ABSTRACT
The deep hydrophobic pocket on the N trimer of HIV-1 gp41 has been considered an ideal drug target. On the basis of the M-T hook structure, we recently developed short-peptide-based HIV-1 fusion inhibitors (MTSC22 and HP23), which mainly target the pocket site and possess highly potent antiviral activity. In this study, we focused on investigating their resistance pathways and mechanisms by escape HIV-1 mutants to SC22EK, a template peptide for MTSC22 and HP23. Two substitutions, E49K and N126K, located, respectively, at the N- and C-heptad repeat regions of gp41, were identified as conferring high resistance to the inhibitors targeting the pocket and cross-resistance to enfuvirtide (T20) and sifuvirtide (SFT). The underlying mechanisms of SC22EK-induced resistance include the following: (i) significantly reduced binding affinity of the inhibitors, (ii) dramatically enhanced interaction of the viral six-helix bundle, and (iii)severely damaged functionality of the viral Env complex. Our data have provided important information for the structure-function relationship of gp41 and the structure-activity relationship of viral fusion inhibitors.
IMPORTANCE Enfuvirtide (T20) is the only HIV-1 fusion inhibitor in clinical use, but the problem of resistance significantly limits its use, calling for new strategies or concepts to develop next-generation drugs. On the basis of the M-T hook structure, short-peptide HIV-1 fusion inhibitors specifically targeting the gp41 pocket site exhibit high binding and antiviral activities. Here, we investigated the molecular pathway of HIV-1 resistance to the short inhibitors by selecting and mapping the escape mutants. The key substitutions for resistance and the underlying mechanisms have been finely characterized. The data provide important information for the structure-function relationship of gp41 and its inhibitors and will definitely help our future development of novel drugs that block gp41-dependent fusion.
INTRODUCTION
Entry of human immunodeficiency virus type 1 (HIV-1) into target cells is mediated by its trimeric envelope (Env) glycoprotein, which consists of the gp120 surface subunit and the noncovalently associated gp41 transmembrane subunit (1, 2). In a generally accepted model, gp120 binding to cellular CD4 and a chemokine receptor (CCR5 or CXCR4) triggers a cascade of conformational changes in the Env complex and activates the fusion machinery of gp41. First, the N-terminal hydrophobic fusion peptide of gp41 is released from the gp120 grip and inserts into the cell membrane. Subsequently, its three C-terminal heptad repeats (CHR) fold in an antiparallel orientation onto the trimeric N-terminal repeats (N trimer), resulting in the formation of a hairpin-like six-helix bundle (6-HB) that pulls the viral and cellular membranes together for the fusion reaction (3–5). Prominently, a deep hydrophobic pocket at the basis of the N-trimer groove is docked by several hydrophobic residues from the pocket-binding domain (PBD) of the CHR helix, which stabilizes the 6-HB structure and determines the fusogenic activity of gp41.
Peptides corresponding to the N-terminal heptad repeat (NHR, or N peptide) and CHR (C peptide) regions of gp41 possess potent anti-HIV activities in vitro and in vivo (6, 7). Both the N and C peptides act as decoys to block 6-HB formation in a dominant negative manner during fusion-inducing conformational changes of gp41. The drug T20 (enfuvirtide), a 36-residue peptide derived from the native CHR sequence, is the first and only clinically approved HIV-1 fusion inhibitor; however, drug resistance is a significant clinical problem (8–14). The mutations responsible for T20 resistance are predominantly mapped to the inhibitor-binding sites on the NHR region, particularly within the hydrophobic amino acid Gly36-Leu45 stretch. In addition to primary NHR mutations, some substitutions on CHR also contribute to the resistance as secondary mutations, such as N126K and S138A (15–17). In addition to the low resistance barrier of T20, its use in a clinical setting is limited by several additional factors, which include the requirement for twice-daily injections at high dosage, severe injection site reactions, and the availability of some alternative options with more favorable resistance and convenience profiles. Therefore, a number of next-generation peptide fusion inhibitors with significantly improved pharmaceutical properties have been developed based on the CHR sequence, including sifuvirtide (SFT; 36 residues) (18), TRI-1144 (T2635; 38 residues) (19), and SC34EK (34 residues) (20). In an advanced stage, SFT has been approved for clinical phase III trials in China and will hopefully become the second HIV-1 fusion inhibitor for clinical use. However, SFT has a low genetic barrier to developing drug resistance during in vitro selection, and the resulting HIV-1 mutants display high cross-resistance to T20 (17). Similarly, several other inhibitors, such as T1249 and SC34EK, can easily select drug-resistant viruses with the mutations largely overlapping T20- and SFT-resistant sites (15, 16, 21). Therefore, new strategies and concepts are urgently required to develop next-generation drugs that target the HIV-1 fusion process.
The deep hydrophobic pocket of gp41 is less prone to drug-resistant mutations as the pocket-forming residues are extremely conserved during HIV-1 evolution so that the virus may not tolerate any induced substitutions (22, 23). Unfortunately, short peptides or small molecules that target the pocket site often lack high anti-HIV activity, most likely due to their weak binding affinity (24–29). Our recent findings on the M-T hook structure have provided a new strategy to meet the challenge (30, 31). We demonstrated that two M-T hook residues can dramatically enhance the binding affinity and antiviral activity of C peptides (31–35). On the basis of the M-T hook strategy, we first created a 24-residue peptide inhibitor named MTSC22, which mainly targets the deep pocket rather than the T20- or SFT-resistant site (32). Based on the structure-activity relationship (SAR) of MTSC22, we recently designed a 23-residue peptide named HP23, which exhibited the highest binding stability and the most potent antiviral activity among a panel of peptide fusion inhibitors (35). More promisingly, HP23 had dramatically improved activity to inhibit diverse drug-resistant HIV-1 mutants and a high genetic barrier to developing resistance. In contrast, the template peptide SC22EK, which lacks the M-T hook structure, easily induced high-level HIV-1 resistance in a parallel in vitro selection (33). In this study, we focused on investigating the molecular pathway of SC22EK-induced resistance in order to gain new insights into the structure-activity relationship of HIV-1 fusion inhibitors. Surprisingly, we found that replacement of Glu49 by lysine (E49K), located near the hydrophobic pocket on the N trimer, conferred high resistance to the pocket-targeting short-peptide inhibitors and mediated mild cross-resistance to the first- and next-generation fusion inhibitors, while the N126K substitution in the CHR helix might serve as a second mutation to boost E49K-mediated resistance. The underlying molecular mechanisms were finely characterized.
MATERIALS AND METHODS
Peptide synthesis.
The C peptides including, C34, C34N126K, T20, SFT, SC22EK, MTSC22, MTSC21, MTSC19, HP23, HP23E, and HP22, and the N peptides, including N36 and N36E49K (Fig. 1), were synthesized using a standard solid-phase FMOC (9-fluorenylmethoxy carbonyl) method as described previously (31). All peptides were acetylated at the N terminus and amidated at the C terminus and purified by reversed-phase high-performance liquid chromatography (HPLC). They were verified for purity of >95% and correct amino acid composition by mass spectrometry. Peptide concentrations were determined using UV absorbance and a theoretically calculated molar extinction coefficient ε (280 nm) of 5,500 M−1 · cm−1 and 1.490 M−1 · cm−1, based on the number of tryptophan and tyrosine residues, respectively (36).
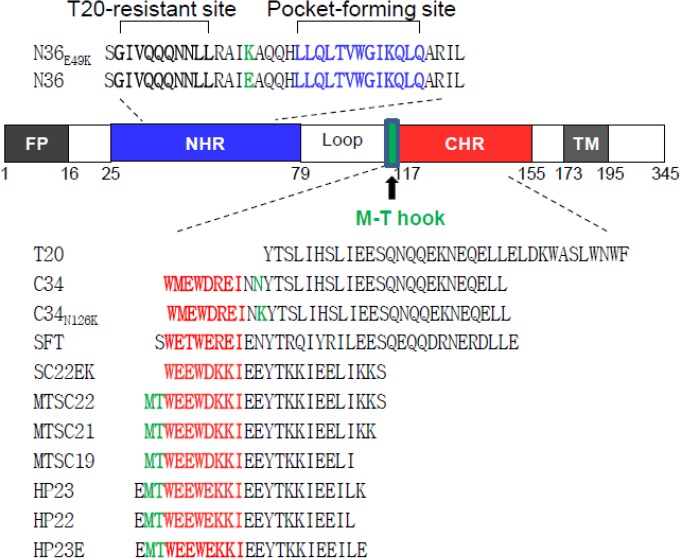
FIG 1.
Schematic illustration of HIV-1 gp41 and peptide fusion inhibitors. The gp41 numbering of HIV-1HXB2 is used. FP, fusion peptide; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; TM, transmembrane domain. The sequences corresponding to the T20-resistant site are highlighted in bold, the sequences corresponding to the NHR pocket region are shown in blue, and the sequences corresponding to the CHR pocket-binding domain (PBD) are shown in red. The position and sequence of the M-T hook structure are shown in green. The peptide inhibitors and their sequences are listed, while the E49K substitution in the N peptide N36 and the N126K substitution in the C peptide C34 are highlighted in green.
Selection of SC22EK-resistant viruses.
The in vitro selection of HIV-1 resistance to the peptide inhibitor SC22EK was performed as described previously (17). Briefly, viral stocks of HIV-1NL4-3 were generated by transfecting 293T cells with an encoding plasmid. MT-4 cells were seeded at 1 × 104 in RPMI 1640 medium containing 10% fetal bovine serum (FBS) on 12-well plates. The virus was used to infect the cells in the presence or absence of diluted SC22EK. Cells were incubated at 37°C with 5% CO2 until an extensive cytopathic effect was observed. Culture supernatants were harvested and used for the next passage on fresh MT-4 cells with a 1.5- to 2-fold increase in peptide concentrations. Cells and supernatant were harvested at regular time points and stored at −80°C.
Site-directed mutagenesis.
HIV-1NL4-3 Env mutants were generated using double-stranded DNA templates and selection of mutants with a restriction enzyme (DpnI) as described previously (37). Briefly, two primers contained the desired mutation and occupied the same starting and ending positions on opposite strands of the plasmid. DNA synthesis was conducted by PCR in a 50-μl reaction volume using 1 ng of denatured plasmid template, 50 pM upper and lower primers, and 5 U of the high-fidelity polymerase PrimeStar (TaKaRa, Dalian, China). PCR amplification was carried out for one cycle of denaturation at 98°C for 5 min, followed by 18 cycles of 98°C for 15 s and 68°C for 15 min, with a final extension at 72°C for 10 min. The amplicons were treated with DpnI for 1 h at 37°C, and DpnI-resistant molecules were recovered by transforming Escherichia coli strain DH5α to antibiotic resistance. The successful mutations were confirmed by sequencing.
Single-cycle infection assay.
HIV-1 entry and its inhibition were determined by single-cycle infection assay as described previously (38). Briefly, HIV-1 pseudovirus was generated by cotransfecting 293T cells with an Env-expressing plasmid and a backbone plasmid, pSG3Δenv, that encodes an Env-defective, luciferase-expressing HIV-1 genome. Supernatants were harvested 48 h after transfection, and 50% tissue culture infectious doses (TCID50) were determined in TZM-bl cells. Peptides were prepared in 3-fold dilutions and mixed with 100 TCID50 of viruses and then incubated for 1 h at room temperature. The mixture was added to TZM-bl cells (104 cells/well) and incubated for 48 h at 37°C. The luciferase activity was measured using luciferase assay reagents and a luminescence counter (Promega, Madison, Wisconsin, USA).
CD spectroscopy.
Circular dichroism (CD) spectroscopy was performed according to our previously described protocols (39). Briefly, a CHR peptide was incubated with an equal molar concentration of N36 or its mutant (N36E49K) at 37°C for 30 min in phosphate-buffered saline (PBS; pH 7.2). CD spectra were acquired on a Jasco spectropolarimeter (model J-815) using a 1-nm bandwidth with a 1-nm step resolution from 195 to 260 nm at room temperature. Spectra were corrected by subtraction of a solvent blank. The α-helical content was calculated from the CD signal by dividing the mean residue ellipticity [θ] at 222 nm by the value expected for 100% helix formation (−33,000° · cm2 · dmol−1). Thermal denaturation was performed by monitoring the ellipticity change at 222 nm from 20°C to 98°C at a rate of 1.2°C/min.
Detection of 6-HBs by ELISA.
Two 6-HB conformation-specific monoclonal antibodies (NC-1 and 17C8), which react with the N36 and C34 complex but not the isolated peptides, were described previously (40, 41). To detect the reactivity of 6-HBs with monoclonal antibodies (MAbs), the wells of an enzyme-linked immunosorbent assay (ELISA) plate were coated with the isolated or mixed peptides at 10 μg/ml and blocked by 3% bovine serum albumin (BSA). The anti-6-HB MAb at 5 μg/ml was added to the wells and incubated at 37°C for 1 h. After three washes, the bound antibodies were detected by horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Sigma). The reaction was visualized by addition of 3,3,5,5-tetramethylbenzidine, and the A450 was measured.
RESULTS
Selection of SC22EK-resistant HIV-1 mutants.
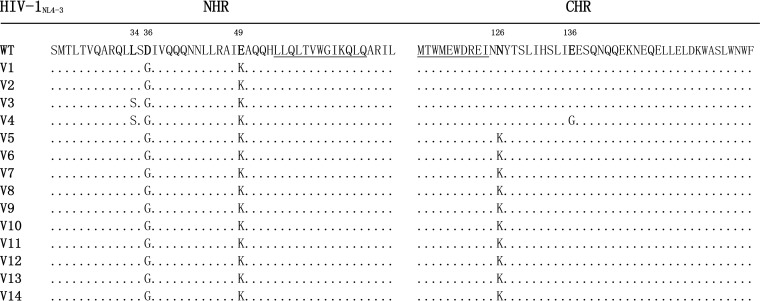
To characterize the structure-function relationship of HIV-1 fusion inhibitors, we previously conducted in vitro selection of escape HIV-1 mutants to the short C peptide SC22EK (33). As shown previously, the concentration of SC22EK could be raised to 8,000 nM after 22 generations of virus passage. To determine the mechanism of SC22EK-induced resistance, we continued to culture the virus for an additional five passages, wherein the peptide concentration was raised as high as 48,000 nM. The entire env genes of resistant viruses were PCR amplified and sequenced. As shown in Fig. 2, all 14 Env clones (V1 to V14) possess D36G and E49K substitutions in the NHR region of gp41. Of them, two Envs (V3 to V4) carry an additional L34S substitution in NHR, and 10 Envs (V5 to V14) carry an N126K substitution in CHR. In addition, the V4 Env clone has an E136G substitution. No consistent substitutions were observed in the other regions of gp41 or in gp120 sequence.
FIG 2.
Alignment of the amino acid sequence of NHR and CHR of HIV-1NL4-3 and its selected mutants. The positions of selected substitutions are in bold, and numbering is according to that of HIV-1HXB2 gp41. The pocket-forming sequence in NHR and the pocket-binding domain in CHR are underlined. WT, wild type.
It is worth noting that the majority of circulating HIV-1 strains carry a glycine at position 36 (Gly36), but the virus HIV-1NL4-3 used in the escape selection harbors an aspartic acid (Asp36), which is known to confer natural resistance to T20. A conversion of D36G has been frequently observed during virus culture even in the absence of peptide inhibitors (10, 15), indicating that it is an HIV-1NL4-3-specific phenotype rather than specifically induced by SC22EK. Second, the N126K substitution readily emerges both in vitro and in vivo, and it is recognized as a secondary mutation.
Resistance profiles of SC22EK-based short-peptide fusion inhibitors.
To characterize the residues responsible for SC22EK resistance, we introduced substitutions into the Env of HIV-1NL4-3 either singly or in combinations, resulting in a panel of 12 mutant Envs (Table 1). The substitutions were verified by DNA sequencing, and the expression of HIV-1 Env glycoprotein was confirmed by Western blot analysis (data not shown). The corresponding HIV-1NL4-3 pseudoviruses were then generated, and their susceptibility to SC22EK was first measured by a single-cycle infection assay. Among the single substitutions, E49K conferred high resistance to SC22EK (21.86-fold), while N126K mediated modest resistance to the inducer (2.52-fold). Apparently, the combined substitution of E49K with N126K or E136G resulted in significant increases in the fold change in resistance to SC22EK, such as the mutants with E49K/N126K (44.74-fold), L34S/D36G/E49K/N126K (51.71-fold), and L34S/D36G/E49K/E136G (52.73-fold). Next, we focused on comparing the resistance profiles of the short-peptide inhibitors MTSC22 and HP23, which were developed using SC22EK as a template and possess high binding and inhibitory activities. As shown in Table 1, the single E49K substitution resulted in 49.25-fold resistance to MTSC22 and 11.24-fold resistance to HP23. Similarly, E49K in combination with N126K or E136G could significantly escalate the level of resistance. In comparison, the D36G and L34S substitutions had little or no contribution to SC22EK-induced resistance. As a control, no characterized HIV-1 mutants were resistant to the viral reverse transcriptase inhibitor zidovudine (AZT) (data not shown).
TABLE 1.
Resistance profiles of HIV-1 mutants to diverse HIV-1 fusion inhibitors
| HIV-1NL4-3 virus or mutant | Resistance to the indicated short-peptide fusion inhibitora |
Cross-resistance to the indicated large fusion inhibitorb |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SC22EK |
MTSC22 |
HP23 |
T20 |
SFT |
C34 |
|||||||
| IC50c | Fold changed | IC50 | Fold change | IC50 | Fold change | IC50 | Fold change | IC50 | Fold change | IC50 | Fold change | |
| Wild type | 50.25 ± 8.4 | 1 | 1.37 ± 0.43 | 1 | 0.45 ± 0.16 | 1 | 66.9 ± 0.16 | 1 | 1.28 ± 0.32 | 1 | 1.09 ± 0.12 | 1 |
| L34S | 58.47 ± 2.02 | 1.12 | 1.69 ± 0.26 | 1.23 | 0.58 ± 0.08 | 1.29 | 228.12 ± 51.5 | 3.41 | 2.55 ± 0.15 | 1.99 | 1.87 ± 0.33 | 1.72 |
| D36G | 48.11 ± 5.82 | 0.96 | 1.4 ± 0.07 | 1.02 | 0.55 ± 0.15 | 1.22 | 2.11 ± 0.2 | 0.03 | 1.14 ± 0.34 | 0.89 | 0.6 ± 0.06 | 0.55 |
| E49K | 1,098.67 ± 2.89 | 21.86 | 67.47 ± 10.18 | 49.25 | 5.06 ± 0.63 | 11.24 | 197.5 ± 13.49 | 2.95 | 3.69 ± 0.48 | 2.88 | 3.11 ± 0.83 | 2.85 |
| N126K | 126.7 ± 3.96 | 2.52 | 2.6 ± 0.09 | 1.9 | 1.43 ± 0.17 | 3.16 | 236.9 ± 15.2 | 3.54 | 2.78 ± 0.16 | 2.17 | 3.19 ± 0.24 | 2.93 |
| L34S/D36G | 85.84 ± 15.2 | 1.71 | 1.83 ± 0.56 | 1.34 | 1.01 ± 0.23 | 2.24 | 38.13 ± 2.67 | 0.57 | 1.95 ± 0.26 | 1.52 | 1.81 ± 0.17 | 1.66 |
| D36G/E49K | 1,032.37 ± 147.63 | 20.54 | 77.18 ± 4.48 | 56.33 | 4.95 ± 0.48 | 10.95 | 10.8 ± 0.42 | 0.16 | 2.58 ± 0.36 | 2.02 | 1.95 ± 0.25 | 1.79 |
| D36G/N126K | 131.1 ± 16.66 | 2.61 | 3.32 ± 0.66 | 2.42 | 1.33 ± 0.11 | 2.96 | 8.71 ± 1.96 | 0.13 | 2.66 ± 0.35 | 2.08 | 2.53 ± 0.19 | 2.32 |
| E49K/N126K | 2,248.17 ± 240.03 | 44.74 | 150.5 ± 3.82 | 109.85 | 14.64 ± 0.51 | 32.53 | 204.2 ± 50.81 | 3.15 | 14.78 ± 1.03 | 11.55 | 19.29 ± 1.01 | 17.69 |
| D36G/E49K/N26K | 1,414.67 ± 141.36 | 28.15 | 157.25 ± 19.77 | 114.78 | 14.68 ± 2.63 | 32.62 | 34.14 ± 2.58 | 0.51 | 10.11 ± 0.57 | 7.9 | 10.6 ± 0.37 | 9.72 |
| L34S/D36G/E49K | 635.89 ± 195.76 | 12.66 | 31.95 ± 2.9 | 23.32 | 3.79 ± 0.35 | 8.42 | 23.5 ± 4.71 | 0.35 | 5.38 ± 0.46 | 4.2 | 2.75 ± 0.15 | 2.52 |
| L34S/D36G/E49K/N126K | 2,598.33 ± 452.81 | 51.71 | 172.13 ± 19.88 | 125.64 | 21.72 ± 1.47 | 48.27 | 99.16 ± 59.42 | 1.67 | 13.82 ± 1.65 | 10.8 | 7.39 ± 2.77 | 6.78 |
| L34S/D36G/E49K/E136G | 2,649.85 ± 154.38 | 52.73 | 177.28 ± 18.34 | 129.4 | 35.58 ± 3.91 | 79.07 | 110.67 ± 11.75 | 1.65 | 23.48 ± 1.46 | 18.34 | 18.86 ± 1.57 | 17.3 |
SC22EK and SC22EK-based short-peptide HIV-1 fusion inhibitors.
T20 is a first-generation HIV-1 fusion inhibitor, SFT is a next-generation inhibitor, and C34 is a template peptide for SFT and many other inhibitors.
IC50, 50% inhibitory concentration.
Fold change in the IC50 was determined relative to the wild-type level.
Cross-resistance to the first- and next-generation inhibitors.
The above results demonstrated that the glutamic acid-to-lysine substitution (E49K) plays a key role in SC22EK-induced resistance and mediates high cross-resistance to the short-peptide-based fusion inhibitors that mainly target the gp41 pocket. We asked whether the E49K substitution and its combinations rendered cross-resistance to the first-generation fusion inhibitor T20 and the next-generation inhibitor SFT. The C peptide C34 was also included in experiments because it was used as a design template for SFT and many other inhibitors. The data are shown in Table 1. As expected, the D36G mutant was highly sensitive to T20 compared to the sensitivity of the wild-type virus, but no significant effect was observed on other inhibitors. Notably, a single substitution (L34S, E49K, or N126K) just produced mild cross-resistance to the three large inhibitors; however, a combination of E49K with N126K or E136G could significantly boost the fold change in resistance against C34 and SFT. Similarly, other combinations had only a marginal role or no role in the observed resistance. Taking these observations together, we concluded that a single E49K substitution can confer high cross-resistance to three short-peptide inhibitors targeting the deep pocket but that significant resistance against the large inhibitors (C34 and SFT) requires its combination with the N126K or E136G substitution.
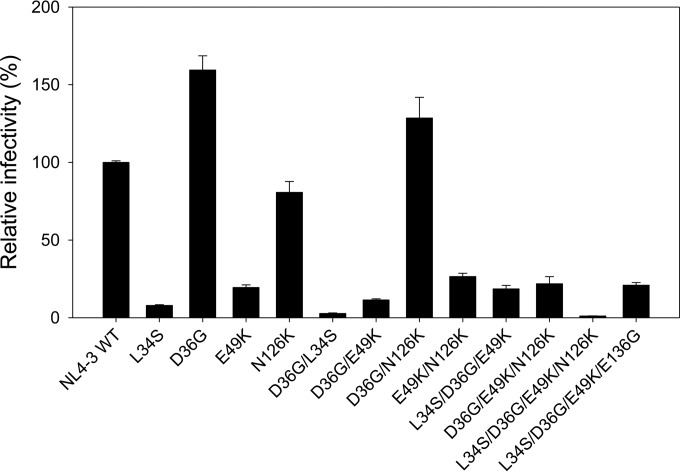
Effects of the SC22EK-induced substitutions on HIV-1 Env-mediated cell entry.
Previous studies demonstrated that HIV-1 resistance to a peptide fusion inhibitor often results in a partial or dramatic loss of function of the target Env protein (15–17). To address whether the acquisition of HIV-1 mutants resistant to SC22EK coincided with a loss of gp41 function, we evaluated the efficiency of viral entry into susceptible target cells in a single-cycle reporter assay (Fig. 3). The infectivity of HIV-1NL4-3 wild-type virus was normalized to 100%, and the relative infectivity of the other mutants was calculated accordingly. It was found that a single substitution of L34S or E49K resulted in a substantial decrease in virus infectivity, while the D36G substitution caused a significant increase, as expected. Consistently, the infectivity of viruses carrying the E49K substitution combined with one or multiple other substitutions was dramatically reduced, with the mutant of L34S/D36G/E49K/N126K showing the lowest entry (1.1%). These data suggested that the residues Leu34 and Glu49 of the NHR helix play a critical role in gp41-mediated HIV-1 fusion and entry.
FIG 3.
Relative infectivity of HIV-1NL4-3 and its mutant viruses. The wild-type HIV-1NL4-3 and mutant pseudoviruses were normalized to a fixed amount by p24 antigen, and viral infectivity was tested in TZM-bl cells using a single-cycle infection assay. The luciferase activity was measured and corrected for background. The luciferase activity of HIV-1NL4-3 was treated as 100%, and the relative infectivity of other mutant viruses was calculated accordingly. Data were derived from the results of three independent experiments and are expressed as means ± standard deviations.
Effects of the E49K substitution on the binding affinity of fusion inhibitors.
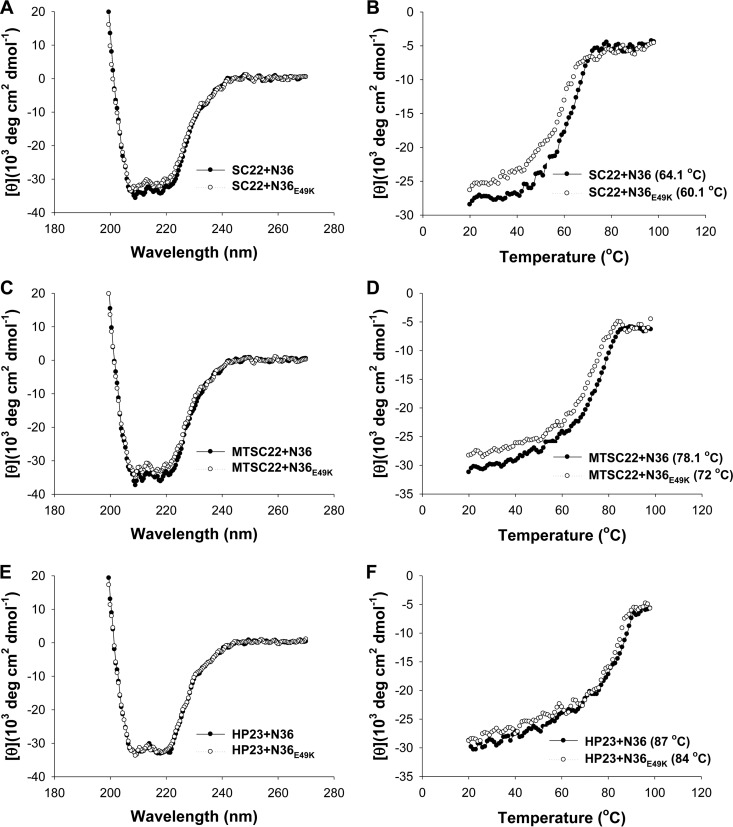
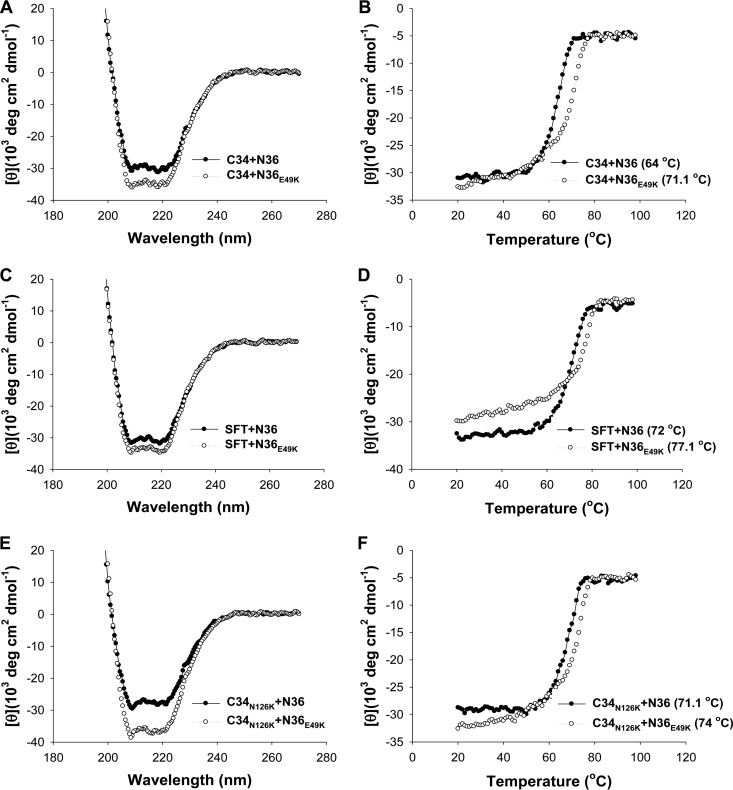
Our above results indicated that the E49K substitution, located near the deep pocket site, plays a key role in SC22EK-induced resistance. To explore the underlying mechanisms, we sought to determine the effect of the E49K substitution on the binding affinity of each inhibitor. To this end, the N peptide N36 with a natural sequence or with an E49K mutation (N36E49K) was synthesized as a target surrogate, and the α-helicity and thermostability of 6-HBs formed by N36 or N36E49K and a C-peptide inhibitor (SC22EK, MTSC22, HP23, C34, or SFT) were measured by CD spectroscopy. The N peptide and C peptide were mixed at equal molar concentrations and incubated at 37°C for 30 min. As shown in Fig. 4 and 5, the CD spectra of all five peptide pairs displayed double minima at 208 and 222 nm, which indicated the formation of α-helical secondary structures. Obviously, the 6-HBs of N36E49K and C34 or SFT exhibited significantly increased α-helicity. The thermostability of each 6-HB, defined as the midpoint of the thermal unfolding transition (Tm) value, was then determined. Compared to the wild-type N36, the mutant peptide N36E49K resulted in significantly decreased Tm values for three short-peptide inhibitors (SC22EK, MTSC22, and HP23), which reflected their reduced binding affinity (Fig. 4). Interestingly, the binding thermostability of C34 and SFT to N36E49K markedly increased compared to the binding to N36 (Fig. 5A to D). The peptides N36 and C34 are, respectively, derived from the NHR and CHR core sequences; thus, the increased binding stability between C34 and N36E49K could be translated into an enhanced interaction between the viral NHR and CHR helices during the fusion process.
FIG 4.
Binding stability of short-peptide inhibitors determined by CD spectroscopy. The α-helicity and thermostability of 6-HBs formed by SC22EK (A and B), MTSC22 (C and D), or HP23 (E and F) with the N peptide N36 or its E49K mutant were measured. The Tm value was defined as the midpoint of the thermal unfolding transition. The final concentration of each peptide in PBS is 10 μM. The experiments were repeated at least three times, and representative data are shown.
FIG 5.
Binding stability of C34, SFT, and a C34 mutant determined by CD spectroscopy. The α-helicity and thermostability of 6-HBs formed by C34 (A and B) or SFT (C and D) with N36 or N36E49K were measured. The Tm value was defined as the midpoint of the thermal unfolding transition. The final concentration of each peptide in PBS is 10 μM. The binding stability of C34 carrying an N126K mutation (C34N126K) with N36 or N36E49K was similarly determined by CD spectroscopy (E and F). The experiments were repeated at least three times, and representative data are shown.
The E49K and N126K substitutions synergistically enhance thermostability of the 6-HB.
The N126K substitution has been widely recognized as a secondary mutation that can enhance the interaction between the NHR and CHR helices (10, 15–17). Here, we found that the N126K substitution can boost E49K-mediated resistance and enhance the interaction of N36 and C34. We were interested to know whether the N126K substitution could further enhance the interaction of the NHR and CHR helices in the context of the E49K substitution. Thus, the C peptide C34 with an N126K substitution (C34N126K) was generated, and the α-helicity and thermostability of N36E49K- and C34N126K-based 6-HBs were determined by CD spectroscopy (Fig. 5E and F). As expected, the 6-HBs of N36 and C34N126K had markedly increased Tm values (71.1°C) relative to the value of the wild-type 6-HBs formed by N36 and C34 (64°C). More interestingly, the combination of two mutant peptides (N36E49K and C34N126K) created a 6-HB with further increased α-helical content and thermostability (74°C). Thus, two substitutions contributed a synergistic effect to enhance the interaction of NHR and CHR helices, which might change the fusion kinetics of the virus.
The E49K and N126K substitutions interfere with the conformation of the 6-HB.
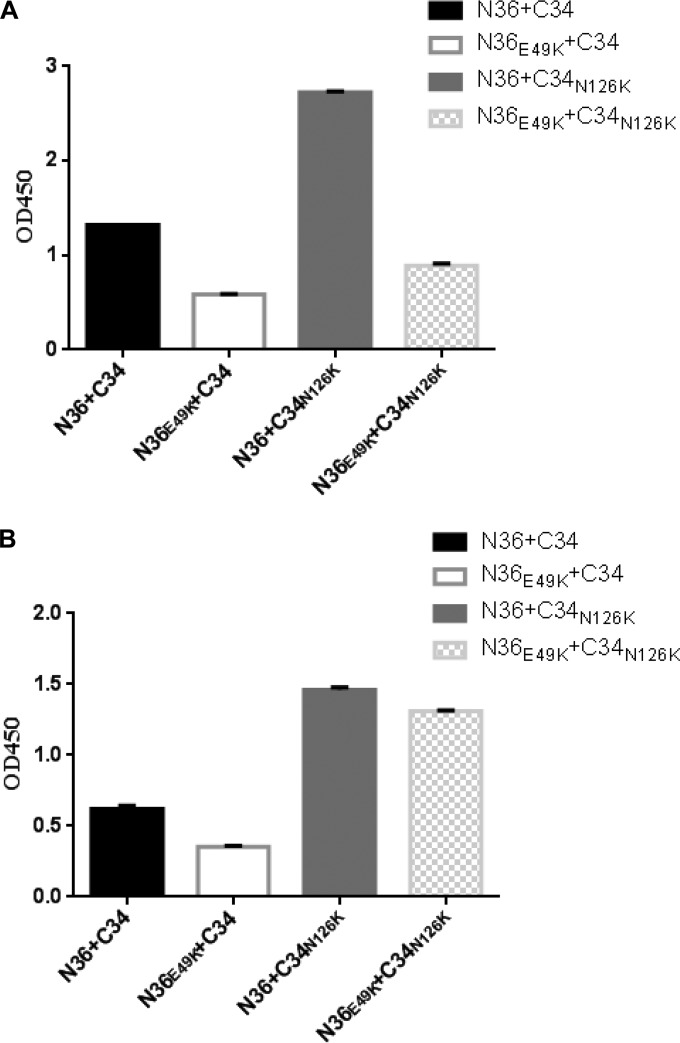
The significantly increased thermostability of 6-HBs formed by N36E49K and/or C34N126K peptides implied that two substitutions might affect the conformation of the 6-HB structure. To prove this hypothesis, an N peptide and a C peptide were mixed at equal concentrations and coated on the ELISA plate wells; then, two conformation-specific antibodies (NC-1 and 17C8) were used to detect the 6-HBs formed by the wild-type or mutant peptides. As shown in Fig. 6, two antibodies reacted strongly with the 6-HB formed by the wild-type N36 and C34; however, the mutant peptide with the E49K substitution resulted in a greatly decreased reactivity for both antibodies, suggesting a conformational change to the 6-HB structure. Surprisingly, the N126K substitution produced a 6-HB with dramatically increased reactivity in the context of either N36 or N36E49K. Taken together, both the E49K and N126K substitutions can directly or indirectly result in a structural rearrangement for the 6-HB conformation, which can decrease or increase the exposure of antibody-recognized epitopes.
FIG 6.
Effects of the E49K and N126K substitutions on the conformation of 6-HB structure. The reactivity of the 6-HBs formed by N36 and C34 or their mutants with conformation-dependent MAbs NC-1 (A) and 17C8 (B) was tested by ELISA. The peptide mixture was coated on the plate wells at 10 μg/ml, and the final concentration of a tested MAb was 5 μg/ml. Data were derived from three independent experiments and are expressed as means ± standard deviations. OD450, optical density at 450 nm.
Structural basis of E49K-mediated resistance to short-peptide inhibitors.
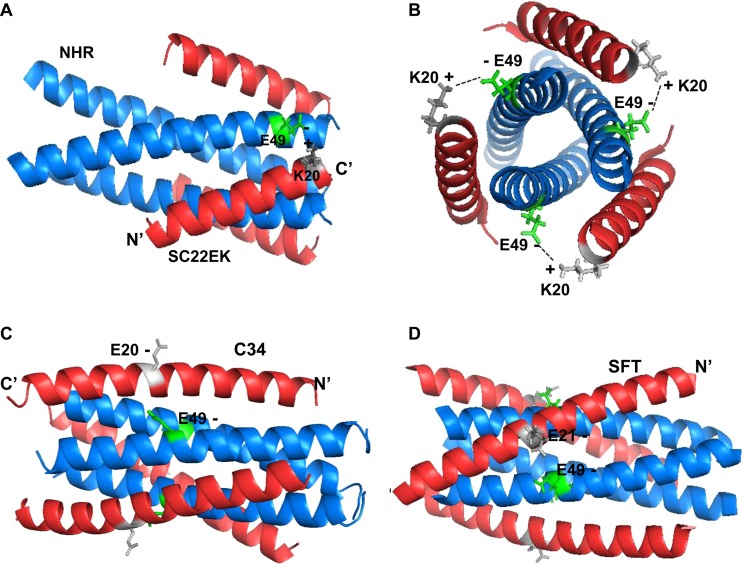
We previously determined the crystal structures of SC22EK and MTSC22 in complexes with a target N peptide (T21), which provided a structural basis to the inhibitors (32). To gain insights into how the E49K substitution impaired the binding of short-peptide-based inhibitors (SC22EK, MTSC22, and HP23) but greatly enhanced the binding of C34 and SFT, we here analyzed the atomic interactions based on the 6-HB structure (Fig. 7). Clearly, the residue Glu49 together with the residues Ala47 and Ile48 is involved in the formation of a hydrophobic patch on an NHR helix, which has extensive interactions with the position 18 to 19 residues (Leu-Ile) of the inhibitors SC22EK and MTSC22. Notably, the negatively charged Glu49 might also mediate a salt bridge interaction with the positively charged lysine at position 20 (Lys20) of the inhibitors. Thus, the E49K substitution disrupts the salt bridge, and an electrostatic repulsion is generated between the inhibitor and targeting NHR helix. On the other hand, the peptides C34 and SFT carry a natural glutamic acid corresponding to the viral CHR sequence. Thus, the E49K substitution might introduce a new salt bridge, which would favor the interaction of endogenous NHR and CHR helices while disfavoring inhibitor binding. Regarding the E49K-mediated mild resistance to C34 and SFT, it is speculated that the enhanced interaction of viral hairpins might have an advantage over the increased binding affinity of such inhibitors.
FIG 7.
Modeling of the E49K-mediated electrostatic interactions on the 6-HB by the program PyMOL. (A) Structural analysis of the E49K substitution-mediated electrostatic repulsion to the short-peptide inhibitors (Protein Data Base identification [ID] code 3VU5), in which a C-terminal lysine at position 20 (K20) of SC22EK may form a salt bridge interaction with glutamic acid at position 49 (E49) of the viral NHR helix. Thus, the E49K substitution would introduce an electrostatic repulsion to the inhibitors. (B) Axial view of the salt bridge between E49 and K20 in the 6-HB. (C) 6-HB structure of N36 and C34 (Protein Data Base ID code 1AIK). Position 20 of peptide C34 is a native glutamic acid (E20) corresponding to the viral CHR sequence. Thus, the E49K substitution might introduce an electrostatic attraction between viral NHR and CHR helices. (D) 6-HB structure of N36 and SFT (Protein Data Base ID code 3VIE). Similarly, position 21 of the SFT inhibitor carries a glutamic acid (E21), similar to the C34 and viral sequence; thus, the E49K substitution would enhance the binding affinity of SFT via an introduced salt bridge interaction.
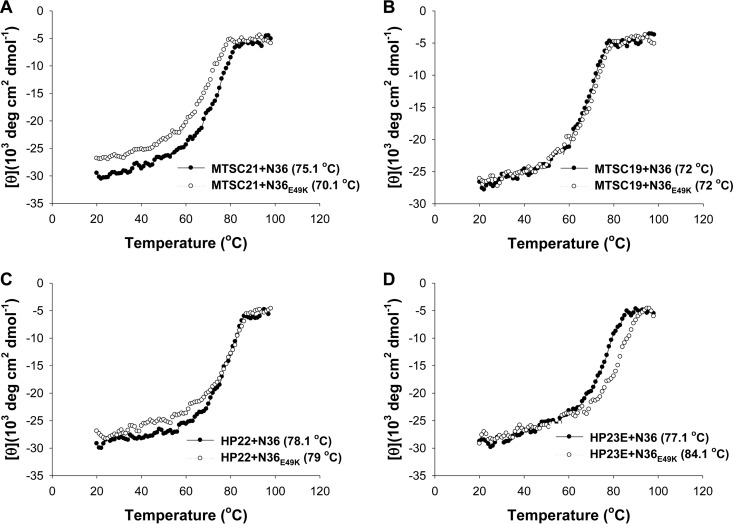
To further verify the results drawn from the structural modeling, we synthesized three additional peptides lacking the C-terminal lysine (Fig. 1). The peptides MTSC19 and HP22 were generated by using MTSC22 and HP23, respectively, as templates, which would prevent electrostatic interactions between the inhibitor and the target site, while the peptide HP23E was created by mutating the lysine to glutamic acid in HP23, which would disrupt the salt bridge with the wild-type target site but establish a salt bridge with the mutant site. In addition, the peptide MTSC21, which lacked the C-terminal serine but retained two lysine residues, was generated as a control. We compared their binding stability with that of N36 and N36E49K by CD spectroscopy (Fig. 8). As expected, the E49K substitution could significantly weaken the binding of MTSC21, similar to its effect on the template MTSC22; however, this substitution did not affect the binding of MTSC19 and HP22 as they interacted with the wild-type and mutant N36 with similar levels of thermostability. More interestingly, HP23E bound to N36 with a sharply decreased thermostability (77.1°C) compared to that of HP23 (87°C), but it bound to N36E49K with a largely restored thermostability (84.1°C), implying an introduced electrostatic interaction between N36E49K and HP23E. These results verified the above structural analysis and suggested that the E49K substitution-mediated electrostatic repulsion might dictate the reduced binding of short-peptide inhibitors, including SC22EK, MTSC22, and HP23, thereby conferring resistance.
FIG 8.
Binding stability of truncated short-peptide inhibitors determined by CD spectroscopy. The thermostability of 6-HBs formed by MTSC21 (A), MTSC19 (B), HP22 (C), or HP23E (D) with N36 and N36E49K was measured. The Tm value was defined as the midpoint of the thermal unfolding transition. The final concentration of each peptide in PBS is 10 μM. The experiments were repeated at least three times, and representative data are shown.
DISCUSSION
Since its discovery, the deep hydrophobic pocket on the N-trimer groove of HIV-1 gp41 has been considered an attractive drug target because of its high degree of conservation, poor tolerance to substitution, and essential role in viral entry (22–24). Indeed, the first-generation drug T20 that binds to the NHR groove just N-terminal to the pocket is significantly more susceptible to resistance mutations than next-generation C-peptide inhibitors containing the pocket-binding sequence (6, 7, 26, 42, 43). The electrostatically constrained short C peptide SC22EK mainly targets the pocket site, but it has relatively low inhibitory activity (44); however, the M-T hook structure-modified SC22EK (MTSC22 and HP23) has strikingly improved binding and inhibitory profiles, especially against diverse drug-resistant HIV-1 mutants (32, 35). Promisingly, MTSC22 and HP23 possess dramatically increased genetic barriers to the development of drug resistance (33–35). In this study, we have finely characterized the resistance pathway and mechanisms of short-peptide-based pocket-targeting inhibitors by the selection of HIV-1 escape mutants. Our studies have demonstrated that the glutamine acid-to-lysine substitution at position 49 of the NHR helix (E49K) critically determines the resistance profiles of SC22EK-based inhibitors and that the asparagine-to-lysine substitution at position 126 of the CHR helix (N126K) can synergistically enhance the resistance phenotype. We have further demonstrated that the underlying mechanisms of SC22EK-induced resistance include the following: (i) significantly reduced binding affinity of the inhibitors, (ii) dramatically enhanced interaction of viral NHR and CHR helices, and (iii) impaired function of the HIV-1 Env protein. These data provide new insights into the structure-function relationship of the gp41 fusion protein and the structure-activity relationship of its fusion inhibitors. The presented information will definitely help our development of next-generation HIV-1 fusion inhibitors, especially of short peptides or small molecules that can specifically target the deep-pocket site.
The in vitro selection of HIV-1 resistance to the first- and next-generation peptide fusion inhibitors has identified a number of substitutions in gp41, which are largely located at the inhibitor-binding sites on the NHR region (15–17, 21, 45). For example, the resistance to T20 and SFT has been predominantly mapped to amino acids 36 to 45, with the GIV motif being a hot spot (17, 45). The resistance to T2635 and SC34EK has been mapped to multiple sites outside the NHR region, including the hot substitutions in the gp41 loop and gp120 (15, 16). Actually, the E49K substitution was rarely observed in previous studies in both in vitro and in vivo settings (10, 13, 15–17, 45–47); however, it did emerge during the selection by a short d-peptide inhibitor (PIE7-dimer) that specifically targeted the deep pocket (27). Although the effect of a single substitution was not evaluated, the authors showed that HIV-1HXB2 pseudovirions carrying E49K/V59I double substitutions conferred ∼400-fold resistance to PIE7-dimer. Interestingly, the E49K substitution was also selected by NHR-derived peptides (N44, N36, and IZN36) that might target both NHR and CHR sites (48, 49), but the studies showed that the single E49K substitution mediated only mild resistance. In our case, this substitution repeatedly resulted in an ∼1.5-fold change in resistance to peptide N36 (data not shown). Therefore, we speculate that the E49K substitution, located closely upstream of the deep pocket, might be a specific response to the inhibitors that specifically bind to the pocket region.
On the basis of mutational analysis and molecular docking, Eggink et al. described four resistance mechanisms for peptide HIV-1 fusion inhibitors, including reduced contact, steric obstruction, electrostatic repulsion, and electrostatic attraction (50). Based on the structure of SFT and its resistance mutations, we also proposed several additional mechanisms, such as hydrogen bond disruption and hydrophobic contact disruption (51). Apparently, our present studies demonstrated that the electrostatic interactions may play key roles in the E49K-mediated resistance pathway. The electrostatic repulsion would occur between a short-peptide inhibitor and the NHR target, resulting in significantly increased binding affinity, while the electrostatic attraction can occur between the viral NHR and CHR (i.e., 6-HB) that greatly enhances the ability of virus to compete against the inhibitors. A body of evidence has already indicated that the bundle-stabilizing mutations can accelerate fusion kinetics and thus decrease the time frame for inhibitor interference (50, 52, 53). Furthermore, HIV-1 resistance to drugs often results in a partial or dramatic loss of the target protein, as described for reverse transcriptase, protease, and Env proteins (15–17). This is particularly the case for gp41-dependent peptide fusion inhibitors (16, 17, 21, 54). Recently, Eggink et al. demonstrated a strong inverse correlation between the level of T2635-induced resistance and viral activity in cell entry (16). Our data have also demonstrated that the E49K mutant virus has strikingly decreased infectivity compared with that of the wild-type virus (Fig. 3), further supporting the impaired Env function being a critical factor in resistance.
The N126K substitution, which is located near the pocket-binding domain (PBD) of the CHR helix, was quite predominant among HIV-1 variants. Actually, this substitution has been frequently observed during escape selection against diverse C-peptide inhibitors (10, 15–17, 21, 46) and in prolonged T20-containing clinical therapy (11–14, 52, 55, 56). Similar to the observed S138A and E154K substitutions (52, 57), the N126K substitution has been considered a secondary mutation that can compensate the delayed fusion kinetics caused by primary resistance substitutions. Several studies also described an improved thermal stability and/or improved free energy of the 6-HB with the secondary substitutions in the CHR region (8, 53, 58, 59). Interestingly, the removal of the glycan at position 126 can also enhance the thermal stability of the NHR-CHR complex (60). In this study, we found again that a single N126K substitution could render the virus mildly resistant to diverse inhibitors (∼3-fold), which emphasized that the mechanism of N126K-mediated resistance is an enhanced viral 6-HB rather than direct binding. Our biophysical data indicated that the N126K substitution did enhance the interaction between the NHR and CHR helices strikingly. Notably, the N126K substitution could further enhance the thermal stability of the 6-HB in the presence of the E49K substitution. In other words, two substitutions might synergistically fortify the interactions of the endogenous NHR and CHR helices, which would favor the virus to compete against the inhibitors. Further, our studies with two conformation-dependent antibodies have also revealed that both the E49K and N126K substitutions may significantly rearrange the conformation of the 6-HB structure. It is conceivable that structural changes, binding enhancement, and viral fusion kinetics are highly coordinated for resistance. In an evolutionary setting, the virus would like to find a balance between the resistance, the viral fitness, and the ease of emergence of the codon change required for escape. Combined, these data expand our knowledge from different angles to understand the mechanisms of HIV-1 resistance to the fusion inhibitors and will guide our future design of next-generation drugs that specifically target the gp41 pocket.
ACKNOWLEDGMENTS
We thank Shibo Jiang at the Lindsley F. Kimball Research Institute of New York Blood Center (New York, NY, USA) for providing the 6-HB-specific MAb NC-1 and Yinghua Chen at the College of Life Science of Tsinghua University (Beijing, China) for providing the 6-HB-specific MAb 17C8.
This work was supported by grants from the National Science Foundation of China (81473255, 81271830, and 81371809).
REFERENCES
- 1.Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 2.Colman PM, Lawrence MC. 2003. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol 4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 3.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 4.Tan K, Liu J, Wang J, Shen S, Lu M. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci U S A 94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 6.Eggink D, Berkhout B, Sanders RW. 2010. Inhibition of HIV-1 by fusion inhibitors. Curr Pharm Des 16:3716–3728. doi: 10.2174/138161210794079218. [DOI] [PubMed] [Google Scholar]
- 7.He Y. 2013. Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Curr Pharm Des 19:1800–1809. doi: 10.2174/1381612811319100004. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin CE, Sanders RW, Deng Y, Jurriaans S, Lange JM, Lu M, Berkhout B. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J Virol 78:12428–12437. doi: 10.1128/JVI.78.22.12428-12437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg ML, Cammack N. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J Antimicrob Chemother 54:333–340. doi: 10.1093/jac/dkh330. [DOI] [PubMed] [Google Scholar]
- 10.Nameki D, Kodama E, Ikeuchi M, Mabuchi N, Otaka A, Tamamura H, Ohno M, Fujii N, Matsuoka M. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J Virol 79:764–770. doi: 10.1128/JVI.79.2.764-770.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu L, Pozniak A, Wildfire A, Stanfield-Oakley SA, Mosier SM, Ratcliffe D, Workman J, Joall A, Myers R, Smit E, Cane PA, Greenberg ML, Pillay D. 2005. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob Agents Chemother 49:1113–1119. doi: 10.1128/AAC.49.3.1113-1119.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrera C, Marfil S, Garcia E, Martinez-Picado J, Bonjoch A, Bofill M, Moreno S, Ribera E, Domingo P, Clotet B, Ruiz L. 2006. Genetic evolution of gp41 reveals a highly exclusive relationship between codons 36, 38 and 43 in gp41 under long-term enfuvirtide-containing salvage regimen. AIDS 20:2075–2080. doi: 10.1097/QAD.0b013e3280102377. [DOI] [PubMed] [Google Scholar]
- 13.Ray N, Harrison JE, Blackburn LA, Martin JN, Deeks SG, Doms RW. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J Virol 81:3240–3250. doi: 10.1128/JVI.02413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svicher V, Aquaro S, D'Arrigo R, Artese A, Dimonte S, Alcaro S, Santoro MM, Di Perri G, Caputo SL, Bellagamba R, Zaccarelli M, Visco-Comandini U, Antinori A, Narciso P, Ceccherini-Silberstein F, Perno CF. 2008. Specific enfuvirtide-associated mutational pathways in HIV-1 Gp41 are significantly correlated with an increase in CD4+ cell count, despite virological failure. J Infect Dis 197:1408–1418. doi: 10.1086/587693. [DOI] [PubMed] [Google Scholar]
- 15.Shimura K, Nameki D, Kajiwara K, Watanabe K, Sakagami Y, Oishi S, Fujii N, Matsuoka M, Sarafianos SG, Kodama EN. 2010. Resistance profiles of novel electrostatically constrained HIV-1 fusion inhibitors. J Biol Chem 285:39471–39480. doi: 10.1074/jbc.M110.145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggink D, Bontjer I, Langedijk JP, Berkhout B, Sanders RW. 2011. Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J Virol 85:10785–10797. doi: 10.1128/JVI.05331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Shan M, Li L, Lu L, Meng S, Chen C, He Y, Jiang S, Zhang L. 2011. In vitro selection and characterization of HIV-1 variants with increased resistance to sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem 286:3277–3287. doi: 10.1074/jbc.M110.199323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, Lu H, Jing W, Jiang S, Zhang L. 2008. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem 283:11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 19.Dwyer JJ, Wilson KL, Davison DK, Freel SA, Seedorff JE, Wring SA, Tvermoes NA, Matthews TJ, Greenberg ML, Delmedico MK. 2007. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc Natl Acad Sci U S A 104:12772–12777. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otaka A, Nakamura M, Nameki D, Kodama E, Uchiyama S, Nakamura S, Nakano H, Tamamura H, Kobayashi Y, Matsuoka M, Fujii N. 2002. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew Chem Int Ed Engl 41:2937–2940. doi:. [DOI] [PubMed] [Google Scholar]
- 21.Eggink D, Baldwin CE, Deng Y, Langedijk JP, Lu M, Sanders RW, Berkhout B. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J Virol 82:6678–6688. doi: 10.1128/JVI.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan DC, Chutkowski CT, Kim PS. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci U S A 95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan DC, Kim PS. 1998. HIV entry and its inhibition. Cell 93:681–684. doi: 10.1016/S0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 24.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. 1999. Inhibiting HIV-1 entry: discovery of d-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103–115. doi: 10.1016/S0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 25.Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. 2007. Potent d-peptide inhibitors of HIV-1 entry. Proc Natl Acad Sci U S A 104:16828–16833. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffen I, Pohlmann S. 2010. Peptide-based inhibitors of the HIV envelope protein and other class I viral fusion proteins. Curr Pharm Des 16:1143–1158. doi: 10.2174/138161210790963751. [DOI] [PubMed] [Google Scholar]
- 27.Welch BD, Francis JN, Redman JS, Paul S, Weinstock MT, Reeves JD, Lie YS, Whitby FG, Eckert DM, Hill CP, Root MJ, Kay MS. 2010. Design of a potent d-peptide HIV-1 entry inhibitor with a strong barrier to resistance. J Virol 84:11235–11244. doi: 10.1128/JVI.01339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu F, Lu L, Liu Q, Yu X, Wang L, He E, Zou P, Du L, Sanders RW, Liu S, Jiang S. 2014. ADS-J1 inhibits HIV-1 infection and membrane fusion by targeting the highly conserved pocket in the gp41 NHR-trimer. Biochim Biophys Acta 1838:1296–1305. doi: 10.1016/j.bbamem.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Jiang S, Lu H, Liu S, Zhao Q, He Y, Debnath AK. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob Agents Chemother 48:4349–4359. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong H, Yao X, Qiu Z, Qin B, Han R, Waltersperger S, Wang M, Cui S, He Y. 2012. Discovery of critical residues for viral entry and inhibition through structural insight of HIV-1 fusion inhibitor CP621-652. J Biol Chem 287:20281–20289. doi: 10.1074/jbc.M112.354126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chong H, Yao X, Sun J, Qiu Z, Zhang M, Waltersperger S, Wang M, Cui S, He Y. 2012. The M-T hook structure is critical for design of HIV-1 fusion inhibitors. J Biol Chem 287:34558–34568. doi: 10.1074/jbc.M112.390393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong H, Yao X, Qiu Z, Sun J, Zhang M, Waltersperger S, Wang M, Liu SL, Cui S, He Y. 2013. Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1. FASEB J 27:1203–1213. doi: 10.1096/fj.12-222547. [DOI] [PubMed] [Google Scholar]
- 33.Chong H, Qiu Z, Sun J, Qiao Y, Li X, He Y. 2014. Two M-T hook residues greatly improve the antiviral activity and resistance profile of the HIV-1 fusion inhibitor SC29EK. Retrovirology 11:40. doi: 10.1186/1742-4690-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong H, Yao X, Qiu Z, Sun J, Qiao Y, Zhang M, Wang M, Cui S, He Y. 2014. The M-T hook structure increases the potency of HIV-1 fusion inhibitor sifuvirtide and overcomes drug resistance. J Antimicrob Chemother 69:2759–2769. doi: 10.1093/jac/dku183. [DOI] [PubMed] [Google Scholar]
- 35.Chong H, Qiu Z, Su Y, Yang L, He Y. 2015. Design of a highly potent HIV-1 fusion inhibitor targeting the gp41 pocket. AIDS 29:13–21. doi: 10.1097/QAD.0000000000000498. [DOI] [PubMed] [Google Scholar]
- 36.Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 37.Chong H, Xu S, Zhang C, Nie J, Wang Y. 2009. Mutation L33M in the HR1 region of HIV-1 gp41 may play a role in T20 resistance. J Clin Virol 45:255–258. doi: 10.1016/j.jcv.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Chong H, Yao X, Zhang C, Cai L, Cui S, Wang Y, He Y. 2012. Biophysical property and broad anti-HIV activity of albuvirtide, a 3-maleimimidopropionic acid-modified peptide fusion inhibitor. PLoS One 7:e32599. doi: 10.1371/journal.pone.0032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y, Liu S, Li J, Lu H, Qi Z, Liu Z, Debnath AK, Jiang S. 2008. Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J Virol 82:11129–11139. doi: 10.1128/JVI.01060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang S, Lin K, Lu M. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol 72:10213–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Chen X, Huang J, Jiang S, Chen YH. 2009. Identification of critical antibody-binding sites in the HIV-1 gp41 six-helix bundle core as potential targets for HIV-1 fusion inhibitors. Immunobiology 214:51–60. doi: 10.1016/j.imbio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Lobritz MA, Ratcliff AN, Arts EJ. 2010. HIV-1 entry, inhibitors, and resistance. Viruses 2:1069–1105. doi: 10.3390/v2051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashkenazi A, Wexler-Cohen Y, Shai Y. 2011. Multifaceted action of Fuzeon as virus-cell membrane fusion inhibitor. Biochim Biophys Acta 1808:2352–2358. doi: 10.1016/j.bbamem.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Naito T, Izumi K, Kodama E, Sakagami Y, Kajiwara K, Nishikawa H, Watanabe K, Sarafianos SG, Oishi S, Fujii N, Matsuoka M. 2009. SC29EK, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Antimicrob Agents Chemother 53:1013–1018. doi: 10.1128/AAC.01211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rimsky LT, Shugars DC, Matthews TJ. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol 72:986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohrengel S, Hermann F, Hagmann I, Oberwinkler H, Scrivano L, Hoffmann C, von Laer D, Dittmar MT. 2005. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J Virol 79:10237–10246. doi: 10.1128/JVI.79.16.10237-10246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mink M, Mosier SM, Janumpalli S, Davison D, Jin L, Melby T, Sista P, Erickson J, Lambert D, Stanfield-Oakley SA, Salgo M, Cammack N, Matthews T, Greenberg ML. 2005. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J Virol 79:12447–12454. doi: 10.1128/JVI.79.19.12447-12454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, De Feo CJ, Zhuang M, Vassell R, Weiss CD. 2011. Selection with a peptide fusion inhibitor corresponding to the first heptad repeat of HIV-1 gp41 identifies two genetic pathways conferring cross-resistance to peptide fusion inhibitors corresponding to the first and second heptad repeats (HR1 and HR2) of gp41. J Virol 85:12929–12938. doi: 10.1128/JVI.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhuang M, Wang W, De Feo CJ, Vassell R, Weiss CD. 2012. Trimeric, coiled-coil extension on peptide fusion inhibitor of HIV-1 influences selection of resistance pathways. J Biol Chem 287:8297–8309. doi: 10.1074/jbc.M111.324483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eggink D, Langedijk JP, Bonvin AM, Deng Y, Lu M, Berkhout B, Sanders RW. 2009. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J Biol Chem 284:26941–26950. doi: 10.1074/jbc.M109.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao X, Chong H, Zhang C, Waltersperger S, Wang M, Cui S, He Y. 2012. Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J Biol Chem 287:6788–6796. doi: 10.1074/jbc.M111.317883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray N, Blackburn LA, Doms RW. 2009. HR-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by HR-1 mutations that cause clinical resistance to enfuvirtide. J Virol 83:2989–2995. doi: 10.1128/JVI.02496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Feo CJ, Wang W, Hsieh ML, Zhuang M, Vassell R, Weiss CD. 2014. Resistance to N-peptide fusion inhibitors correlates with thermodynamic stability of the gp41 six-helix bundle but not HIV entry kinetics. Retrovirology 11:86. doi: 10.1186/s12977-014-0086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berkhout B, Sanders RW. 2011. Molecular strategies to design an escape-proof antiviral therapy. Antiviral Res 92:7–14. doi: 10.1016/j.antiviral.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Loutfy MR, Raboud JM, Montaner JS, Antoniou T, Wynhoven B, Smaill F, Rouleau D, Gill J, Schlech W, Brumme ZL, Mo T, Gough K, Rachlis A, Harrigan PR, Walmsley SL. 2007. Assay of HIV gp41 amino acid sequence to identify baseline variation and mutation development in patients with virologic failure on enfuvirtide. Antiviral Res 75:58–63. doi: 10.1016/j.antiviral.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 56.Poveda E, Rodes B, Labernardiere JL, Benito JM, Toro C, Gonzalez-Lahoz J, Faudon JL, Clavel F, Schapiro J, Soriano V. 2004. Evolution of genotypic and phenotypic resistance to enfuvirtide in HIV-infected patients experiencing prolonged virologic failure. J Med Virol 74:21–28. doi: 10.1002/jmv.20141. [DOI] [PubMed] [Google Scholar]
- 57.Sivaraman V, Zhang L, Meissner EG, Jeffrey JL, Su L. 2009. The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 Env. J Virol 83:11715–11725. doi: 10.1128/JVI.00649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Izumi K, Kodama E, Shimura K, Sakagami Y, Watanabe K, Ito S, Watabe T, Terakawa Y, Nishikawa H, Sarafianos SG, Kitaura K, Oishi S, Fujii N, Matsuoka M. 2009. Design of peptide-based inhibitors for human immunodeficiency virus type 1 strains resistant to T-20. J Biol Chem 284:4914–4920. doi: 10.1074/jbc.M807169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desmezieres E, Gupta N, Vassell R, He Y, Peden K, Sirota L, Yang Z, Wingfield P, Weiss CD. 2005. Human immunodeficiency virus (HIV) gp41 escape mutants: cross-resistance to peptide inhibitors of HIV fusion and altered receptor activation of gp120. J Virol 79:4774–4781. doi: 10.1128/JVI.79.8.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LX, Song H, Liu S, Lu H, Jiang S, Ni J, Li H. 2005. Chemoenzymatic synthesis of HIV-1 gp41 glycopeptides: effects of glycosylation on the anti-HIV activity and alpha-helix bundle-forming ability of peptide C34. Chembiochem 6:1068–1074. doi: 10.1002/cbic.200400440. [DOI] [PubMed] [Google Scholar]