (NP) in influenza A virus replication. PLoS One 9:e105081. doi: 10.1371/journal.pone.0105081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Q, Krug RM, Tao YJ. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 37.Kistner O, Muller K, Scholtissek C. 1989. Differential phosphorylation of the nucleoprotein of influenza A viruses. J Gen Virol 70:2421–2431. doi: 10.1099/0022-1317-70-9-2421. [DOI] [PubMed] [Google Scholar]