Abstract
Replacement of the herpes simplex virus 1 small capsid protein VP26 phosphorylation site Thr-111 with alanine reduced viral replication and neurovirulence to levels observed with the VP26 null mutation. This mutation reduced VP26 expression and mislocalized VP26 and its binding partner, the major capsid protein VP5, in the nucleus. VP5 mislocalization was also observed with the VP26 null mutation. Thus, we postulate that phosphorylation of VP26 at Thr-111 regulates VP26 function in vitro and in vivo.
TEXT
Viral capsids encase and protect viral genomes, and they enable release of viral genomes into newly infected cells (1). In infected cells, viral capsid proteins also perform important additional roles in regulating viral replicative processes, such as viral gene expression, genome replication, trafficking and maturation of virions, and inhibiting host defenses (i.e., immune response and apoptosis), and in modifying host cellular machinery to establish an environment for effective viral replication, i.e., host cellular signaling and cell cycle control (2–6). The fact that the capsid proteins of various viruses are phosphorylated in infected cells is well established (7–13). It has been reported that phosphorylation of viral capsid proteins regulates both subcellular localization and functions including transcriptional regulation, viral uncoating, and nucleic acid chaperone activity (7–13). These observations hold true in the life cycle of various viruses, including human immunodeficiency virus, hepatitis B virus, hepatitis C virus, and severe acute respiratory syndrome virus (7–13).
Herpes simplex virus 1 (HSV-1), the subject of this study, is one of the best-studied members of the Herpesviridae family (herpesviruses) (14). All herpesvirus capsids, which contain genomic DNA and are able to mature into infectious virus (C capsids), share common structural features: an icosahedral shape with a diameter of 125 nm and consist of 161 capsomers (150 hexons and 11 pentons), a portal complex which has an axial channel through which viral genome DNA enters and exits the capsid, 320 triplexes which connect each of the capsomers and the portal complex, small capsomere-interacting proteins (SCP), and capsid vertex-specific components (CVSC) (15, 16). In HSV-1, the hexons and pentons consist of 6 and 5 molecules of VP5, respectively; the triplexes consist of one VP19C and two VP23 molecules, the portal complex consists of 12 molecules of UL6, the SCP is VP26 (which interacts with each of the VP5 molecules in the hexon), and the CVSC consists of one molecule of UL17 and one of UL25 (15–21). All of these HSV-1 capsid proteins are conserved in the Herpesviridae family, and all have been reported to be phosphorylated in HSV-1-infected cells (22–25). However, there is a lack of information on the significance of phosphorylation in HSV-1 capsid proteins, and those of other herpesviruses, in viral replication and pathogenesis.
Recently, Bell et al. reported that a large-scale, phosphoproteomic analysis of HSV-1-infected murine macrophage BMA3.1A7 cells identified 90 novel phosphorylation sites in 37 HSV-1 proteins (24). These included VP26 threonine 111 (Thr-111), VP19C Thr-110, UL17 Thr-272, and UL25 Thr-383 (24). We previously reported the results of a similar, large-scale, phosphoproteomic analysis of HSV-1-infected human carcinoma HEp-2 cells and, among the 4 phosphorylation sites in HSV-1 capsid proteins identified by Bell et al., we were only able to identify VP26 Thr-111 (25). Therefore, as a first step in clarifying the biological significance of herpesvirus capsid phosphorylation, we focused on an investigation into the effects at VP26 Thr-111.
To investigate the biological significance of VP26 Thr-111 phosphorylation in vitro and in vivo, we constructed recombinant viruses (Fig. 1) as follows. A VP26 null mutant virus, YK490(ΔVP26), in which the UL35 gene encoding VP26 was disrupted by replacing VP26 codons 10 to 102 with a kanamycin resistance gene, was constructed using the Red-mediated mutagenesis procedure. Escherichia coli GS1783 carrying pYEbac102, which contains a full-length infectious HSV-1 clone (26), was used as described previously (27, 28) but with the following primers: 5′-ACCTCCGGTCCCGATGGCCGTCCCGCAATTTCACCGCCCCAGGATGACGACGATAAGTAGGG-3′ and 5′-CTGGGCCTCACGGGGTCCCGGGCGTCGAAGGTTCTCGAACCAACCAATTAACCAATTCTGATTAG-3′. YK491(ΔVP26/EGFP) with the VP26 null mutation and expressing enhanced green fluorescent protein (EGFP) was constructed by coinfection of simian kidney epithelial Vero cells (29) with YK490(ΔVP26) and YK333(EGFP) expressing EGFP (Fig. 1A) as described previously (30). YK333(EGFP) carries an expression cassette consisting of the Egr-1 promoter, the EGFP open reading frame (ORF), and bidirectional polyadenylation signals of the HSV-1 UL21 and UL22 genes in the intergenic region between the UL3 and UL4 genes (31) and shows growth properties identical to those of wild-type HSV-1(F) (32). YK492(VP26T111A/EGFP), with VP26 Thr-111 replaced by alanine (VP26T111A) and expressing EGFP, was constructed by cotransfection of rabbit skin cells with a transfer plasmid, pBS-VP26T111A, and intact YK491(ΔVP26/EGFP) DNA as described previously (29). YK493(VP26Δ/TA-repair/EGFP), a virus in which both YK491(ΔVP26/EGFP) and YK492(VP26T111A/EGFP) are repaired and which expresses EGFP, was constructed by cotransfection of rabbit skin cells with transfer plasmid pBS-VP26-kpn+ and intact YK492(VP26T111A/EGFP) DNA as described previously (29). YK494(VP26T111D/EGFP), in which VP26 Thr-111 is replaced by aspartic acid and which expresses EGFP, was constructed by cotransfection of rabbit skin cells with transfer plasmid pBS-VP26T111D-bam+ and intact YK492(VP26T111A/EGFP) DNA as described previously (29). To enrich YK494(VP26T111D/EGFP), infectious viruses obtained from the cotransfection were passaged two times in human neuroblastoma SK-N-SH cells (33); YK494(VP26T111D/EGFP) grew more efficiently than YK492(VP26T111A/EGFP), as described below (Fig. 1B to D). The infected SK-N-SH cells were harvested and subjected to freeze-thawing and sonication. The cell lysates were diluted and inoculated on Vero cells. Plaques were isolated and screened for the T111D amino acid substitution in VP26 as described previously (29). It is known that an acidic amino acid such as glutamic acid or aspartic acid can mimic the negative charge produced by phosphorylation (34–37). The genotype of each recombinant virus was confirmed by sequencing (data not shown). These recombinant viruses expressing EGFP enabled us to monitor HSV-1-infected cells by using fluorescence microscopy and thus to investigate the ability of the recombinant viruses to form plaques in cases where HSV-1 is unable to produce plaques that are detectable in classical plaque assays (29). pBS-VP26-kpn+, pBS-VP26T111A, and pBS-VP26T111D-bam+, described above, were constructed as follows: pBS-VP26 was constructed by PCR cloning the domain containing the VP26 ORF (the 967-bp upstream sequence flanking the VP26 start codon and the 691-bp downstream sequence flanking the VP26 stop codon), from pYEbac102 into pBluescript II KS(+) (Stratagene). pBS-VP26-kpn+, into which a silent mutation in the wobble base of VP26 glycine-110 within pBS-VP26 was introduced to create a KpnI restriction site, pBS-VP26T111A with the VP26T111A mutation, and pBS-VP26T111D-bam+ with a VP26T111D mutation that created a BamHI restriction site, were constructed as described previously (34) (Fig. 1). The engineered KpnI or BamHI restriction sites were useful for screening recombinant viruses YK493(VP26Δ/TA-repair/EGFP) or YK494(VP26T111D/EGFP), respectively, as described previously (38).
FIG 1.
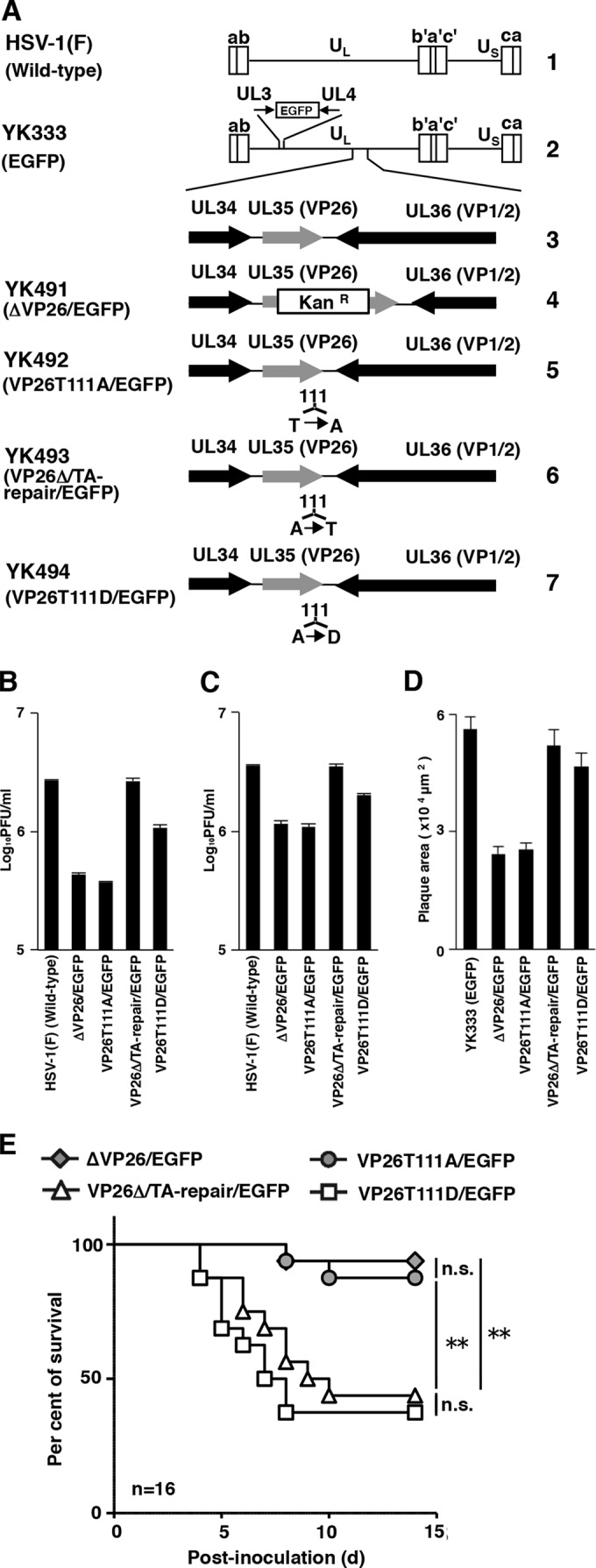
(A) Schematic diagrams of the genome structure of the wild-type and recombinant HSV-1 viruses used in this study. Line 1, the wild-type HSV-1(F) genome; line 2, the wild-type HSV-1(YK333) genome carrying an expression cassette for EGFP in the intergenic region between UL3 and UL4; line 3, the domain carrying the UL34, UL35(VP26), and UL36(VP1/2) open reading frames; lines 4 to 7, genomes of recombinant viruses with a mutation in the UL35(VP26) gene. (B and C) Effects of mutations in VP26 on progeny virus yield in SK-N-SH cells. SK-N-SH cells were infected with each of the indicated viruses at an MOI of 0.01 (B) or MOI of 5 (C). Total virus from the cell culture supernatants and infected cells was harvested at 48 h postinfection (B) or 24 h postinfection (C) and assayed on Vero cells. Each value is the mean ± standard error of triplicate experiments. Data are representative of three independent experiments. (D) Effect of mutations in VP26 on virus plaque diameter. SK-N-SH cells were infected with each of the indicated viruses at an MOI of 0.00001 under plaque assay conditions. The areas of 25 single plaques for each of the indicated viruses were determined 48 h postinfection. Each value is the mean ± standard error of the measured plaque sizes. Data are representative of three independent experiments. (E) Effect of mutations in VP26 on mortality of mice following intracranial inoculation. Sixteen 3-week-old female ICR mice were inoculated with 500 PFU of the indicated viruses intracranially, and survival was monitored for 14 days. Asterisks indicate significant differences: **, P < 0.0083 (0.05/6) by the log-rank test with Bonferroni adjustment for the four comparison analyses; n.s., not significant.
In this study, we investigated the biological significance of VP26 Thr-111 phosphorylation in vitro by using SK-N-SH cells. Since phosphorylation of VP26 Thr-111 has only been detected in HSV-1-infected BMA3.1A7 and HEp-2 cells (24, 25), we examined first whether this phosphorylation did in fact occur in HSV-1-infected SK-N-SH cells via a phosphoproteomic analysis of titanium dioxide affinity chromatography-enriched phosphopeptides from SK-N-SH cells infected with wild-type HSV-1(F) at a multiplicity of infection (MOI) of 5 for 24 h with high-accuracy mass spectrometry as described previously (39, 40). This analysis detected several VP26 phosphopeptides, including phosphorylated Thr-111 (data not shown), thus confirming that VP26 Thr-111 is phosphorylated in HSV-1-infected SK-N-SH cells.
To examine the effect of VP26 Thr-111 phosphorylation on viral replication in cell culture, we analyzed progeny yields in SK-N-SH cells infected with wild-type HSV-1(F), YK491(ΔVP26/EGFP), YK492(VP26T111A/EGFP), YK493(VP26Δ/TA-Repair/EGFP), or YK494(VP26T111D/EGFP) at an MOI of 0.01 for 48 h (Fig. 1B) or at an MOI of 5 for 24 h (Fig. 1C). As shown in Fig. 1B, the progeny virus yield in SK-N-SH cells infected with YK491(ΔVP26/EGFP) at an MOI of 0.01 was 6.2-fold less than that in cells infected with wild-type HSV-1(F). Also, the level of progeny virus yield in cells infected with YK493(VP26Δ/TA-Repair/EGFP) was restored to that in cells infected with wild-type HSV-1(F). Similarly, the progeny virus yields in cells infected with YK492(VP26T111A/EGFP) were 7.1-fold less than that in cells infected with wild-type HSV-1(F) or YK493(VP26Δ/TA-Repair/EGFP) (Fig. 1B). Thus, the T111A mutation in VP26 reduced HSV-1 replication to levels similar to those observed with the VP26 null mutation. Notably, the progeny virus yield in SK-N-SH cells infected with YK494(VP26T111D/EGFP) carrying a phosphomimetic mutation at VP26 Thr-111 was 2.9-fold higher than that in cells infected with YK492(VP26T111A/EGFP) (Fig. 1B). Progeny virus yields in cells infected with YK494(VP26T111D/EGFP), however, were 2.5-fold lower than those in cells infected with wild-type HSV-1(F) (Fig. 1B). Thus, the VP26 T111D mutation, which mimics constitutive phosphorylation at VP26 Thr-111, in part restored the progeny virus yield reduced by the VP26 T111A mutation, which prevents phosphorylation of VP26 Thr-111. Similar results were observed with SK-N-SH cells infected at an MOI of 5 (Fig. 1C).
We also analyzed plaque sizes in SK-N-SH cell monolayers infected with YK333(EGFP), YK491(ΔVP26/EGFP), YK492(VP26T111A/EGFP), YK493(VP26Δ/TA-Repair/EGFP), or YK494(VP26T111D/EGFP) under plaque assay conditions described previously (41). In agreement with the growth properties of these viruses demonstrated in this and previous studies (42), YK491(ΔVP26/EGFP) and YK492(VP26T111A/EGFP) produced plaques similar in size but that were smaller than those observed with YK333(EGFP) and YK493(VP26Δ/TA-Repair/EGFP) (Fig. 1D). The plaque sizes for cells infected with YK494(VP26T111D/EGFP) were restored to those observed in cells infected with YK333(EGFP) or YK493(VP26Δ/TA-Repair/EGFP) (Fig. 1D).
Taken together, these results suggest that phosphorylation of VP26 Thr-111 is required for efficient HSV-1 replication and cell-cell spread in HSV-infected SK-N-SH cells.
To examine the effect of VP26 Thr-111 phosphorylation on HSV-1 pathogenesis, 3-week-old female mice (Charles River Laboratories) were infected intracranially with 500 PFU of YK491(ΔVP26/EGFP), YK492(VP26T111A/EGFP), YK493(VP26Δ/TA-Repair/EGFP), or YK494(VP26T111D/EGFP), as described previously (40, 43), and monitored daily for 14 days. This experiment was carried out in accordance with the Guidelines for Proper Conduct of Animal Experiments, Science Council of Japan, and the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Medical Science, University of Tokyo (IACUC protocol approval number 19-26). As shown in Fig. 1E, survival was significantly higher in mice infected with YK491(ΔVP26/EGFP) or YK492(VP26T111A/EGFP) than in mice infected with YK493(VP26Δ/TA-Repair/EGFP) (Fig. 1E). Notably, the survival of mice infected with YK492(VP26T111A/EGFP) was similar to that of mice infected with YK491(ΔVP26/EGFP). Furthermore, the survival of mice infected with YK494(VP26T111D/EGFP) was significantly lower than that of mice infected with YK492(VP26T111A/EGFP). These results suggest that VP26 and its phosphorylation at Thr-111 are required for efficient viral neurovirulence in mice following intracranial inoculation.
To further clarify the effect of VP26 Thr-111 phosphorylation in HSV-1 replication, SK-N-SH cells infected with wild-type HSV-1(F), YK491(ΔVP26/EGFP), YK492(VP26T111A/EGFP), YK493(VP26Δ/TA-Repair/EGFP), or YK494(VP26T111D/EGFP) at an MOI of 5 were harvested 24 h postinfection and analyzed by immunoblotting with antibodies to VP26 (30) and α-tubulin (DM1A; Sigma). The amount of protein present in immunoblot bands was quantified using the ImageQuant LAS 4000 system with ImageQuant TL7.0 analysis software (GE Healthcare Life Sciences). As shown in Fig. 2A and B, the level of VP26 expression in cells infected with YK492(VP26T111A/EGFP) was significantly lower than that in cells infected with wild-type HSV-1(F) or YK493(VP26Δ/TA-Repair/EGFP). The level of VP26 expression in cells infected with YK494(VP26T111D/EGFP) was significantly higher than that in cells infected with YK492(VP26T111A/EGFP) but lower than that in cells infected with wild-type HSV-1(F) (Fig. 2C and D). Thus, the VP26 T111D mutation in part restored the VP26 expression downregulated by the VP26 T111A mutation.
FIG 2.
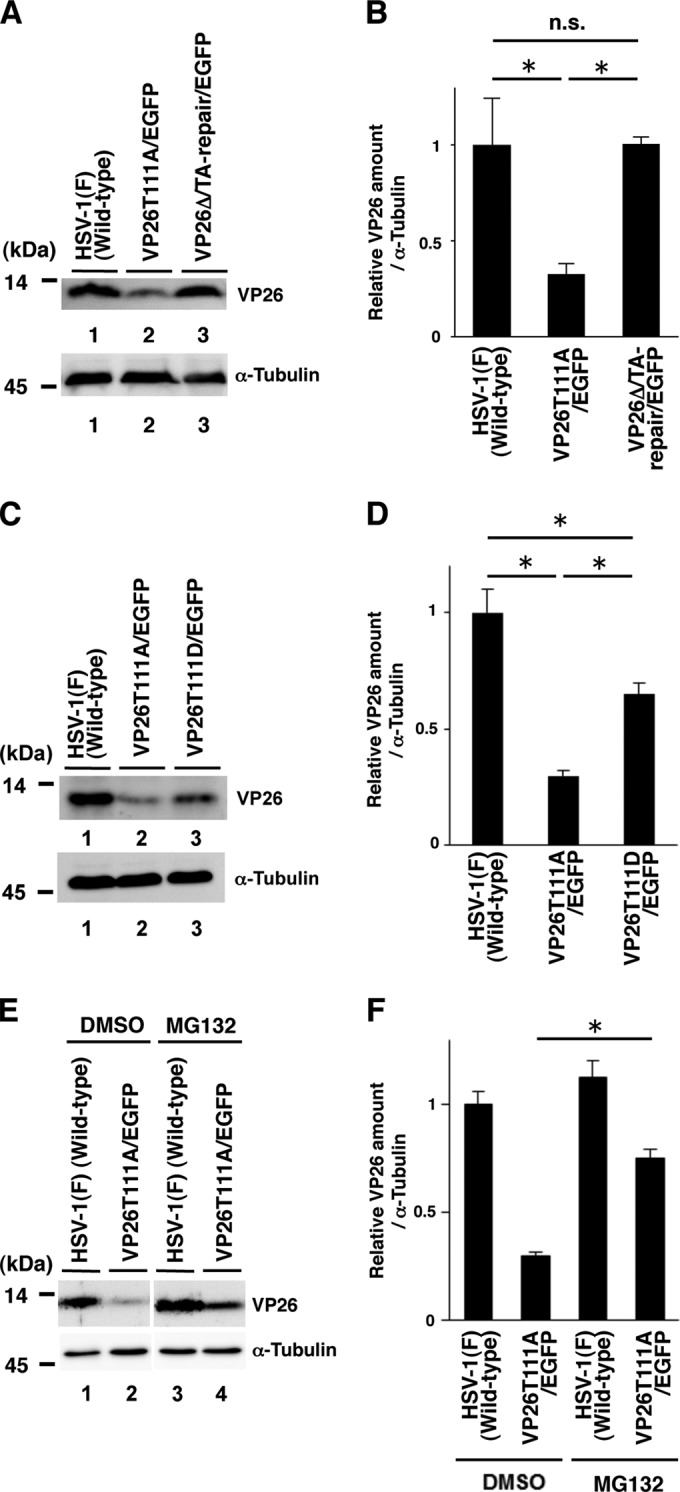
Effects of the mutations in VP26 on its expression in SK-N-SH cells. (A and C) SK-N-SH cells infected with each of the indicated viruses at an MOI of 5 were harvested 24 h postinfection and analyzed by immunoblotting with an antibody to VP26 or α-tubulin. (B and D) The amount of VP26 in the SK-N-SH cells shown in panels A and C (top) relative to those of α-tubulin shown in panels A and C (bottom). Each value is the mean ± standard error of triplicate experiments and is expressed relative to the mean value of wild-type HSV-1(F)-infected SK-N-SH cells, which was normalized to 1. Asterisks indicate significant differences: *, P < 0.05 (by analysis of variance and Tukey's test); n.s., not significant. (E) SK-N-SH cells infected with each of the indicated viruses at an MOI of 5 were treated with DMSO or 20 μM MG132 6 h postinfection, and infection proceeded for an additional 18 h in the presence of DMSO or MG132. Infected cells were then harvested and analyzed by immunoblotting with an antibody to VP26 or α-tubulin. (F) Amount of VP26 in the SK-N-SH cells shown in top blot in panel E relative to that of α-tubulin, shown in the bottom blot in panel E. Each value is the mean ± standard error of triplicate experiments and is expressed relative to the mean value of wild-type HSV-1(F)-infected SK-N-SH cells treated with DMSO, which was normalized to 1. Asterisks indicate significant differences: *, P < 0.05 (by two-tailed Student's t test).
Next we examined the effect of the proteasome inhibitor MG132 on VP26 accumulation in cells infected with wild-type HSV-1(F) or YK492(VP26T111A/EGFP). In agreement with the results shown in Fig. 2A to D, the level of VP26 expression in cells infected with YK492(VP26T111A/EGFP) and treated with dimethyl sulfoxide (DMSO) was lower than that in cells infected with wild-type HSV-1(F) and treated with DMSO (Fig. 2E). In contrast, MG132 treatment significantly increased VP26 expression in cells infected with YK492(VP26T111A/EGFP) (Fig. 2E and F). However, we should note that the level of VP26 expression in YK492(VP26T111A/EGFP)-infected cells treated with MG132 was still lower than that in cells infected with wild-type HSV-1(F) (Fig. 2E). These results suggest that phosphorylation of VP26 Thr-111 is required for proper accumulation of VP26 in HSV-1-infected cells and in part exerts its effect by inhibiting VP26 degradation by the proteasome. At present, it remains uncertain whether this is a direct or an indirect effect of proteasome inhibition.
We next examined whether phosphorylation of VP26 Thr-111 regulates the subcellular localization of both it and its binding partner, VP5, in HSV-1-infected cells. SK-N-SH cells were infected with wild-type HSV-1(F), YK491(ΔVP26/EGFP), YK492(VP26T111A/EGFP), YK493(VP26Δ/TA-repair/EGFP), or YK494(VP26T111D/EGFP) at an MOI of 5. Infected cells were analyzed 12 h postinfection via immunofluorescence with antibodies to VP5 (3B6; Virusys) and VP26 (30) and secondary antibodies Alexa Fluor 680 and Alexa Fluor 546 (Invitrogen), respectively, using a Nikon confocal A1 laser scanning microscope. The secondary antibodies Alexa Fluor 680 and Alexa Fluor 546 were imaged using 637.5- and 561.2-nm excitation wavelengths, respectively. In agreement with previous reports (44), VP5 and VP26 were localized diffusely throughout the nucleus in most cells (86 and 95%) infected with wild-type HSV-1(F) or YK493(VP26Δ/TA-repair/EGFP) and appeared to colocalize in the nucleus (Fig. 3). In contrast, in most cells (91%) infected with YK491(ΔVP26/EGFP), VP5 was detected as punctate structures in the nucleus, suggesting that VP26 is required for correct localization of VP5 in HSV-1-infected cells. Interestingly, in cells infected with YK492(VP26T111A/EGFP), VP5 (60%) and VP26 (50%) were detected as punctate structures in the nucleus and mostly colocalized in these nuclear structures. Furthermore, the frequencies of cells showing diffuse nuclear staining of VP5 and VP26 were restored to 86 and 89%, respectively, in cells infected with YK494(VP26T111D/EGFP) (Fig. 3B). These results suggest that localization of VP5 and VP26 is regulated by phosphorylation at VP26 Thr-111 in HSV-1-infected cells.
FIG 3.
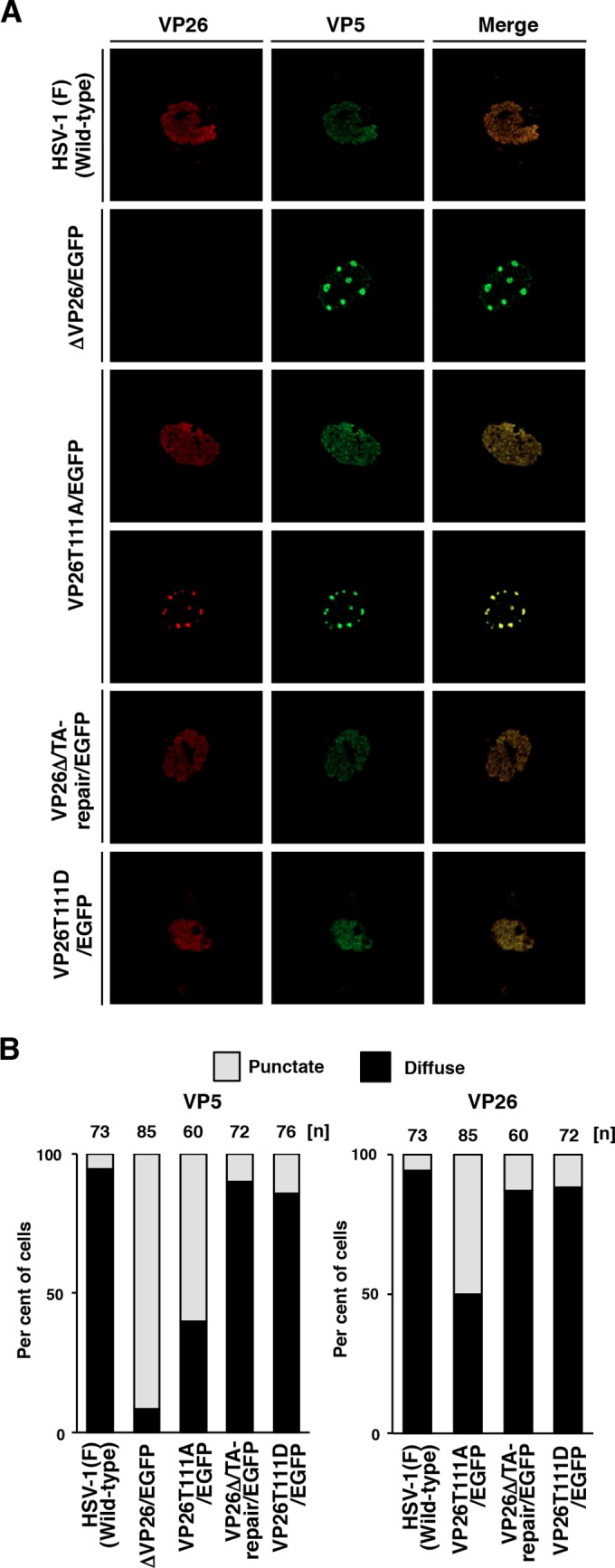
Effects of mutations in VP26 on the subcellular localization of VP26 and VP5 in SK-N-SH cells. (A) SK-N-SH cells were infected with each of the indicated viruses at an MOI of 5, fixed at 12 h postinfection, permeabilized, stained with antibody to VP26 (red) and VP5 (far red, pseudocolored in green), and examined by confocal microscopy. (B) Quantitation of localization patterns of VP26 and VP5 in the nucleus of the infected SK-N-SH cells shown in panel A. The subcellular localizations of VP26 and VP5, classified into “diffuse” and “punctate,” and frequencies of their localization in infected cells for each of the indicated viruses. The numbers above the columns describe the number of cell samples analyzed. Each data point is representative of the results from three independent experiments.
Although all capsid proteins of HSV-1 have previously been reported to be phosphorylated in infected cells, as described above, the biological significance of HSV-1 capsid phosphorylation, as well as that of other herpesvirus capsid proteins, remains to be fully elucidated (22–25). In this study, we have shown that the T111A mutation in VP26, which abolishes the phosphorylation of VP26 Thr-111, reduced HSV-1 replication and cell-cell spread in SK-N-SH cells and reduced HSV-1 neurovirulence in mice following intracranial inoculation. Furthermore, these reductions were restored by a phosphomimetic mutation at VP26 Thr-111 (T111D at VP26). From these observations, we conclude that VP26 Thr-111 phosphorylation is required for both efficient HSV-1 replication and cell-cell spread in SK-N-SH cells and HSV-1 neurovirulence in mice. To our knowledge, this is the first report showing the significance of phosphorylation of a herpesvirus capsid protein in viral replication and pathogenesis. Notably, the T111A mutation in VP26 impaired HSV-1 replication in SK-N-SH cells and neurovirulence in mice at levels similar to those observed with the VP26 null mutation. These results suggest that phosphorylation of VP26 at Thr-111 is critical for efficient regulation of HSV-1 replication and pathogenesis.
In this study, we have furthered our understanding of the biological significance of VP26 and identified previously unknown VP26 functions. (i) We have shown that the VP26 null mutation misdirects VP5 to nuclear punctate structures, indicating that VP26 regulates VP5 localization in infected cells. Since VP5 was diffusely distributed throughout the nucleus in wild-type HSV-1-infected cells, VP26 appears to prevent the aggregation of VP5 into punctate structures. (ii) Desai et al. previously reported that VP26 is important for HSV-1 replication in murine trigeminal ganglia following ocular inoculation (42). However, the role of VP26 in HSV-1 pathogenesis was not investigated in that study. Indeed, the relevance of VP26 in HSV-1 pathogenesis in vivo has not yet been reported. In this study, we showed that VP26 is required for efficient viral neurovirulence in mice following intracranial inoculation, indicating that VP26 is an HSV-1 neurovirulence factor. This observation is in agreement with a previous report by Krautwald et al. which showed that VP26 homologs of pseudorabies virus (PRV) were required for efficient neuroinvasion in mice following intranasal inoculation (45).
At present, the mechanism(s) by which VP26 Thr-111 phosphorylation regulates HSV-1 replication and pathogenesis remains unclear. In this study, we have describe two effects of VP26 Thr-111 phosphorylation in HSV-1-infected SK-N-SH cells: the stabilization of VP26 by direct or indirect inhibition of its degradation by the proteasome and regulation of nuclear localization of both itself and its binding partner, VP5. Based upon the requirement for VP26 in efficient HSV-1 replication and pathogenesis shown in this and previous studies (42), it is reasonable to hypothesize that proper expression of VP26, regulated by its phosphorylation at Thr-111, is important for HSV-1 replication and pathogenesis. In addition, Nagel et al. reported that tagging VP26 with fluorescent proteins (FPs) at the N terminus produced punctate structures in the nucleus containing FP-tagged VP26, VP5, UL25, and UL17, but not often capsids, and impaired viral nuclear egress and replication (44). These nuclear punctate structures appear not to be sites for capsid assembly, but they prevent proper capsid assembly and nuclear egress by sequestering capsid proteins (44). Notably, these nuclear punctate structures, induced by tagging of VP26 with FPs, resembled those containing VP5 and/or mutated VP26 induced by the VP26 null mutation or the VP26 T111A mutation. Tagging the 12-kDa VP26 with an approximately 25-kDa FP may have caused steric hindrance in VP26, which may in turn have contributed to VP26 loss of function. Support for this hypothesis comes from a report that FP tagging of a PRV VP26 homolog impairs viral replication and neuroinvasiveness at levels similar to those observed with the null mutation of PRV VP26 (45). Furthermore, as described above, the T111A mutation in HSV-1 VP26 reduced HSV-1 replication and neurovirulence at levels similar to those caused by the null mutation of HSV-1 VP26. It is well-documented that phosphorylation of a protein can cause conformational changes (46). Taken together, these observations suggest that in the absence of functional VP26, HSV-1 capsid proteins tend to accumulate in punctate structures in the nucleus. VP26 may prevent the aggregation of these HSV-1 capsid proteins, as suggested above, thereby distributing the capsid proteins throughout the nucleus and facilitating efficient capsid assembly. It is likely that phosphorylation of VP26 Thr-111 is required in order for VP26 to adopt the correct conformation required to prevent the aggregation of the HSV-1 capsid proteins in the nucleus, thus promoting not only efficient capsid assembly but also nuclear egress and viral replication.
It is not known why the T111A mutation in VP26 affected its own and VP5 localization only in a fraction of infected cells. However, we previously reported a similar observation that alanine substitution in the phosphorylation site of HSV-1 UL47 impaired nuclear localization only in some cells (34). One possible explanation of these observations may be that the subcellular localization of UL47 and VP26 is regulated by multiple factors, and the subcellular localization of the viral proteins may be determined by the balance among the factors. In agreement with this hypothesis, it has been reported that HSV-2 UL14 has the ability to regulate the subcellular localization of VP26 (47). Blocking the phosphorylation of VP26 Thr-111 may cause a subtle imbalance among the factors that determine the localization of VP26 in infected cells, resulting in an aberrant accumulation of VP26 and VP5 in punctate structures in the nucleus in only a fraction of infected cells.
Although the amino acid sequence of VP26 is conserved in all Herpesviridae subfamilies, VP26 Thr-111 is only conserved in HSV-2. Thr-111 is located in the C-terminal domain of VP26 in HSV-1 and, interestingly, it has also been reported that the VP26 homolog in Epstein-Barr virus is phosphorylated in the C-terminal domain (48). Therefore, there is a possibility that the function(s) of the EBV VP26 homolog, as well as those of other herpesviruses, is also regulated by phosphorylation of the C-terminal domain. As described above, phosphorylation of the capsid proteins of various other viruses contributes to their efficient viral life cycles (7–13). Thus, it will be of interest to investigate whether phosphorylation of other HSV-1 capsid proteins regulates their function. We are currently pursuing this possibility.
ACKNOWLEDGMENTS
We thank Tomoko Ando, Noriko Tokai, and Shihoko Koyama for excellent technical assistance.
This study was supported by the Funding Program for Next Generation World-Leading Researchers and Grants for Scientific Research from the Japan Society for the Promotion of Science (JSPS), a contract research fund of the Program of Japan Initiative for Global Research Network on Infectious Diseases (J-GRID), a grant for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan, and grants from the Takeda Science Foundation, the Ichiro Kanehara Foundation, and the Motida Memorial Foundation.
REFERENCES
- 1.Harrison SC. 2013. Principles of virus structure, p 52–86. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott-Williams &Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Almazan F, Galan C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J Virol 78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Haussinger D. 2003. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J 17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 4.Urbanowski MD, Hobman TC. 2013. The West Nile virus capsid protein blocks apoptosis through a phosphatidylinositol 3-kinase-dependent mechanism. J Virol 87:872–881. doi: 10.1128/JVI.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surjit M, Liu B, Chow VT, Lal SK. 2006. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem 281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni P, Cheng Kao C. 2013. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 446:123–132. doi: 10.1016/j.virol.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartier C, Sivard P, Tranchat C, Decimo D, Desgranges C, Boyer V. 1999. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J Biol Chem 274:19434–19440. [DOI] [PubMed] [Google Scholar]
- 8.Dochi T, Nakano T, Inoue M, Takamune N, Shoji S, Sano K, Misumi S. 2014. Phosphorylation of human immunodeficiency virus type 1 capsid protein at serine 16, required for peptidyl-prolyl isomerase-dependent uncoating, is mediated by virion-incorporated extracellular signal-regulated kinase 2. J Gen Virol 95:1156–1166. doi: 10.1099/vir.0.060053-0. [DOI] [PubMed] [Google Scholar]
- 9.Deroubaix A, Osseman Q, Cassany A, Begu D, Ragues J, Kassab S, Laine S, Kann M. 2015. Expression of viral polymerase and phosphorylation of core protein determine core and capsid localization of the human hepatitis B virus. J Gen Virol 96:183–195. doi: 10.1099/vir.0.064816-0. [DOI] [PubMed] [Google Scholar]
- 10.Kock J, Kann M, Putz G, Blum HE, Von Weizsacker F. 2003. Central role of a serine phosphorylation site within duck hepatitis B virus core protein for capsid trafficking and genome release. J Biol Chem 278:28123–28129. doi: 10.1074/jbc.M300064200. [DOI] [PubMed] [Google Scholar]
- 11.Chu TH, Liou AT, Su PY, Wu HN, Shih C. 2014. Nucleic acid chaperone activity associated with the arginine-rich domain of human hepatitis B virus core protein. J Virol 88:2530–2543. doi: 10.1128/JVI.03235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W, Ou J-H. 2002. Phosphorylation of hepatitis C virus core protein by protein kinase A and protein kinase C. Virology 300:20–30. doi: 10.1006/viro.2002.1524. [DOI] [PubMed] [Google Scholar]
- 13.Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, Lal SK. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol 79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 1823–1897. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott-Williams &Wilkins, Philadelphia, PA. [Google Scholar]
- 15.Baines JD. 2011. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol 19:606–613. doi: 10.1016/j.tim.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Homa FL, Brown JC. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol 7:107–122. doi:. [DOI] [PubMed] [Google Scholar]
- 17.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol 232:499–511. [DOI] [PubMed] [Google Scholar]
- 19.Booy FP, Trus BL, Newcomb WW, Brown JC, Conway JF, Steven AC. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc Natl Acad Sci U S A 91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wingfield PT, Stahl SJ, Thomsen DR, Homa FL, Booy FP, Trus BL, Steven AC. 1997. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol 71:8955–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol 75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaster S, Roizman B. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J Virol 35:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNabb DS, Courtney RJ. 1992. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J Virol 66:4839–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell C, Desjardins M, Thibault P, Radtke K. 2013. Proteomics analysis of herpes simplex virus type 1-infected cells reveals dynamic changes of viral protein expression, ubiquitylation, and phosphorylation. J Proteome Res 12:1820–1829. doi: 10.1021/pr301157j. [DOI] [PubMed] [Google Scholar]
- 25.Kato A, Tsuda S, Liu Z, Kozuka-Hata H, Oyama M, Kawaguchi Y. 2014. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells. J Virol 88:655–666. doi: 10.1128/JVI.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 28.Kato A, Tanaka M, Yamamoto M, Asai R, Sata T, Nishiyama Y, Kawaguchi Y. 2008. Identification of a physiological phosphorylation site of the herpes simplex virus 1-encoded protein kinase Us3 which regulates its optimal catalytic activity in vitro and influences its function in infected cells. J Virol 82:6172–6189. doi: 10.1128/JVI.00044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi Y, Van Sant C, Roizman B. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol 71:7328–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J Virol 82:5198–5211. doi: 10.1128/JVI.02681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka M, Kodaira H, Nishiyama Y, Sata T, Kawaguchi Y. 2004. Construction of recombinant herpes simplex virus type I expressing green fluorescent protein without loss of any viral genes. Microbes Infect 6:485–493. doi: 10.1016/j.micinf.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 33.Kato A, Yamamoto M, Ohno T, Tanaka M, Sata T, Nishiyama Y, Kawaguchi Y. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J Virol 80:1476–1486. doi: 10.1128/JVI.80.3.1476-1486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato A, Liu Z, Minowa A, Imai T, Tanaka M, Sugimoto K, Nishiyama Y, Arii J, Kawaguchi Y. 2011. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J Virol 85:9599–9613. doi: 10.1128/JVI.00845-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato A, Arii J, Shiratori I, Akashi H, Arase H, Kawaguchi Y. 2009. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral envelope glycoprotein B and regulates its expression on the cell surface. J Virol 83:250–261. doi: 10.1128/JVI.01451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mou F, Wills E, Baines JD. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J Virol 83:5181–5191. doi: 10.1128/JVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imai T, Sagou K, Arii J, Kawaguchi Y. 2010. Effects of phosphorylation of herpes simplex virus 1 envelope glycoprotein B by Us3 kinase in vivo and in vitro. J Virol 84:153–162. doi: 10.1128/JVI.01447-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka M, Nishiyama Y, Sata T, Kawaguchi Y. 2005. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology 341:301–312. doi: 10.1016/j.virol.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Kozuka-Hata H, Nasu-Nishimura Y, Koyama-Nasu R, Ao-Kondo H, Tsumoto K, Akiyama T, Oyama M. 2012. Phosphoproteome of human glioblastoma initiating cells reveals novel signaling regulators encoded by the transcriptome. PLoS One 7:e43398. doi: 10.1371/journal.pone.0043398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato A, Shindo K, Maruzuru Y, Kawaguchi Y. 2014. Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery. J Virol 88:2775–2785. doi: 10.1128/JVI.03300-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujii H, Kato A, Mugitani M, Kashima Y, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2014. The UL12 protein of herpes simplex virus 1 is regulated by tyrosine phosphorylation. J Virol 88:10624–10634. doi: 10.1128/JVI.01634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desai P, DeLuca NA, Person S. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 43.Imai T, Arii J, Minowa A, Kakimoto A, Koyanagi N, Kato A, Kawaguchi Y. 2011. Role of the herpes simplex virus 1 Us3 kinase phosphorylation site and endocytosis motifs in the intracellular transport and neurovirulence of envelope glycoprotein B. J Virol 85:5003–5015. doi: 10.1128/JVI.02314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagel CH, Dohner K, Binz A, Bauerfeind R, Sodeik B. 2012. Improper tagging of the non-essential small capsid protein VP26 impairs nuclear capsid egress of herpes simplex virus. PLoS One 7:e44177. doi: 10.1371/journal.pone.0044177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krautwald M, Maresch C, Klupp BG, Fuchs W, Mettenleiter TC. 2008. Deletion or green fluorescent protein tagging of the pUL35 capsid component of pseudorabies virus impairs virus replication in cell culture and neuroinvasion in mice. J Gen Virol 89:1346–1351. doi: 10.1099/vir.0.83652-0. [DOI] [PubMed] [Google Scholar]
- 46.Groban ES, Narayanan A, Jacobson MP. 2006. Conformational changes in protein loops and helices induced by post-translational phosphorylation. PLoS Comput Biol 2:e32. doi: 10.1371/journal.pcbi.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamauchi Y, Wada K, Goshima F, Takakuwa H, Daikoku T, Yamada M, Nishiyama Y. 2001. The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J Gen Virol 82:321–330. [DOI] [PubMed] [Google Scholar]
- 48.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]


