ABSTRACT
Influenza A viruses (IAVs) express the PB1-F2 protein from an alternate reading frame within the PB1 gene segment. The roles of PB1-F2 are not well understood but appear to involve modulation of host cell responses. As shown in previous studies, we find that PB1-F2 proteins of mammalian IAVs frequently have premature stop codons that are expected to cause truncations of the protein, whereas avian IAVs usually express a full-length 90-amino-acid PB1-F2. However, in contrast to other avian IAVs, recent isolates of highly pathogenic H5N1 influenza viruses had a high proportion of PB1-F2 truncations (15% since 2010; 61% of isolates in 2013) due to several independent mutations that have persisted and expanded in circulating viruses. One natural H5N1 IAV containing a mutated PB1-F2 start codon (i.e., lacking ATG) was 1,000-fold more virulent for BALB/c mice than a closely related H5N1 containing intact PB1-F2. In vitro, we detected expression of an in-frame protein (C-terminal PB1-F2) from downstream ATGs in PB1-F2 plasmids lacking the well-conserved ATG start codon. Transient expression of full-length PB1-F2, truncated (24-amino-acid) PB1-F2, and PB1-F2 lacking the initiating ATG in mammalian and avian cells had no effect on cell apoptosis or interferon expression in human lung epithelial cells. Full-length and C-terminal PB1-F2 mutants colocalized with mitochondria in A549 cells. Close monitoring of alterations of PB1-F2 and their frequency in contemporary avian H5N1 viruses should continue, as such changes may be markers for mammalian virulence.
IMPORTANCE Although most avian influenza viruses are harmless for humans, some (such as highly pathogenic H5N1 avian influenza viruses) are capable of infecting humans and causing severe disease with a high mortality rate. A number of risk factors potentially associated with adaptation to mammalian infection have been noted. Here we demonstrate that the protein PB1-F2 is frequently truncated in recent isolates of highly pathogenic H5N1 viruses. Truncation of PB1-F2 has been proposed to act as an adaptation to mammalian infection. We show that some forms of truncation of PB1-F2 may be associated with increased virulence in mammals. Our data support the assessment of PB1-F2 truncations for genomic surveillance of influenza viruses.
INTRODUCTION
Highly pathogenic avian influenza viruses (HPAIVs) of the H5N1 subtype are now endemic in many countries (1). H5N1 became a public health concern in 1997 when a poultry outbreak in Hong Kong resulted in 18 human infections with six fatalities (2). Subsequent reassortment events with other avian influenza viruses were associated with the spread of new genotypes of H5N1 over much of Asia, the Middle East, and Europe and with localized evolution and antigenic variation of the H5 and N1 surface proteins (3, 4). H5N1 viruses remain capable of infecting humans with high mortality, with over 700 confirmed human cases with a case fatality rate of nearly 60% (5). Unlike seasonal influenza A viruses (IAVs) that circulate among humans, H5N1 HPAIVs do not readily transmit from human to human. One of the key reasons for inefficient human-to-human transmission of H5N1 viruses is the preference of their hemagglutinin (HA) for α-2,3-linked sialic acids, which are uncommon in the upper respiratory tract of humans (6). In addition to the HA gene, other IAV genes may also contribute to transmission and virulence.
The PB1 gene segment is of particular interest, since this was the only internal gene from an avian source that reassorted with the surface glycoprotein genes encoding HA and neuraminidase (NA) in the pandemic virus of 1957 and with the HA gene in the 1968 pandemic virus (7). One possible factor in the role of PB1 in pathogenicity and transmission is the presence of the PB1-F2 protein, encoded by a +1 reading frame (8), which was present as a full-length protein in the 1918, 1957, and 1968 human pandemic viruses but not in the 2009 H1N1 pandemic virus.
PB1-F2 is a short (87- to 90-amino-acid) IAV protein discovered serendipitously in 2001 during the search for CD8 T cell epitopes encoded by alternative reading frames (8). It was found to localize in mitochondria and cause cell death (8, 9). It is dispensable for viral replication but has been associated with viral pathogenesis in mice and ducks (8, 10–13), and truncation of PB1-F2 in the 2009 pandemic influenza virus (H1N1pdm09) is hypothesized to be one cause of the less severe pathology and relatively low case fatality rate of this pandemic compared to previous pandemics (14, 15). The effect of PB1-F2 on viral phenotype has been demonstrated to be strain specific (16–18), particularly among viruses with a PB1 gene of recent avian origin (10). This may be because other pathogenic determinants are able to mask the influence of PB1-F2 (11).
The mechanisms underlying the pathogenic effects of PB1-F2 remain unclear. Several possible mechanisms have been suggested, including mitochondrial targeting and proapoptotic activity (8, 9, 19), enhancing PB1 function (20), and inhibition (21–27) or enhancement (28) of the IFN-β response. PB1-F2 has also been linked to bacterial infections following IAV infection (29–32). However, most of these phenotypes have been difficult to generalize to all strains of virus and across host models tested.
A striking feature of PB1-F2 among global IAVs is the frequent presence of premature stop codons. For example, almost all 2009 H1N1pdm viruses have truncated 11-amino-acid PB1-F2 proteins. In 2007, analysis of publicly available PB1-F2 sequences indicated that a large proportion (19%) of mammalian IAVs had truncated PB1-F2, but such truncations were rare among avian IAVs (only 4%) (33). Similarly, in 2013, Pasricha et al. determined that only 5% of avian IAVs analyzed contained truncated PB1-F2, compared to 41% of human and swine IAVs (34). These observations suggest that truncation of PB1-F2 may play a role in adaptation of IAVs to mammalian hosts.
Although PB1-F2 truncations have been sporadically observed in avian influenza viruses, until recently it has been unusual for groups of related avian influenza virus subtypes to persistently possess truncated variants. However, since 2009, an increasing number of H5N1 HPAIV lineages have maintained PB1-F2 truncations over several years. Based on previous suggestions that viruses with these changes may be better adapted to mammalian replication, we tested these viruses for their ability to cause disease in mice and to alter cellular functions in mammalian and avian cells.
MATERIALS AND METHODS
Database analysis of PB1-F2 truncations.
We retrieved full-length PB1 sequences with collection dates prior to 2014 from the GISAID (see Table S1 in the supplemental material) and the NCBI Influenza databases (as of 31 December 2013) and merged the sets, eliminating duplicates based on virus strain name. The host species (human, swine, or avian) and year of collection were also noted. Highly pathogenic H5N1 viruses were considered a separate subset and were excluded from the avian and human subsets. PB1 sequences were aligned using MAFFT (35), and PB1-F2 sequences were translated from each PB1 gene. The numbers of truncated versions present in each subset, per year of collection, were calculated. In cases where no initiating ATG was present at the appropriate position, the length of PB1-F2 was considered to be zero.
Distribution of PB1-F2 truncations in H5N1 viruses.
For H5N1 viruses with PB1-F2 truncations, we obtained the corresponding HA sequences from the appropriate database, annotating HA sequences with the length of the truncated PB1-F2. We also included HA sequences from the report of the WHO/OIE/FAO H5N1 Evolution Working Group (36) in order to identify clades. We aligned HA sequences using MAFFT and constructed a maximum likelihood tree (generalized time-reversible model of nucleotide evolution; gamma, 10,000 bootstraps) with FastTree (37), using FigTree (38) for annotations.
Identification of related PB1-F2 full-length/truncated H5N1 viruses.
We analyzed the set of virus isolates available to us to identify a pair of similar viruses differing in PB1-F2 length. Viral RNAs were isolated by using the QIAamp Viral RNA minikit (catalog no. 52904; Qiagen Inc., USA), reverse transcribed, and amplified by the AccessQuick reverse transcription (RT)-PCR system (catalog no. A1702; Promega Corporation, USA). Sequencing reactions were performed by using the BigDye Terminator v3.1 cycle sequencing kit (catalog no. 4337456; Life Technologies, Grand Island, NY) and run in the Applied Biosystems 3730xl DNA Analyzer (Life Technologies). RT-PCR and sequencing primers are available upon request. For each H5N1 virus containing a PB1-F2 truncation, the amino acid sequence of each viral protein was compared to that of the most closely related virus containing full-length PB1-F2.
Preparation of viral stocks.
Clade 2.3.4.2 H5N1 viruses A/chicken/Vietnam/NCVD-281/2009 (here referred to as VN/281; GISAID isolate ID EPI_ISL_80619) and A/chicken/Vietnam/NCVD-296/2009 (here referred to as VN/296; EPI_ISL_80637) were grown in 10- to 11-day-old embryonated chicken eggs incubated at 37°C for 24 h. Allantoic fluids from eggs, infected with the highest dilution of virus inoculum that gave a hemagglutination inhibition titer of 256 or more, were harvested, clarified by centrifugation, and frozen at −80°C as viral stocks.
To determine the 50% egg infectious dose (EID50) of virus stocks, dilutions (10−5 to 10−10) were made from each virus and inoculated into five 10- to 11-day-old embryonated hen eggs per dilution. Eggs were incubated at 37°C for 24 h and then chilled overnight at 4°C. Egg infections were detected by standard hemagglutination assays, and EID50 was calculated by the method of Reed and Muench (39).
Plaque assay.
Confluent MDCK-London cells were infected with six dilutions (10−5 to 10−10) of each virus, overlaid with 0.8% agarose medium, and incubated at two different temperatures (37°C and 40°C). Seventy-two hours postinfection, agarose was gently removed from wells and cell monolayers were stained with crystal violet-formalin mix. Plaques were counted in appropriate wells, and PFU titers were calculated.
Pathogenicity in mice.
All animal experiments were done in an animal biosafety level 3 (ABSL3E) facility with enhancements required by the U.S. Department of Agriculture and the Select Agent Program (40) under CDC's IACUC-approved protocols. Six- to 8-week-old female BALB/cJ mice (Jackson Laboratory, Bar Harbor, ME) were used in this study. Five mice each were intranasally infected with 1 of 5 virus doses (100 to 104 PFU in 50-μl volume) of each virus; 5 mice were mock infected with phosphate-buffered saline (PBS). Mice were observed daily for weight loss and other clinical signs for a period of 16 days. Mice were euthanized if they lost 25% or more of their original body weight or if neurological signs were observed. Mouse 50% lethal dose (LD50) values were calculated using the Reed and Muench method (39).
Cloning and mutagenesis of PB1-F2.
Wild-type PB1-F2 from VN/281 and VN/296 were RT-PCR amplified, using oligonucleotide primers with added KpnI and XbaI restriction sites. Amplified products were cloned into a pTracer-CMV2-GFP vector (Life Technologies, Grand Island, NY). Additional PB1-F2 clones were made with insertion of C-terminal HA epitope tags. Mutants of cloned PB1-F2 (summarized in Fig. 1 and Table 1) were made by site-directed mutagenesis using the Quick Change Mutagenesis kit (Agilent Technologies, Santa Clara, CA) and mutagenesis primers. Cloned PB1-F2 plasmids were amplified in competent Escherichia coli cells and were purified using the Qiagen plasmid purification kit (Qiagen Inc., Valencia, CA). The sequences of all plasmid clones were confirmed before use in experiments.
FIG 1.
Schematic diagrams of PB1-F2 mutants. Seven PB1-F2 variants from VN/296 (shown here) and VN/281 (not shown) were cloned into expression plasmids. Red text indicates stop codons or mutation of the initiating start codon. HA, an in-frame C-terminal HA epitope tag. All in-frame ATGs in the PB1-F2 ORF are indicated. F2FL, full-length PB1-F2, 90 aa; F2NO, 1st ATG is mutated to ACG as in wild-type VN/296; F2TR, truncated PB1-F2 (24 amino acids) due to stop codons at positions 25 and 26. Please note that there are two variants of F2TR with an HA tag. F2TR-HA has an HA tag at the end of the protein and so can detect only C-terminal protein expressed from in-frame downstream ATGs. F2HA-TR has the tag immediately upstream from the double stops (25, 26) and thus can be used to detect N-terminal protein (24 aa).
TABLE 1.
Details of PB1-F2 mutants made in this studya
| Source virus | Name of mutant | PB1-F2 status or size (aa) | Mutagenesis primer(s) |
|---|---|---|---|
| VN/281 | VN/281/PB1-F2FL | Full length (90) | Kpn1PB1F76-94 and Xba1PB1R384-67 |
| VN/281/PB1-F2TR | Truncatedb (24) | PB1C167T_C170T_F | |
| VN/281/PB1-F2NO | Deletedc | PB1T96C _F | |
| VN/281/PB1-F2FL-HA | Full length (90) | Kpn1PB1F68-90 and Xba1HAtagPB1R364-43 | |
| VN/281/PB1-F2HA-TR | Truncatedb (24) | PB1F151-185 (HA_C167T_C170T) | |
| VN/281/PB1-F2TR-HA | Truncatedd | PB1C167T_C170T_F and Xba1HAtagPB1R364-43 | |
| VN/281/PB1-F2NO-HA | Deletedc | PB1T96C _F | |
| VN/296 | VN/296/PB1-F2FL | Full length (90) | PB1C96T_F |
| VN/296/PB1-F2TR | Truncated (24) | PB1C167T_C170T_F | |
| VN/296/PB1-F2NO | Deletedc | Kpn1PB1F76-94 and Xba1PB1R384-67 | |
| VN/296/PB1-F2FL-HA | Full length (90) | PB1C96T_F | |
| VN/296/PB1-F2HA-TR | Truncatedb (24) | PB1F151-185 (HA_C167T_C170T) | |
| VN/296/PB1-F2TR-HA | Truncatedd | PB1C167T_C170T_F and Xba1HAtagPB1R364-43 | |
| VN/296/PB1-F2NO-HA | Deletedc | Kpn1PB1F68-90 and Xba1HAtagPB1R364-43 |
A total of 6 mutants were made from each virus: 3 with a C-terminal HA tag and 3 without the HA tag. See Fig. 1 for schematic diagrams.
Stop codons were added at positions 25 and 26; the plasmid is expected to express a truncated protein (24 aa). One-half of these mutants have the HA tag immediately upstream from the stop codons.
In this variant, only the first ATG is mutated to ACG, leaving the downstream in-frame ATGs intact.
This variant has stops at positions 25 and 26, but the HA tag is at the C terminus.
Confocal microscopy.
Posttransfection A549 cells were washed with PBS, fixed in 4% paraformaldehyde for 15 min, permeabilized with 0.5% Tween 20 in PBS for 10 min, washed with PBST (PBS plus 0.2% Tween 20), and blocked with 2% bovine serum albumin (BSA) in PBST for 2 h. After blocking, cells were incubated with HA-Tag (C29F4) rabbit monoclonal antibody (MAb; 1:1,000; Cell Signaling) and anti-MTCO2 (mitochondrially encoded cytochrome c oxidase II) antibody (ab91317; 1:2,000; Abcam) overnight at 4°C. Then, cells were washed (2.5% fetal bovine serum [FBS] in PBST, 6 times for 5 min) and incubated with Alexa Fluor 546 donkey anti-mouse IgG (1:1,000) and Alexa Fluor 633 goat anti-rabbit IgG (H+L; 1:1,000; Life Technologies), for 2 hours at room temperature. Nonspecific secondary antibodies were removed by washing with PBST (6 times for 5 min), followed by a final soak in PBS. Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole), and coverslips were mounted with Prolong gold antifade mounting medium (Invitrogen). Since the plasmids expressing PB1-F2 expressed green fluorescent protein (GFP) under a separate promoter, we limited analysis to cells expressing GFP (green). Images were captured using the Zeiss invert confocal microscope LSM 710 with a 63× objective. Images were processed using Zen 2010 (Zeiss) and Adobe Photoshop (Adobe Inc.).
qRT-PCR for IFN-β expression.
Human A549 (ATCC CCL-185) and chicken DF1 (ATCC CRL-12203) cells grown in 12-well cell culture plates were transfected with 1.0 μg of plasmid DNA using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI). As a positive control, cells were treated with in vitro-transcribed 5′ triphosphate RNA (TP-RNA) (41). Cells were lysed, and lysates were used directly for quantitative RT-PCR (qRT-PCR) using the CellsDirect One-Step qRTPCR kit (Life Technologies, Grand Island, NY) and TaqMan gene expression assays for human and avian beta interferon (IFN-β; Life Technologies, Grand Island, NY). The ΔΔ Cq method in Bio-Rad CFX Manager 2.1 software was used to analyze the expression data. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression was measured in each sample as a control.
Flow cytometry for cell apoptosis.
A549 and DF1 cells were transfected as described above. As a positive control, cells were treated with camptothecin (Sigma-Aldrich, St. Louis, MO, USA) at final concentration of 20 μM for 24 h. After harvesting, cells were stained for apoptosis markers using the violet chromatin condensation/dead cell apoptosis kit with Vybrant DyeCycle Violet and Sytox AADvanced for flow cytometry (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. Flow cytometry data were acquired within 1 h of staining. Data were acquired on a FACSCanto II flow cytometer (BD, Franklin Lakes, NJ) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Statistical analyses.
To compare weight loss, a linear mixed model with repeated measures was used, using as a cutoff P values of <0.05 for statistical significance. Kaplan-Meier survival curves were compared by the log rank test. Effects on apoptosis and IFN-β expression were analyzed by the Kruskal-Wallis test using GraphPad Prism software (La Jolla, CA, USA).
RESULTS
Evolution of PB1-F2 truncations in avian and mammalian viruses.
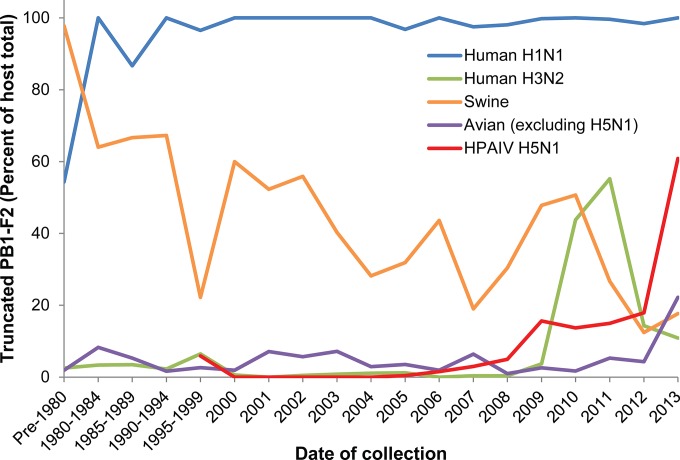
Previous studies (33, 34) have shown that a large fraction of mammalian influenza viruses express truncated PB1-F2. In contrast, avian influenza viruses usually express full-length PB1-F2 of 90 amino acids (aa). To assess changes in this trend since these previous studies, we retrieved 21,984 PB1 sequences that included the full PB1-F2 region from the combined GISAID and NCBI Influenza databases (removing duplicates based on strain name) as of 31 December 2013: 10,498 human, 1,966 HPAIV H5N1, 7,392 avian (excluding H5N1), and 2,128 swine sequences. We identified 23 different truncations of PB1-F2, resulting in PB1-F2 lengths ranging from 0 to 87 aa. A small number of viruses (53, 0.24% of total) had PB1-F2 proteins that were 101 aa; most of these were human H3N2 viruses from 1997 to 1998 (24 viruses) and non-H5N1 avian viruses from the 2000s (16 viruses) (data not shown). Of all PB1-F2s, 64.3% were considered to be functionally full length (≥79 aa [33]). The prevalence of PB1-F2 truncations (78 amino acids or shorter) was 63.1% for human viruses (H1N1 and H3N2), 6.7% for H5N1, 3.8% for avian viruses excluding H5N1, and 39.7% for swine viruses. The prevalence of PB1-F2 truncations by year of collection, subtype, and host is shown in Fig. 2, and a summary of the PB1-F2 truncations in H5N1 viruses is shown in Table 2.
FIG 2.
Prevalence of PB1-F2 truncations. PB1 sequences from human, swine, and avian (not including H5N1) influenza viruses and from highly pathogenic H5N1 influenza viruses were retrieved from the NCBI, GISAID, and CDC databases. Human H1N1 viruses include both seasonal H1N1 and 2009 H1N1pdm viruses. Sequences were aligned using MAFFT, and PB1-F2 sequence lengths were inferred from the sequences. Sequences were sorted by year of isolation after 2000. Because sequences were less abundant pre-2000, sequences from 1980 to 1999 were pooled in 5-year groups, and all sequences pre-1980 were pooled. Data are shown as the percentage of truncated PB1-F2 among all sequences for that host and year.
TABLE 2.
Summary of PB1-F2 truncations in H5N1 virusesa
| Yr of isolation | No. of viruses with PB1-F2 protein of indicated putative length (aa) |
Total no. of viruses | % carrying truncated protein | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Truncated protein (≤78 aa) |
Nontruncated protein (≥79 aa) |
||||||||||||||
| 0 | 8 | 11 | 24 | 25 | 34 | 57 | 63 | 79 | 81 | 87 | 90 | 101 | |||
| 1996 | 1 | 1 | 0 | ||||||||||||
| 1997 | 2 | 17 | 19 | 10.5 | |||||||||||
| 1998 | 2 | 2 | 0.0 | ||||||||||||
| 1999 | 11 | 11 | 0.0 | ||||||||||||
| 2000 | 11 | 11 | 0.0 | ||||||||||||
| 2001 | 39 | 39 | 0.0 | ||||||||||||
| 2002 | 3 | 31 | 34 | 0.0 | |||||||||||
| 2003 | 61 | 1 | 62 | 0.0 | |||||||||||
| 2004 | 2 | 136 | 1 | 139 | 0.0 | ||||||||||
| 2005 | 1 | 242 | 243 | 0.4 | |||||||||||
| 2006 | 1 | 2 | 1 | 1 | 1 | 316 | 322 | 1.6 | |||||||
| 2007 | 3 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 323 | 336 | 3.0 | ||||
| 2008 | 2 | 1 | 2 | 4 | 2 | 3 | 167 | 1 | 182 | 4.9 | |||||
| 2009 | 4 | 16 | 106 | 2 | 128 | 15.6 | |||||||||
| 2010 | 1 | 12 | 7 | 1 | 1 | 2 | 129 | 153 | 13.7 | ||||||
| 2011 | 1 | 2 | 3 | 11 | 12 | 1 | 164 | 194 | 14.9 | ||||||
| 2012 | 2 | 3 | 7 | 2 | 52 | 1 | 67 | 17.9 | |||||||
| 2013 | 14 | 9 | 23 | 60.9 | |||||||||||
| Totals | 10 | 7 | 4 | 16 | 41 | 1 | 43 | 1 | 6 | 5 | 9 | 1,817 | 6 | 1,966 | |
| 123 | 1,843 | 1,966 | 6.7 | ||||||||||||
PB1 sequences from highly pathogenic H5N1 influenza viruses were retrieved from the NCBI, GISAID, and CDC databases. Sequences were aligned using MAFFT, and PB1-F2 sequence lengths were inferred from the sequences. Sequences were sorted by year of isolation and PB1-F2 protein length. The sequences expressing a protein of 78 aa or smaller were annotated as truncated.
The most common truncations were 11 and 57 amino acids (23.2% and 7.6% of total, respectively), with no other truncation exceeding 1% of the total. Swine influenza viruses include multiple subtypes (reviewed in reference 42), and PB1-F2 truncations were most common in the H1N1 subtypes (data not shown). Among avian viruses, 57- and 79-residue versions were more prevalent before 2005, after which variants containing 24 or 25 aa became more prevalent. (We group these together since in H5N1 viruses the 24-aa variant arose by adding an extra stop codon to the 25-aa variant.) Similar mutants have arisen sporadically in swine and human viruses since 2001 and 2007, respectively. The prevalence of these variants has been less than 3% in avian, human, and swine viruses separately and combined.
Evolution of PB1-F2 truncations in H5N1 viruses.
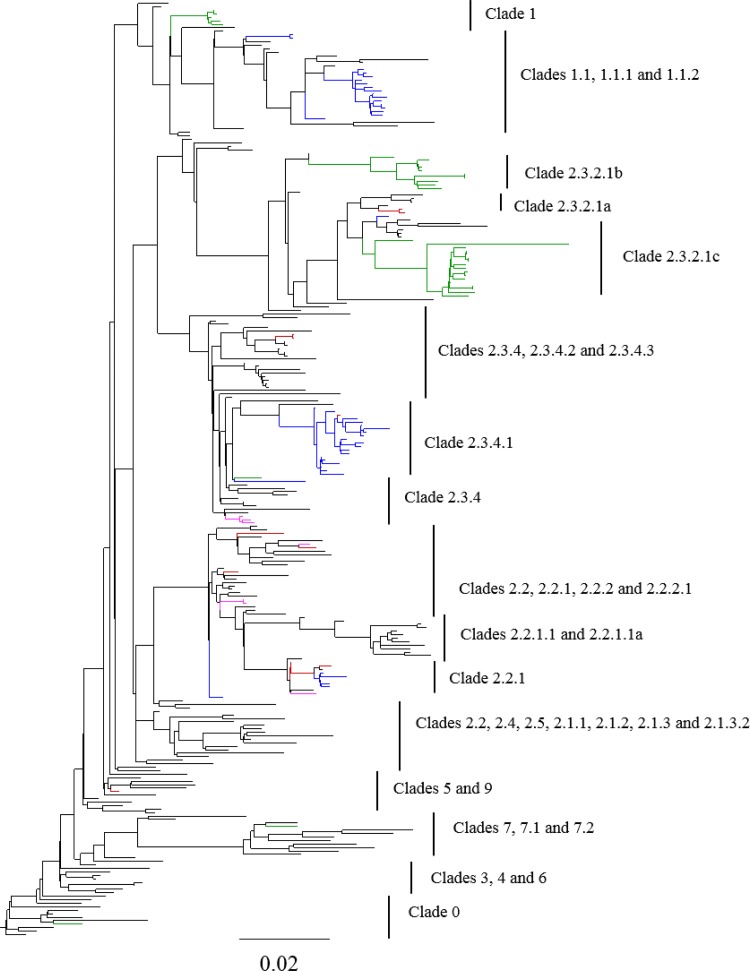
HPAIV H5N1 viruses have been categorized into multiple clades based on their HA sequences (36). We constructed phylogenetic trees, based on HA sequence (Fig. 3; see also Fig. S1 in the supplemental material), to analyze the distribution of PB1-F2 truncations in H5N1 clades. Until 2009, PB1-F2 truncations in H5N1 viruses were rare (∼2% of sequences [Fig. 2 and Table 2]), with only a few instances in which truncations persisted in a particular lineage for more than several months (e.g., clade 1 in Thailand from 2007 to 2008; see below). However, in 2009, 15.6% of the H5N1 viruses had truncated PB1-F2 (Fig. 2 and Table 2). In particular, clade 2.3.4.1 viruses isolated in 2009 and 2010 in Vietnam and China consistently had a PB1-F2 truncation of 24 or 25 amino acids. These viruses included both poultry isolates and human cases (A/Guizhou/1/2009, A/Hunan/2/2009). In 2010 and 2011, clade 2.3.4.1 was effectively replaced in Vietnam by clade 2.3.2.1 and has not been isolated from Vietnam since 2010. In 2011-2012, a new PB1-F2 truncation of 25 amino acids arose in the clade 1.1.1 lineage in Vietnam. Since PB1 in the clade 1.1.1 viruses is phylogenetically distinct from that of the 2.3.4.1 lineage (data not shown), this mutation appears to have arisen independently and not via reassortment. To date, these truncations in the clade 1.1.1 lineage have been detected only in viruses from poultry. A number of other viruses containing a 24-/25-aa truncation of PB1-F2 have been detected (Fig. 3), but these appear to have been sporadic mutants with short-lived circulation.
FIG 3.
Evolutionary relationship of PB1-F2 truncations in H5N1 viruses. HA gene sequences of H5N1 viruses were aligned using MAFFT, and a maximum likelihood tree was determined using FastTree. The virus strains having PB1-F2 truncations of 24/25, 57, and 8 aa are colored blue, green, and fuchsia, respectively. Viruses with mutated initiating ATG are colored red. Clades were defined by taking representative sequences from the WHO/OIE/FAO H5N1 Classification Working Group.
A PB1-F2 truncation of 57 amino acids arose in the clade 2.3.2.1b lineage in China and Vietnam in 2010, and this variant has persisted through 2013, with the majority of 2.3.2.1b viruses containing a truncated PB1-F2. Furthermore, in 2012 a reassortment event occurred among 2.3.2.1 viruses circulating in Vietnam, in which viruses with HA from 2.3.2.1c acquired the internal genes including PB1 from 2.3.2.1b viruses (43). Since the 2.3.2.1b PB1 included the 57-aa truncation of PB1-F2, most H5N1 viruses isolated in Vietnam in 2012 and 2013 contained this PB1-F2 variant (Fig. 3 and Table 2). Other sporadic instances of 57-aa truncations of PB1-F2 arose independently in several years, including in two of the 1997 viruses, but were not detected over multiple years.
Viruses in which the initiating ATG for PB1-F2 was mutated were detected sporadically from 2006 to 2012, representing 0.3 to 3% of sequenced H5N1 PB1-F2 (Table 2). Although limited persistence of these variants was observed (e.g., in clade 2.3.4 [Fig. 3]), no lineages of viruses containing these variants were detected for multiple years.
Finally, a number of less common PB1-F2 truncations have been isolated in HPAIV(H5N1), including variants that are 63, 34, 11, and 8 amino acids in length. However, since none of these variants have been isolated in a subgroup of viruses for more than 1 year, we consider them to be sporadic mutations.
Construction of PB1-F2 truncation mutants.
To assess the effects of H5N1 PB1-F2 truncations in vitro, we cloned PB1-F2 from A/chicken/Vietnam/NCVD-296/2009 (VN/296), which lacks the initiating ATG, here called F2NO. We made a mutant by restoring the initiating ATG to express a full-length (90-aa) protein (F2FL) and another mutant by introducing mutations into this gene to produce a 24-aa protein (F2TR) (Fig. 1 and Table 1). Each variant was constructed with and without an HA tag at the C terminus (Fig. 1 and Table 1) to facilitate detection by antibodies. In the case of the 24-aa truncation, variants were constructed in which the HA tag was placed at the C terminus of the anticipated truncated protein (F2HA-TR) or at the C terminus of the full-length (90-aa) protein (F2TR-HA) to detect expression from in-frame initiation sites (Fig. 1 and Table 1). Similar PB1-F2 constructs were made from the paired virus VN/281 (Table 1).
PB1-F2 mutants had no effect on apoptosis.
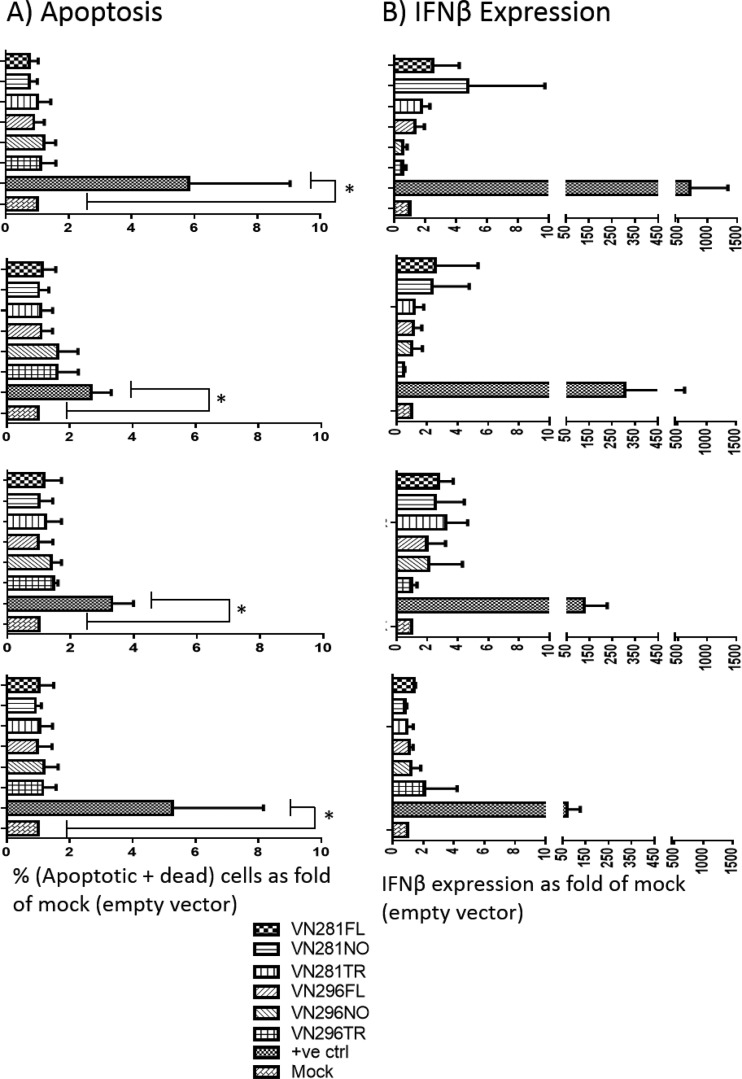
Apoptosis has been associated with expression of PB1-F2 from PR8 and 1918 H1N1 strains (10, 44), but not with PB1-F2 from H5N1, 1957, or 1968 pandemic viruses (10). We transfected A549 (human) and DF1 (chicken) cells with PB1-F2 variants, harvested cells at 12, 24, 36, and 48 h posttransfection, stained for apoptosis and cell death, and analyzed the cells by flow cytometry. Neither the full-length nor the truncated or ATG-less versions of PB1-F2 affected the proportion of apoptotic or dead cells compared to the proportion in control cells (P > 0.05, Kruskal-Wallis test) (Fig. 4A).
FIG 4.
PB1-F2 mutants do not affect cell apoptosis or interferon response. A549 cells were transfected with either empty vector or PB1-F2 mutant plasmids. Cells were harvested at 12 h (top set of two graphs), 24 h (second set from top), 36 h (third set from top), and 48 h (bottom set) posttransfection. (A) Cells were stained using dyes specific for apoptosis and necrosis. Data represent the fold change of numbers of dead and apoptotic cells over CMV2 vector (control). Significant apoptosis induction was detected in positive-control cells (treated with camptothecin) at all time points. (B) IFN-β expression was measured using the TaqMan assay and qRT-PCR. IFN-β expression is normalized to GAPDH (internal control) and presented as fold increase relative to CMV2 vector transfection. For positive control, cells were transfected with triphosphate RNA (TP-RNA) and harvested at the indicated time points. Statistical significance was determined by the Kruskal-Wallis test (all groups) and the Mann-Whitney U test (mock and positive-control pair). Bar graphs represent means, with standard deviations shown as error bars.
PB1-F2 truncations and IFN-β expression.
PB1-F2 from mouse-adapted viruses alters type I interferon production in some cell types (22–24, 28). We transfected A549 and DF1 cells with the PB1-F2 plasmids and measured IFN-β expression at various time points using the TaqMan assay qRT-PCR. We used an empty plasmid vector as a negative control and in vitro-transcribed 5′ triphosphate RNA (TP-RNA) as a positive control for IFN-β induction (41). IFN-β expression levels were normalized to GAPDH and expressed as fold induction over empty-vector transfection. Treatment with TP-RNA consistently induced IFN-β expression at least 100-fold over CMV2 vector transfection. However, transfection with full-length or truncated PB1-F2 did not significantly alter IFN-β expression in A549 cells and DF1 cells (P > 0.05, Kruskal-Wallis test) (Fig. 4B).
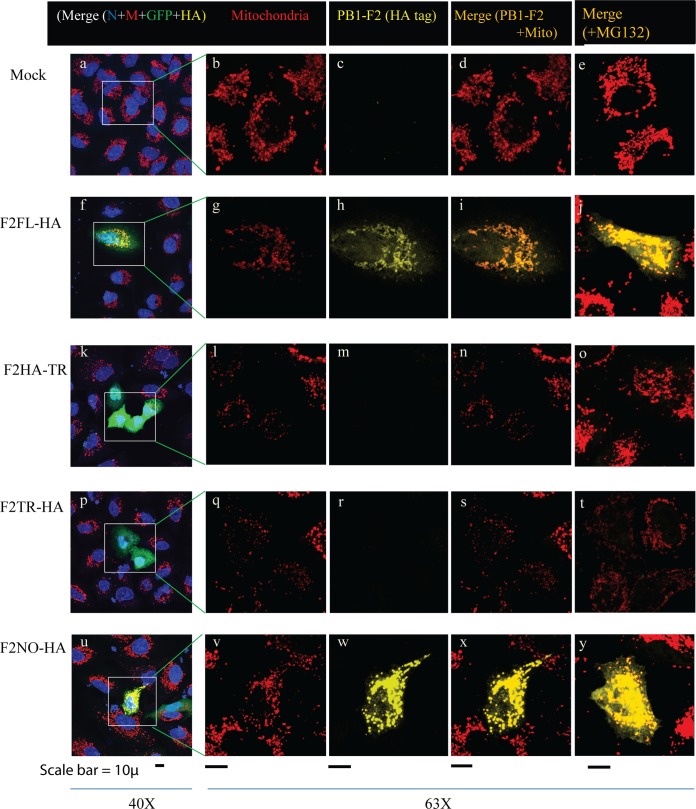
Full-length and C-terminal PB1-F2 from H5N1 viruses localize to mitochondria.
We evaluated the expression and cellular colocalization of each of the PB1-F2 variants containing a C-terminal HA epitope tag after transient transfection in A549 cells and using immunofluorescence. Since the plasmids expressing PB1-F2 also expressed GFP under a separate promoter, we limited analysis to cells expressing GFP (green). Confocal microscopy revealed that the full-length PB1-F2 proteins were expressed and readily detected (Fig. 5). The full-length PB1-F2 localized with cell mitochondria in A549 cells (Fig. 5f to j). The truncated 24-amino-acid version of PB1-F2 was not detectable (Fig. 5k to o) even after treating the cells with the proteasome inhibitor MG-132 (Fig. 5o). Interestingly, the presence of an HA-tagged protein was detectable by confocal microscopy in cells transfected with the ATG-less version of PB1-F2; this was expressed at levels comparable to that of full-length PB1-F2 and also localized to mitochondria (Fig. 5u to y). Since the HA epitope tag is in frame and at the C terminus (Fig. 1), this protein expression presumably originated from an in-frame ATG (residue number 39, 46, or 51 [Fig. 1]) downstream of the authentic start site, leading to expression of an N-terminally truncated version of the protein (c-terminal PB1-F2), as previously described (13). Alternatively, expression of full-length PB1-F2 using ACG as an initiation codon is possible. However, Western blotting assays did not detect expression of a full-length PB1-F2 in cells transfected with this plasmid (data not shown). No protein expression was detected in two truncated variants in which the initiating ATG was intact and two stop codons were inserted at positions 25 and 26, one of which had an HA tag at position 25 (F2HA-TR) and the other had an HA tag at the C terminus (F2TR-HA).
FIG 5.
PB1-F2 expression and cellular localization. A549 cells were transfected with either control plasmids (vector) or PB1-F2-HA plasmids. In some cases (e, j, o, t, y), MG132 (proteasome inhibitor) was added to the medium at 30 h posttransfection for 6 h. Thirty-six hours posttransfection, cells were fixed, permeabilized, and stained using anti-HA antibody (yellow), anti-MTCO2 (mitochondrially encoded cytochrome c oxidase II) (red, to indicate mitochondria), or DAPI (blue, nucleus) and examined by confocal microscopy. Transfected cells were identified by GFP expression (green), expressed under a separate promoter on the plasmid vector. (a to e) Untransfected cells as negative control; (f to j) F2FL-HA; (k to o) F2HA-TR (N-terminal 24 aa); (p to t) F2TR-HA (C terminus); (u to y) F2NO-HA (initiating ATG to ACG). The first column shows the merged images of all stains at low resolution. The 2nd and 3rd columns show mitochondria and HA (marker for PB1-F2), respectively. The 4th and 5th columns show the merge of mitochondria and PB1-F2 in the absence and presence of MG132, respectively.
Paired viruses for PB1-F2 studies.
Although several studies have tested the effects of PB1-F2 truncations on mouse virulence, these studies introduced stop codons into the PB1-F2 open reading frame (ORF) instead of, or as well as, mutating the initiating ATG (10, 11, 13, 25). Since confocal microscopy suggested that mutating the start codon, but not introducing stop codons, allowed abundant expression of a protein from the PB1-F2 ORF, we tested the virulence of H5N1 viruses with full-length or ATG-less PB1-F2 in mice. VN/281 and VN/296 are closely related clade 2.3.4.2 H5N1 viruses, isolated from chickens in Vietnam, with disparate PB1-F2 sequences. VN/281 has full-length PB1-F2, while VN/296 has an ATG-to-ACG mutation in the PB1-F2 start codon. Aside from PB1-F2, these viruses differ by only 16 of a total of 4,440 amino acids across the viral proteome (0.34%) (summarized in Table 3). None of these amino acid differences are known determinants of pathogenicity in H5N1 viruses (45).
TABLE 3.
Amino acid differences between viruses VN/281 and VN/296a
| Protein | Size (aa) | No. of variations | Variant residues (VN/281→VN/296) |
|---|---|---|---|
| HA | 567 | 1 | Q31R |
| M1 | 252 | 0 | |
| M2 | 97 | 2 | G21V, L86V |
| NA | 449 | 4 | I63V, G85S, I203T, T244I |
| NP | 498 | 1 | A373T |
| NS1 | 225b | 1 | D120G |
| NS2 | 121 | 0 | |
| PA | 716 | 2 | S66G, A323V |
| PB1 | 756 | 3 | I181M, P369S, V667I |
| PB1-F2 | 90c | 3 | M1T, Q54R, P67L |
| PB2 | 759 | 2 | D304E, A588V |
| Total | 4,530 | 19 | |
| Total (excluding PB1-F2) | 4,440 | 16 |
Amino acid differences for each protein were calculated from the nucleotide sequences of the respective ORFs using the Cubit application in BioEdit software.
Both viruses have a C-terminal truncation in NS1 that renders it 215 aa long.
PB1-F2 is 90 aa in virus VN/281, while the initiating ATG is mutated to ACG in virus VN/296. The 3 differences mentioned in the table are counted since a protein may express either from alternating start codons or from downstream in-frame ATGs.
Viruses VN/281 and VN/296 replicated to similar titers in eggs and cells, as measured by 50% egg infective dose (EID50)/ml (109.5 and 108.8, respectively) and plaque assay (108.9 and 108.4 PFU/ml, respectively).
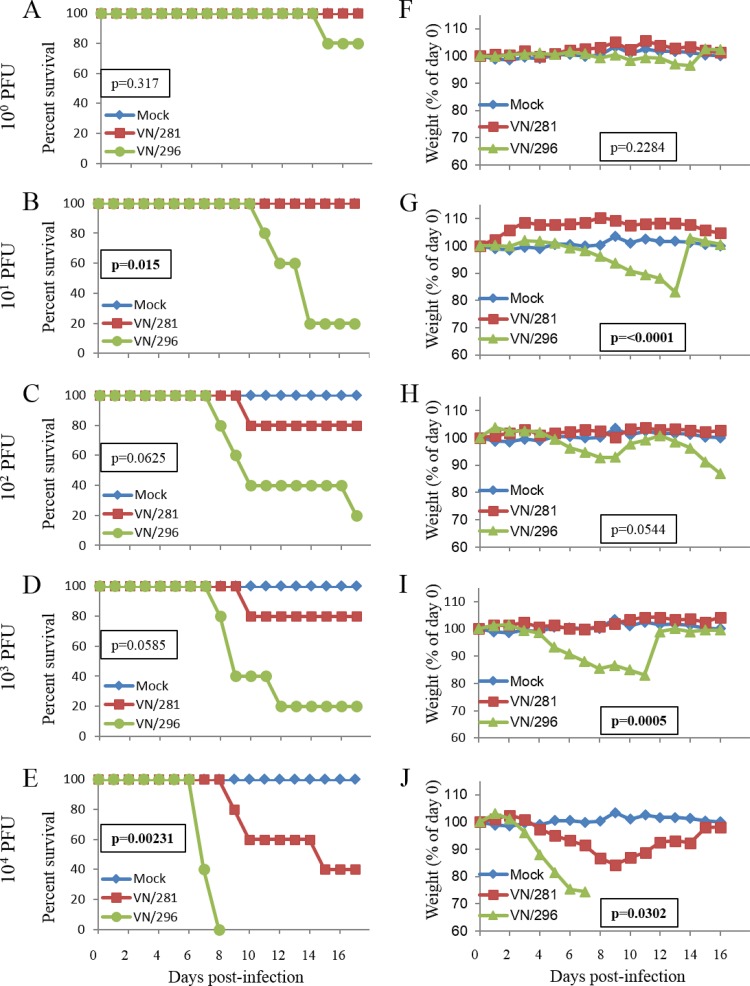
The absence of PB1-F2 initiating ATG is associated with increased virulence in mice.
Mice were infected with various doses (100 to 104 PFU) of either VN/281 or VN/296. The two viruses showed marked differences in their virulence in mice. Mice infected with VN/281 (full-length PB1-F2) showed weight loss only at the highest dose (104 PFU) and yielded a 50% mouse lethal dose (MLD50) of 104.3 PFU (Fig. 6). In contrast, a majority of mice infected with VN/296 virus (lacking the start codon for PB1-F2) showed marked weight loss at all doses of >100. VN/296 was much more lethal than VN/281, with the 50% lethal dose (LD50) of virus VN/296 (101.4 PFU) being approximately 1,000-fold lower than that of the VN/281 virus.
FIG 6.
Mouse virulence of paired viruses with and without PB1-F2. Five mice each were infected intranasally with VN/281 or VN/296 at various doses as indicated on the left or with PBS as a control. Average survival (A to E) and body weight (F to J) of mice are shown as percentages of day 0 values. The statistical significance of the difference between the VN/281 and VN/296 groups (survival, log rank test; body weight, linear mixed model with repeated measures) is indicated on each chart.
DISCUSSION
Previous studies have shown that the great majority of avian influenza viruses contain a full-length PB1-F2 protein, whereas mammalian viruses frequently have premature stop codons resulting in truncation (33, 34). Incorporating more-recent sequences obtained from influenza virus sequence databases, we found a similar picture. In mammalian viruses (swine and human), PB1-F2 truncations (≤78 aa) are common (approximately 63% of sequences in the database). In contrast, in avian viruses over 95% of PB1-F2 sequences are full length. An important exception to this is in the highly pathogenic H5N1 virus group, in which the frequency of PB1-F2 truncations in databases increased to 61% in 2013. In addition, non-H5N1 avian viruses in general also showed an increase in PB1-F2 truncation frequency in 2013, to about 20%. This increase was due mainly to H7N9 and H9N2 AIV in China, in which 24% and 21% of PB1-F2, respectively, were truncated.
It is important to note that these percentages are inevitably skewed due to sampling bias. For example, human H1N1pdm viruses (almost all of which have truncated PB1-F2) were intensively sequenced in 2009. Similarly, the published sequences of H5N1 internal genes are dominated by those from Vietnam and China, where PB1-F2 truncations are increasingly common in endemic, circulating viruses. Nevertheless, the frequent appearance and evolutionary success of viruses bearing truncations in PB1-F2 appear to be a recent phenomenon.
Although multiple studies have demonstrated a role for PB1-F2 in viral virulence (10, 11, 13, 25), the molecular mechanisms involved are not clear. Most studies on the effect of PB1-F2 on virulence have used mouse-adapted strains of H1N1 viruses, e.g., A/Puerto Rico/08/1934(H1N1) (PR8) and WSN/33. The effects of PB1-F2 are dependent on genomic context and the presence of other virulence factors (10, 11, 16–18). Only two studies have looked at the effect of PB1-F2 on H5N1 pathogenicity, and both found that PB1-F2 truncations slightly reduced lethality in mice (12, 46).
Although PB1-F2 has been shown to affect apoptosis, most studies on PB1-F2 effects have used the gene from the mouse-adapted strain PR8 or WSN/33, and the limited studies on PB1-F2 from H5N1 virus have not supported a role in apoptosis (44). Similarly, PB1-F2 in H5N1 was not found to affect IFN levels in most cell types (12). In full-length PB1-F2, the presence of serine rather than arginine at position 66 (N66S) is associated with increased virulence and inhibition of IFN (21, 24, 47), while the PB1-F2 used in our experiments has 66N. It is important to note that the proapoptotic and IFN-β-regulatory roles of PB1-F2 are specific for both virus strain and cell type. These effects have been most consistently (though not solely) seen with PB1-F2 from lab-adapted viruses such as A/Puerto Rico/8/1934 and A/WSN/1933 cells of immune origin. However, PB1-F2 from H5N1 viruses has shown no effect either on IFN-β expression (12) or on apoptosis (44), even in cells of immune origin. Consistent with these observations, in our experiments neither full-length, truncated, nor ATG-mutated versions of PB1-F2 from H5N1 affected apoptosis or IFN-β expression in human or chicken cells.
Previous studies on the effect of PB1-F2 truncation or deletion have modified PB1-F2 by mutating multiple downstream ATGs along with the 1st start codon (12, 28, 46), by introducing a stop codon after the last in-frame ATG (13) or by both mutating the 1st start codon and inserting a stop codon at position 11 (10, 11, 20, 25). Importantly, we observed abundant expression of an in-frame C-terminal fragment of PB1-F2 (as previously described by Zamarin et al. [13]) from a plasmid lacking the initiating ATG but without downstream stop codons, as seen in natural H5N1 isolates. In contrast, only very low level, or no, expression of in-frame proteins was detected from PB1-F2 plasmids containing the initiating ATG with premature stop codons (Fig. 5p to t). This C-terminal fragment presumably was not expressed in the previous studies that inserted one or more stop codons into the open reading frame. This C-terminal fragment, like the full-length PB1-F2, localized to mitochondria, consistent with the mitochondrial targeting signal in PB1-F2 being entirely present in the C-terminal region (9, 48).
To test the effect on virulence of the start codon mutation, we used a pair of naturally occurring viruses, VN/281 and VN/296, which differ by only 0.34% (differences in 16 amino acids of 4,440 in the proteome). Although the viruses replicated similarly in eggs and in MDCK cells, their MLD50 differed markedly, with VN/281 (full-length PB1-F2) being 1,000-fold less pathogenic than VN/296 (ATG-less PB1-F2). Thus, while complete deletion of PB1-F2 was found to reduce H5N1 virulence (12, 46), expression of the C-terminal fragment instead of the full-length protein is correlated with increased lethality in mice.
Previous studies have raised the concern that avian viruses containing truncated PB1-F2 may be “preadapted” to mammalian infection. Despite the increased prevalence of PB1-F2 truncated H5N1 mutants (i.e., clade 2.3.2.1 viruses in Vietnam and China), these viruses continue to circulate in poultry populations and cause outbreaks, suggesting that viral replication is not dramatically altered in the avian host. Expression of a C-terminal version of PB1-F2 may be associated with increased virulence. Further experiments will be needed to determine the mechanism(s) underlying the differential virulence identified, since no effects on apoptosis and type I IFN induction were observed. The study justifies continued monitoring of changes in the prevalence of H5N1 viruses carrying truncated PB1-F2 proteins, including those containing start codon deletions, for their public health significance.
Supplementary Material
ACKNOWLEDGMENTS
We thank the CDC Animal Resource Branch for excellent animal care. We thank Vilma Yuzbasiyan-Gurkan and administrators of the Comparative Medicine and Integrative Biology program at Michigan State University for their unfailing support of R.P.K. during his Ph.D. program.
We thank ORISE for supporting R.P.K. as a guest researcher at CDC.
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03137-14.
REFERENCES
- 1.OIE. 26 February 2015. Update on highly pathogenic avian influenza in animals (type H5 and H7). Accessed 4 March 2015 http://www.oie.int/animal-health-in-the-world/update-on-avian-influenza/2015/.
- 2.Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 3.Duan L, Bahl J, Smith GJ, Wang J, Vijaykrishna D, Zhang LJ, Zhang JX, Li KS, Fan XH, Cheung CL, Huang K, Poon LL, Shortridge KF, Webster RG, Peiris JS, Chen H, Guan Y. 2008. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology 380:243–254. doi: 10.1016/j.virol.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen T, Rivailler P, Davis CT, Hoa do T, Balish A, Dang NH, Jones J, Vui DT, Simpson N, Huong NT, Shu B, Loughlin R, Ferdinand K, Lindstrom SE, York IA, Klimov A, Donis RO. 2012. Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology 432:405–416. doi: 10.1016/j.virol.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 5.WHO. 26 January 2015. Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO, 2003-2015. Accessed 2 March 2015 http://www.who.int/influenza/human_animal_interface/EN_GIP_20150126CumulativeNumberH5N1cases.pdf?ua=1.
- 6.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 7.Kawaoka Y, Krauss S, Webster RG. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol 63:4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 77:7214–7224. doi: 10.1128/JVI.77.13.7214-7224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. 2010. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog 6:e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmolke M, Manicassamy B, Pena L, Sutton T, Hai R, Varga ZT, Hale BG, Steel J, Perez DR, Garcia-Sastre A. 2011. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog 7:e1002186. doi: 10.1371/journal.ppat.1002186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamarin D, Ortigoza MB, Palese P. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan MA, Gramer MR, Goyal SM, Sreevatsan S. 2009. A Serine12Stop mutation in PB1-F2 of the 2009 pandemic (H1N1) influenza A: a possible reason for its enhanced transmission and pathogenicity to humans. J Vet Sci 10:349–351. doi: 10.4142/jvs.2009.10.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Molbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 16.Solbak SM, Sharma A, Bruns K, Roder R, Mitzner D, Hahn F, Niebert R, Vedeler A, Henklein P, Henklein P, Schubert U, Wray V, Fossen T. 2013. Influenza A virus protein PB1-F2 from different strains shows distinct structural signatures. Biochim Biophys Acta 1834:568–582. doi: 10.1016/j.bbapap.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Pena L, Vincent AL, Loving CL, Henningson JN, Lager KM, Lorusso A, Perez DR. 2012. Restored PB1-F2 in the 2009 pandemic H1N1 influenza virus has minimal effects in swine. J Virol 86:5523–5532. doi: 10.1128/JVI.00134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pena L, Vincent AL, Loving CL, Henningson JN, Lager KM, Li W, Perez DR. 2012. Strain-dependent effects of PB1-F2 of triple-reassortant H3N2 influenza viruses in swine. J Gen Virol 93:2204–2214. doi: 10.1099/vir.0.045005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, Ludwig S. 2008. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol 10:1140–1152. doi: 10.1111/j.1462-5822.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 21.Conenello GM, Tisoncik JR, Rosenzweig E, Varga ZT, Palese P, Katze MG. 2011. A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J Virol 85:652–662. doi: 10.1128/JVI.01987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudek SE, Wixler L, Nordhoff C, Nordmann A, Anhlan D, Wixler V, Ludwig S. 2011. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol Chem 392:1135–1144. [DOI] [PubMed] [Google Scholar]
- 23.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. 2011. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog 7:e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varga ZT, Grant A, Manicassamy B, Palese P. 2012. Influenza virus protein PB1-F2 inhibits the induction of type i interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol 86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meunier I, von Messling V. 2012. PB1-F2 modulates early host responses but does not affect the pathogenesis of H1N1 seasonal influenza virus. J Virol 86:4271–4278. doi: 10.1128/JVI.07243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reis AL, McCauley JW. 2013. The influenza virus protein PB1-F2 interacts with IKKbeta and modulates NF-kappaB signalling. PLoS One 8:e63852. doi: 10.1371/journal.pone.0063852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, Latz E, Brown LE, Mansell A. 2013. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog 9:e1003392. doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, Leymarie O, Bourdieu C, Decamps L, Dhorne-Pollet S, Delmas B. 2010. Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells. J Immunol 185:4812–4823. doi: 10.4049/jimmunol.0903952. [DOI] [PubMed] [Google Scholar]
- 29.Weeks-Gorospe JN, Hurtig HR, Iverson AR, Schuneman MJ, Webby RJ, McCullers JA, Huber VC. 2012. Naturally occurring swine influenza A virus PB1-F2 phenotypes that contribute to superinfection with Gram-positive respiratory pathogens. J Virol 86:9035–9043. doi: 10.1128/JVI.00369-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AM, Adler FR, Ribeiro RM, Gutenkunst RN, McAuley JL, McCullers JA, Perelson AS. 2013. Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog 9:e1003238. doi: 10.1371/journal.ppat.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber VC. 2012. Can surveillance of the influenza virus PB1-F2 gene be used to predict the severity of secondary bacterial infections? Virulence 3:523–524. doi: 10.4161/viru.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alymova IV, Green AM, van de Velde N, McAuley JL, Boyd KL, Ghoneim HE, McCullers JA. 2011. Immunopathogenic and antibacterial effects of H3N2 influenza A virus PB1-F2 map to amino acid residues 62, 75, 79, and 82. J Virol 85:12324–12333. doi: 10.1128/JVI.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zell R, Krumbholz A, Eitner A, Krieg R, Halbhuber KJ, Wutzler P. 2007. Prevalence of PB1-F2 of influenza A viruses. J Gen Virol 88:536–546. doi: 10.1099/vir.0.82378-0. [DOI] [PubMed] [Google Scholar]
- 34.Pasricha G, Mishra AC, Chakrabarti AK. 2013. Comprehensive global amino acid sequence analysis of PB1F2 protein of influenza A H5N1 viruses and the influenza A virus subtypes responsible for the 20th-century pandemics. Influenza Other Respir Viruses 7:497–505. doi: 10.1111/j.1750-2659.2012.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. 2014. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses 8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut A, Drummond A. 2014. FigTree v. 1.4.2 ed. http://tree.bio.ed.ac.uk/software/figtree/.
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hygiene 27:493–497. [Google Scholar]
- 40.Chosewood LC, Wilson DE (ed). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed U.S. Department of Health and Human Services publication no. (CDC) 21-1112. U.S. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/biosafety/publications/bmbl5/BMBL.pdf. [Google Scholar]
- 41.Ranjan P, Jayashankar L, Deyde V, Zeng H, Davis WG, Pearce MB, Bowzard JB, Hoelscher MA, Jeisy-Scott V, Wiens ME, Gangappa S, Gubareva L, Garcia-Sastre A, Katz JM, Tumpey TM, Fujita T, Sambhara S. 2010. 5′PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol J 7:102. doi: 10.1186/1743-422X-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health 61:4–17. doi: 10.1111/zph.12049. [DOI] [PubMed] [Google Scholar]
- 43.Creanga A, Thi Nguyen D, Gerloff N, Thi Do H, Balish A, Dang Nguyen H, Jang Y, Thi Dam V, Thor S, Jones J, Simpson N, Shu B, Emery S, Berman L, Nguyen HT, Bryant JE, Lindstrom S, Klimov A, Donis RO, Davis CT, Nguyen T. 2013. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology 444:12–20. doi: 10.1016/j.virol.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Chen CJ, Chen GW, Wang CH, Huang CH, Wang YC, Shih SR. 2010. Differential localization and function of PB1-F2 derived from different strains of influenza A virus. J Virol 84:10051–10062. doi: 10.1128/JVI.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CDC. 2012. H5N1 genetic changes inventory: a tool for influenza surveillance and preparedness. http://www.cdc.gov/flu/pdf/avianflu/h5n1-inventory.pdf.
- 46.Leymarie O, Jouvion G, Herve PL, Chevalier C, Lorin V, Lecardonnel J, Da Costa B, Delmas B, Escriou N, Le Goffic R. 2013. Kinetic characterization of PB1-F2-mediated immunopathology during highly pathogenic avian H5N1 influenza virus infection. PLoS One 8:e57894. doi: 10.1371/journal.pone.0057894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada H, Chounan R, Higashi Y, Kurihara N, Kido H. 2004. Mitochondrial targeting sequence of the influenza A virus PB1-F2 protein and its function in mitochondria. FEBS Lett 578:331–336. doi: 10.1016/j.febslet.2004.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.