ABSTRACT
Influenza A viruses (IAV) replicate their segmented RNA genome in the nucleus of infected cells and utilize caspase-dependent nucleocytoplasmic export mechanisms to transport newly formed ribonucleoprotein complexes (RNPs) to the site of infectious virion release at the plasma membrane. In this study, we obtained evidence that apoptotic caspase activation in IAV-infected cells is associated with the degradation of the nucleoporin Nup153, an integral subunit of the nuclear pore complex. Transmission electron microscopy studies revealed a distinct enlargement of nuclear pores in IAV-infected cells. Transient expression and subcellular accumulation studies of multimeric marker proteins in virus-infected cells provided additional evidence for increased nuclear pore diameters facilitating the translocation of large protein complexes across the nuclear membrane. Furthermore, caspase 3/7 inhibition data obtained in this study suggest that active, Crm1-dependent IAV RNP export mechanisms are increasingly complemented by passive, caspase-induced export mechanisms at later stages of infection.
IMPORTANCE In contrast to the process seen with most other RNA viruses, influenza virus genome replication occurs in the nucleus (rather than the cytoplasm) of infected cells. Therefore, completion of the viral replication cycle critically depends on intracellular transport mechanisms that ensure the translocation of viral ribonucleoprotein (RNP) complexes across the nuclear membrane. Here, we demonstrate that virus-induced cellular caspase activities cause a widening of nuclear pores, thereby facilitating nucleocytoplasmic translocation processes and, possibly, promoting nuclear export of newly synthesized RNPs. These passive transport mechanisms are suggested to complement Crm1-dependent RNP export mechanisms known to occur at early stages of the replication cycle and may contribute to highly efficient production of infectious virus progeny at late stages of the viral replication cycle. The report provides an intriguing example of how influenza virus exploits cellular structures and regulatory pathways, including intracellular transport mechanisms, to complete its replication cycle and maximize the production of infectious virus progeny.
INTRODUCTION
Influenza A viruses (IAV) are RNA viruses that belong to the Orthomyxoviridae family. The viruses pose a major threat to human health, causing severe and potentially fatal respiratory disease if the infection proceeds to the lower airways (1–3). The IAV genome is comprised of eight single-stranded RNA segments of negative polarity (viral RNA [vRNA]), which together encode at least 10 proteins. Viral genome replication and transcription occur in the nucleus of the infected cell and are mediated by a heterotrimeric RNA-dependent RNA polymerase (RdRp) complex comprised of subunits PB1, PB2, and PA. The combination of vRNA with the RdRp and the nucleoprotein (NP) forms the viral ribonucleoprotein complex (RNP). Viral genome replication and transcription take place in the nucleus and therefore depend on regulated bidirectional transport of RNPs across the nuclear envelope. After virus entry via endocytosis and fusion of viral and endosomal membranes, viral RNPs (vRNPs) are released into the cytoplasm and subsequently imported into the nucleus. Following viral replication in the nucleus, newly synthesized RNPs are exported to the cytoplasm, where they reach the viral assembly sites at the plasma membrane (for a recent review, see references 2 and 4).
It is now well established that IAV infection induces apoptosis in cultured epithelial cells and leukocytes, as well as in murine and human pulmonary cells in vivo (5). Apoptosis is an important, signal-mediated form of programmed cell death that is commonly observed in virus-infected cells (6). A variety of cellular signaling pathways are known to be involved in the initiation of the apoptotic cascade in IAV-infected cells (for a review, see reference 5). In the past, apoptosis was generally considered an important host cell defense mechanism and, in accordance with that assumption, IAV and many other viruses have evolved proteins that modulate or counteract this antiviral cellular response (7–10). Interestingly, there is now increasing evidence that IAV may also benefit from proapoptotic signaling, for example, by increasing viral replication efficiency (reviewed in references 5 and 11).
IAV-infected cells, e.g., human alveolar epithelial cells or alveolar macrophages, release proinflammatory and proapoptotic mediators, such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and FasL (12, 13). These mediators bind to their cognate receptors and induce the assembly of the death-inducing signaling complex (DISC) (14). The adaptor molecule, Fas-associated death domain (FADD), translocates to the DISC and recruits procaspases 8 and 10, resulting in their autocatalytic activation. Activated initiator caspases 8 and 10 then trigger a downstream cascade by proteolytic activation of procaspases 3 and 7. Alternatively, these effector caspases can be activated by the mitochondrial amplification loop, also called the intrinsic or Bcl-2-controlled apoptosis pathway. Activation of effector caspases 3/7 leads to cleavage of specific target proteins, to DNA fragmentation, and, finally, to cell death (reviewed in reference 15). Caspases have a variety of cellular substrates. Of special interest in the context of this study is the role of caspases in the degradation of nuclear pore proteins and removal of the nuclear basket that controls active export functions of the nuclear pore complex (NPC) (16). NPCs are large, 125-MDa protein assemblies comprised of more than 30 proteins called nucleoporins (Nups) that together form an octagonal symmetrical structure (17–20). The NPC forms a selective barrier. Small (up to 40-kDa) molecules may shuttle between the nucleus and cytoplasm by passive diffusion, while larger molecules require active transport mechanisms involving specific signal sequences (21, 22).
Previous studies have shown that caspase inhibitors (CI) reduce the production of infectious IAV progeny, with nuclear retention of viral RNP complexes playing a key role in this inhibition (23). A subsequent study showed that NF-κB-dependent induction of TRAIL and FasL enhances IAV replication (24), while, in similarity to the CI data mentioned above, inhibition of NF-κB causes nuclear retention of RNPs. Based on these data, it was suggested that IAV-induced NF-κB activation can lead to auto- and paracrine caspase activation via TRAIL and FasL, which, in turn, might affect NPC function and thereby promote nuclear RNP export (11). To date, however, the underlying structural and functional changes that are triggered by IAV-induced caspase activities and thereby support IAV replication have not been resolved. Also, these observations need to be reconciled with earlier studies showing that nuclear RNP export depends on Crm1, indicating active rather than passive transport mechanisms (25). Inhibition of Crm1 via Leptomycin B (LepB) was previously shown to result in the retention of IAV RNPs in the nucleus, similarly to what was observed in CI-treated cells (26, 27). Crm1 (also called exportin-1) is a member of the importin-β family and is critically involved in the transport of a variety of proteins across the nuclear envelope in both directions (26, 28–30). Notably, Crm1 interacts in a RanGTP-dependent manner with the nucleoporin Nup153, which is a caspase substrate (31). Furthermore, it was also shown that Crm1 is associated with caspase substrate Nup214 (27, 32).
In this report, we provide evidence that, at late stages of the viral infection cycle, IAV-induced caspase activation causes a widening of nuclear pores, thereby facilitating the passive translocation of large protein complexes across the nuclear membrane. Our data support the idea that, at later stages of the replication cycle, IAV employ an additional mode of RNP exit from the nucleus which complements active Crm1-dependent export mechanisms (26) and ensures efficient production of infectious virus progeny.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney II (MDCK-II) and human lung epithelial cells (A549) were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen). The MCF-7 human mammary gland epithelial cell line was grown in RPMI 1640 medium (PAA), and quail fibroblast cell line QT6 was grown in Ham's F10 (c-c-pro), all containing 10% fetal calf serum (FCS; PAA) and antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin [P/S]). Ham's F10 was additionally supplemented with 2 mM l-glutamine (Invitrogen), 1% chicken serum (Sigma), and 2% tryptose phosphate broth (Sigma). MDCK-I cells were grown in minimum essential medium with Earle's salts (MEM; c-c-pro) containing 10% FCS, P/S, and 2 mM l-glutamine. The permanent cell lines were all obtained from the cell culture collection of the Institute of Medical Virology, Giessen.
The following IV strains were used: A/FPV/Bratislava/79 (FPV, H7N7), A/Hamburg/01/09 (H1N1pdm09, H1N1), B/Lee/40, and C/Johannesburg/1/66. All strains were obtained from the IV strain collection at the Institute of Medical Virology, Giessen. Prior to infection, cells were washed with phosphate-buffered saline (PBS) containing 1 mM MgCl2 and 0.9 mM CaCl2 (PBS++) and then inoculated at room temperature (rt) with the appropriate virus diluted in PBS containing 0.2% bovine albumin (BA), 1 mM MgCl2, 0.9 mM CaCl2, and P/S (PBS-BA). After 1 h, the inoculum was replaced with the appropriate medium containing 0.2% BA, P/S, and, if indicated, specific inhibitors (40 μM caspase 3/7 inhibitor, Z-DEVD-FMK [CI], R&D Systems; 2 ng/ml Leptomycin B [LepB], Sigma; 2 μg/ml actinomycin D [ActD], Sigma). Virus titers were determined by a focus assay, using a mouse anti-IAV NP antibody (Ab) (provided by S. Ludwig, Münster, Germany) as described previously (33).
hAEC.
Human lung tissue was obtained from lobectomy specimens distal from tumors. Human alveolar epithelial cells (hAEC) were isolated as described previously (34) and maintained in Ham's F12 medium (Biochrom) supplemented with 10% FCS and antibiotics for 5 days. The purity of human AEC was assessed using anti-CD45, anti-CD326/EpCam, and anti-pro-SP-C antibodies (Biolegend; Millipore). Cell suspensions with 90% purity were used for further experiments.
Western blotting and activity measurement.
For protein detection via the use of an Odyssey system (Li-Cor), cell lysates were separated on 2 precast gradient NuPAGE Novex 4% to 12% Bis-Tris protein gels (Invitrogen, Germany) and transferred onto Immobilon-FL polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Germany) at 150 V/cm2. Following protein transfer, PVDF membranes were blocked using 1× TBS blocking buffer (20 mM Tris-HCl [pH 7.6], 140 mM NaCl, 5% nonfat dry milk) for 1 h at rt. The membranes were washed once using washing buffer (1× TBS-Tween [TBS containing 0.05% Tween 20]). Afterward, detection of Nup153 protein and cellular beta actin was achieved using mouse anti-Nup153 (QE5, detecting Nup153, Nup214, and p62; Abcam) (1:10,000), and rabbit anti-beta actin monoclonal antibody (Abcam) diluted in blocking buffer at a dilution of 1:10,000. One hour later, the membranes were washed three times with washing buffer, incubated with secondary goat anti-mouse IRDye 800 and goat anti-rabbit IRDye 680 (1:10,000 dilution in blocking buffer containing SDS [1:1,000 dilution of a 10% stock solution]), and added to the membranes in the dark for 1 h. After three washes with washing buffer and one with 1× TBS, the proteins were visualized using an Odyssey infrared imaging system and the corresponding application software package (Li-Cor, Germany). To quantify the amount of the protein, basic Quantity One software (Bio-Rad, Germany) was used. Caspase 3 activity was measured using a caspase 3 colorimetric assay kit (Genscript).
Immunofluorescence, CLSM, and SRM.
Cells were seeded on 8 chamber culture slides (BD Falcon) or, for superresolution microscopy (SRM), on high-precision cover slides (Marienfeld). Following infection, cells were washed with PBS++ and fixed with 3.7% paraformaldehyde (PFA)–PBS for 1 h at rt. For confocal laser scanning microscopy (CLSM) and SRM, cells were permeabilized for 45 and 15 min, respectively, using PBS containing 1% Triton X-100. For SRM, the fixed cells were incubated for another 30 min in PBS containing 5% rabbit serum (Sigma). Incubation was done in PBS containing 3% bovine serum albumin (BSA) at rt for 1 h or overnight at 4°C as appropriate with the following primary antibodies: mouse anti-IAV NP (strain FPV monoclonal Ab [MAb], clone 1331 [Biodesign; 1:1,000]; mouse anti-IBV NP MAb [Thermo Scientific; 1:100]; mouse anti-ICV NP, clone F17 [1:5,000]; mouse anti-NPC proteins; Mab414 [recognizing related proteins of the nucleoporin Nup84/Nup107 family] [Covance; 1:300]; mouse anti-Nup153 [QE5] [Abcam; 1:50]; and goat anti-IAV NP [R. Webster, Memphis, TN; 1:800]). Cells were washed with PBS and incubated for 1 h with the following secondary antibodies: Alexa Fluor 594-coupled F(ab′)2 fragment goat anti-mouse antibody (Molecular Probes; 1:200), rabbit anti-mouse Alexa 488 (Life Technologies; 1:4,000), and rabbit anti-goat Alexa 555 (Life Technologies; 1:8,000). Washed cells were stained with DAPI (4′,6-diamidino-2-phenylindole) (Roth; 1:400) for 10 min. Finally, cells were washed in PBS and water, embedded in 0.13 M Tris-HCl (Roth) containing 9.1% Mowiol (Sigma-Aldrich), 22.7% glycerol, and 2.5% DABCO (1,4-diazabicyclo[2.2.2] octane; Merck), and analyzed using a confocal laser scanning microscope (Leica TCS SP5). Superresolution imaging of nuclear pores was performed by structured illumination microscopy (SIM) (Zeiss Elyra, Germany) (objective, Plan Apochromat 63×/1.40 oil differential interference contrast [DIC] M27). Surface rendering for three-dimensional (3D) projections was done using ZEN 2011 software (Carl Zeiss). Minimal image adjustment for each channel was done only when necessary for illustration. No gamma corrections were applied, and all images were acquired using identical settings.
Transfection.
MDCK-II cells were seeded on 8-chamber culture slides (BD Falcon) and transfected with 2 μg of either p3xEGFP-NLS (3xGFP-NLS) or pEGFP-GFP5x (5xGFP) using jetPRIME Polyplus transfection (Peqlab) reagents and protocol. At 36 h posttransfection (p.t.), the cells were either infected with strain FPV (± 40 μM CI) or treated with 2 μg/ml actinomycin D (± 40 μM CI). Fixation, permeabilization, and immunostaining were done as described above.
Electron microscopy and immunogold labeling.
MDCK-II cells grown to 70% confluence were infected with FPV (multiplicity of infection [MOI] = 15) or mock infected. At 4 to 10 h p.i., cells were washed with PBS, detached using Accutase (PAA) for 20 min at 37°C and 5% CO2, and washed again in PBS.
For ultrastructural analysis, cells were fixed overnight at 4°C in 1.25% glutaraldehyde (Roth)–PBS and then for 1 h at rt in 2.5% glutaraldehyde–PBS. After washing and embedding in gelatin (Fluka) were performed, cells were postfixed in 1% osmium tetroxide (Serva), washed, and incubated in 2% aqueous uranyl acetate (Polysciences) overnight at 4°C. Specimens were dehydrated in an ethanol series, followed by propylene oxide (Serva), and embedded in Epon 812 (Serva). From the blocks (cured by heat), ultrathin sections were cut and subsequently contrasted in uranyl acetate (Merck) and lead citrate (Merck) (35).
For immunogold labeling, cells were fixed in 1% paraformaldehyde (PFA; Serva) at 4°C overnight and then in 4% PFA–PBS at rt for 1 h. After they were washed, cells were dehydrated and embedded in LR White (London Resin Company Ltd.) (chemical curing on ice). Ultrathin sections from these specimens were etched with 2% periodic acid (Merck) for 10 min prior to immunogold labeling. For quenching of aldehyde groups, hydrophobic moieties, and positive charges, grids were floated on 0.05 M glycine (Serva)–PBS for 45 min and then for 30 min on blocking solution for goat immune reagents containing normal goat serum, bovine serum albumin, cold-water fish skin gelatin, and PBS (Aurion). Next, grids were rinsed in 0.1% BSA-c (Aurion)–PBS and incubated overnight at 4°C in anti-mouse Nup153 (QE5; Abcam) (1:600) containing 0.1% BSA-c. After repeated washing in 0.1% BSA-c–PBS, specimens were incubated with 6-nm-diameter-gold-particle-labeled goat anti-mouse IgG (Aurion; 1:20) for 2 h at rt followed by thorough washing. Grids were postfixed in 1% glutaraldehyde, washed again, and finally contrasted in 4% uranyl acetate (36).
Ultrathin sections were inspected using transmission electron microscopy (TEM) (EM912a/b; Zeiss) at 120 kV under zero-loss conditions, and images were recorded at slight underfocus using a 1-k-by-1-k slow-scan charge-coupled-device (CCD) camera (Proscan) at 16,000× and ×25,000 magnification (pixel sizes corresponding to 0.9 nm and 0.6 nm, respectively).
Diameters of NPCs were measured using Esivision software (Olympus-SIS), and the distribution of gold particles relative to the nuclear lamina was quantified (37) using the iTEM package (Olympus-SIS).
Statistics.
The NPC diameters and patterns determined for the different test groups by Nup153 immunogold labeling were compared using the Mann-Whitney U test (Sigma-Stat 3.5; Systat). In Western blot analysis and other analyses, the t test was used.
Ethics statement.
Human lung tissue was obtained from patients who underwent a lobectomy after providing informed written consent (Departments of Pathology and Surgery, Justus Liebig University, Giessen). Use of human lung tissue was approved by the Justus Liebig University Giessen Ethics Committee.
Biosafety.
All experiments with infectious virus were performed according to German regulations for the propagation of influenza viruses (IVs). All experiments involving highly pathogenic avian influenza A viruses (HPAIV) were performed in a biosafety level 3 (BSL3) containment laboratory approved for such use by the local authorities (RP, Giessen, Germany).
RESULTS
The critical role of caspase activity for efficient nucleocytoplasmic RNP export is conserved across the three genera influenza virus type A, type B, and type C.
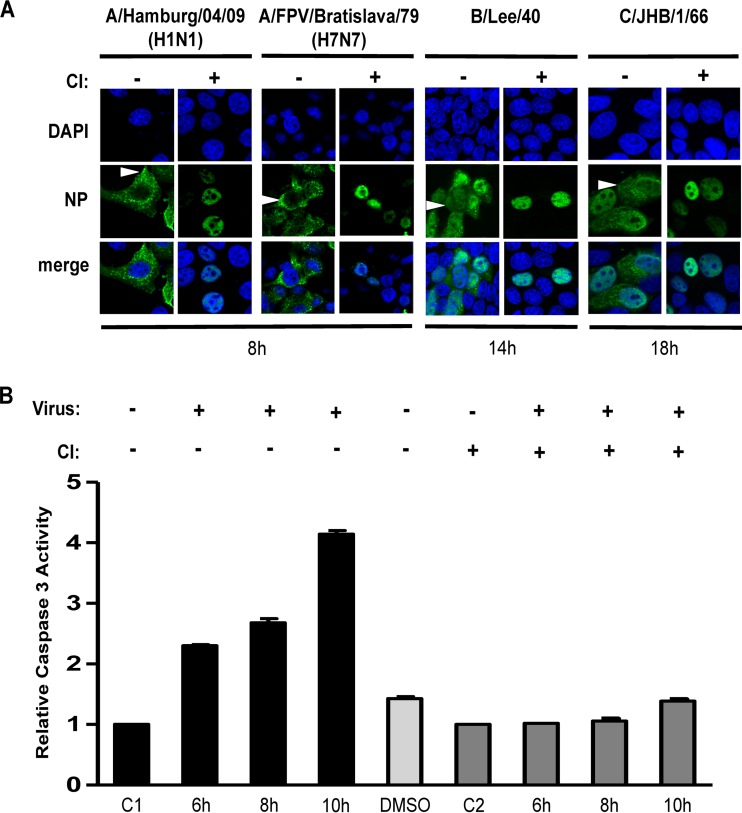
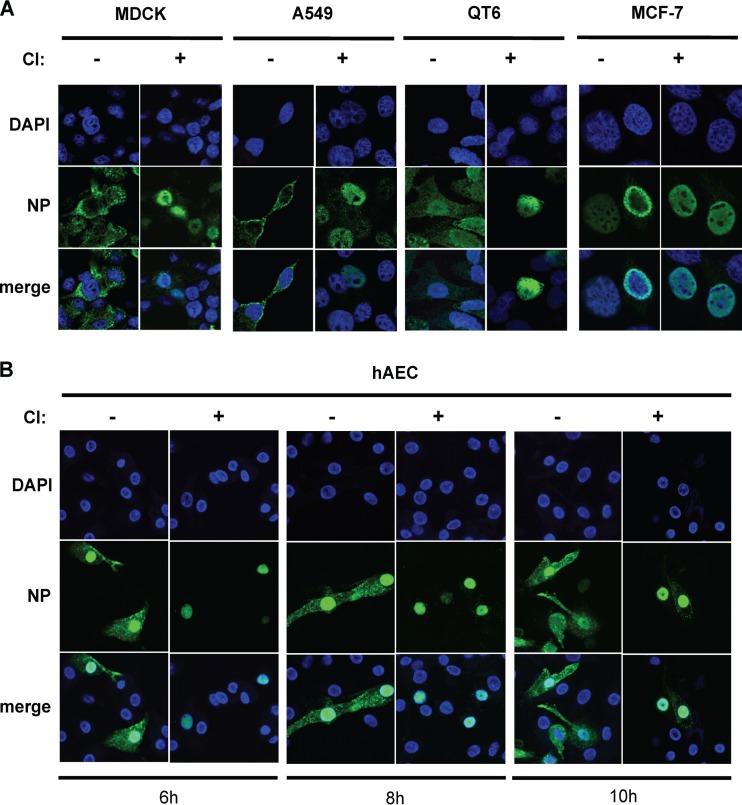
Previously, it was shown that efficient nuclear RNP export in cells infected with the A-type highly pathogenic avian influenza virus (HPAIV) A/FPV/Bratislava/79 (FPV, H7N7) requires cellular caspase 3 activity (23). To test whether this requirement is conserved among human A-, B-, and C-type influenza viruses, we infected MDCK-II cells with FPV, A-type 2009 pandemic human IAV (H1N1pdm09; A/Hamburg/0109, H1N1), or B-type human IV (B/Lee/40). We also infected MDCK-I cells with a C-type human IV (C/JHB/1/66). MDCK-I cells contain receptors that are necessary for influenza C virus infection, while MDCK-II cells lack these receptors (38). To assess the role of caspase activity in IV replication, cells were treated with a commonly used, noncytotoxic, cell-permeative, irreversible caspase 3/7 inhibitor (CI [Z-DEVD-FMK]). Z-DEVD-FMK contains the tetrapeptide recognition motif for caspase 3/7 and inhibits these caspases with high specificity (39). In infected cells treated with CI (Fig. 1A), no cytoplasmatic RNP signal was detectable at 8 h (for IAV), 14 h (for IBV), or 18 h (for ICV) postinfection (p.i.), indicating that, for all IVs included in this study, RNP export requires caspase activation. Using FPV, we extended these observations to other cell types. We found that caspase activity was also required for efficient nucleocytoplasmic export in other cell types, such as the A549 human alveolar epithelial cell line and an avian fibroblast cell line (QT6). Consistent observations were made using H1N1pdm09-infected primary human alveolar epithelial cells (hAEC) (Fig. 2). In line with these observations, we found that, in caspase 3-deficient MCF-7 cells, little (if any) nuclear RNP export occurred and there was no significant difference with respect to viral RNP localization (as judged by NP detection) and virus titers in the absence or presence of caspase 3/7 inhibitor (CI) (Fig. 2 and data not shown). The data provide additional evidence to suggest a major role for caspase 3 activity in the IV replication cycle and indicate that Crm1-dependent nuclear RNP export mechanisms (and caspase 7 activity) may be insufficient to sustain efficient nucleocytoplasmic RNP export in these cells. Still, it remains to be investigated if the lack of caspase 3-induced nuclear RNP export is the only cause for inefficient IV replication in caspase 3-deficient MCF-7 cells. Taken together, our data strongly support the idea of a critical role of caspase 3 activity for efficient nucleocytoplasmic RNP export in many cell types and human IVs from three different virus genera.
FIG 1.
Viral RNP localization and caspase activity in IV-infected cells in the presence or absence of caspase 3/7 inhibitor (CI). (A) MDCK-II cells were infected with the indicated A-, B-, and C-type IVs (MOI = 1) in the presence (+) or absence (−) of CI. Intracellular RNP localization was analyzed at the indicated time points p.i. by confocal laser scanning microscopy using an NP-specific antibody (green). DNA was stained with DAPI (blue). Arrowheads indicate cytoplasmic accumulation of RNPs. (B) Caspase 3 activities in lysates obtained from MDCK-II cells that were mock infected (C1 and C2) or infected with strain FPV (MOI = 15) in the presence (dark gray bars) or absence (black bars) of CI. Caspase activities were determined at the indicated time points p.i. Values were normalized to the mock control, which was set to 1. Experiments were done in duplicate. The lysate of the dimethyl sulfoxide (DMSO) control (light gray bar) was prepared at 10 h.
FIG 2.
RNP localization in different IV-infected cell lines treated with or without caspase 3/7 inhibitor. (A) A range of permanent cell lines were infected with FPV (MOI = 1) and treated with or without caspase 3/7 inhibitor (CI) immediately after infection. Intracellular RNP localization was detected at 8 h p.i. by confocal laser scanning microscopy using anti-NP specific Ab (green). DNA was stained with DAPI (blue). (B) Primary human alveolar epithelial cells (hAEC) were infected with H1N1pdm09 (MOI = 0.5) and treated with or without CI immediately after infection. Intracellular RNP localization was detected at 6, 8, and 10 h p.i. by confocal laser scanning microscopy using anti-NP specific Ab (green). DNA was stained with DAPI (blue).
Influenza virus-induced caspase activity leads to degradation of NPC component Nup153.
To further investigate the role of caspase activities in the nuclear export of viral RNPs, we analyzed the kinetics of IAV-induced caspase 3 activation (Fig. 1B). MDCK-II cells were infected with strain FPV, and caspase activity was determined at different time points p.i. Caspase activity was found to increase by more than 4-fold until 10 h p.i. in IAV-infected (but not mock-infected) cells.
Caspase activation is known to cause degradation of proteins of the NPC and disintegration of the nuclear basket that controls active export functions (16). One of the caspase 3/7 substrates was previously shown to be Nup153, an important component of the NPC nuclear basket and a key player in controlling nuclear import and export functions (31, 32, 40, 41).
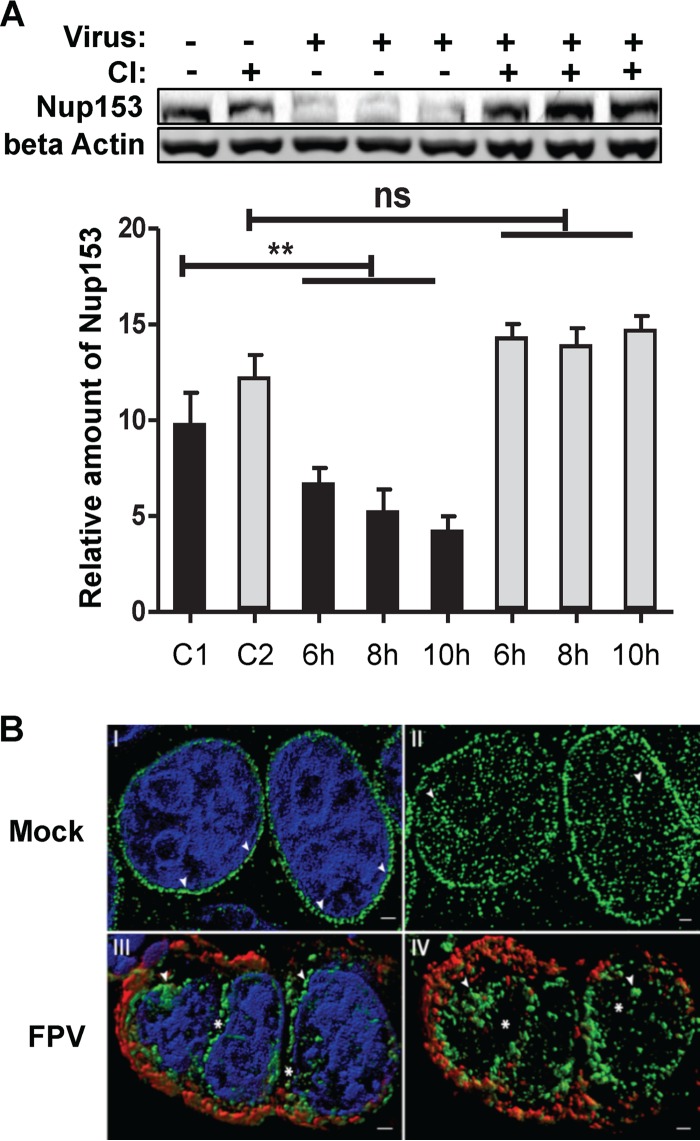
We therefore analyzed whether or not IAV infection affects Nup153 integrity. To this end, MDCK-II cells were infected with FPV (MOI = 3) and total lysates were prepared at time points when caspase activity had been sufficiently induced by the virus infection (Fig. 1B and 3A). Western blot analysis using a MAb specific for Nup153 or a MAb specific for beta actin revealed that Nup153 was degraded and that this degradation correlated with the level of IAV-induced caspase activation (see Fig. 1B and 3A). The idea of the specific degradation of Nup153 by cellular caspases was further supported by data showing that Nup153 degradation in IV-infected cells could be blocked by CI. Taken together, these data led us to conclude that IAV infection induces caspase-dependent Nup153 degradation, especially at late time points postinfection.
FIG 3.
Caspase-dependent Nup153 degradation and redistribution in IV-infected cells. (A) MDCK-II cells were mock infected (C1 and C2) or infected with FPV (MOI = 3) in the presence (gray bars) or absence (black bars) of caspase inhibitor (CI). Intracellular amounts of Nup153 and beta actin were determined by Western blotting, and the amount of Nup153 was quantified in relation to the amount of beta actin (control) at the indicated time points p.i. Column sizes represent relative Nup153 amounts normalized to the amount of beta actin. Experiments were done in triplicate. (C1, noninfected and nontreated control; C2, noninfected and CI-treated control). Significance levels are indicated (n.s., no significant difference; **, P = 0.03). (B) Superresolution microscopy reveals IV-induced aggregation and delocalization of nuclear pore complexes. MDCK-II cells were either mock infected (panels I and II) or infected with FPV (MOI = 0.01) (panels III and IV) for 24 h. MAbs against IV nucleoprotein (red) and Nup153 (green) were used. DNA was counterstained with DAPI (blue). Panels I and III show a surface-rendered 3D projection of five Z-planes acquired from the nuclear center to demonstrate the regular distribution (arrowheads) of NPCs in mock-infected cells (panel I) and aggregation (arrowheads) as well as delocalization (gaps indicated by asterisks) of NPCs in IV-infected cells (panel III). Panels II and IV show projections of the upper halves of the nucleus of mock-treated (panel II) and infected (panel IV) cells without DNA staining. Scale bar, 1 μm.
Influenza virus-induced caspase activity results in Nup153 displacement.
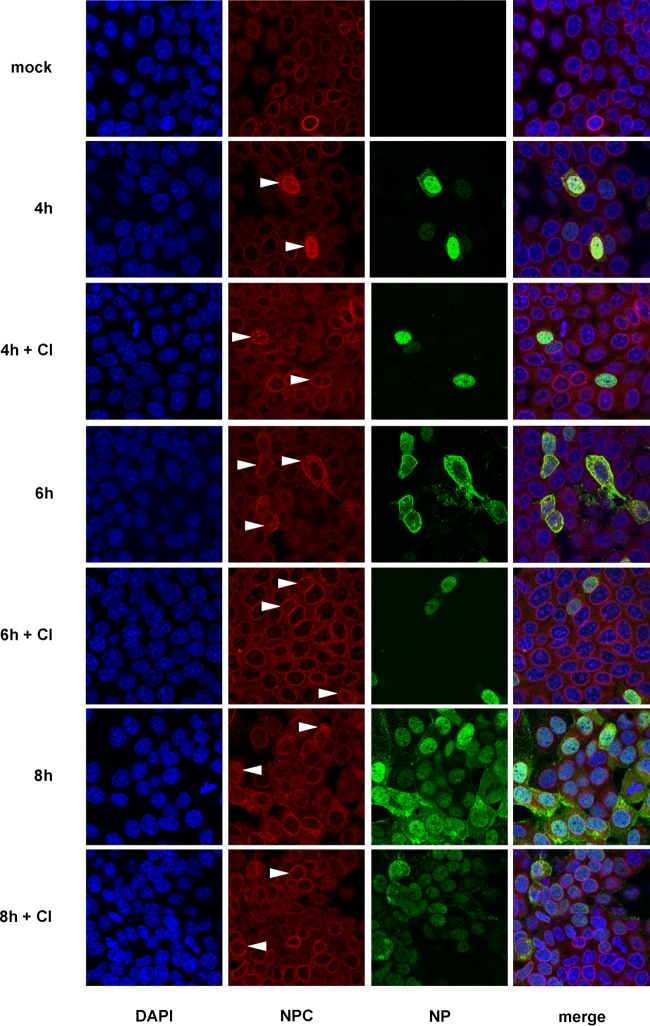
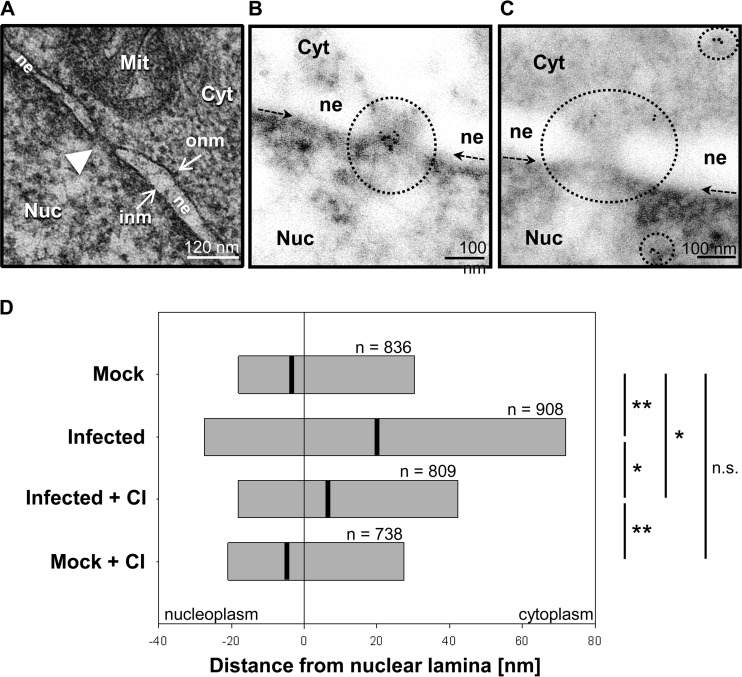
Degradation of Nup153 occurs in a coordinated manner and is linked to a gradual loss of transport functions (and structural integrity) of the NPC (42), illustrating the important role of Nup153 in NPC structure and function(s). To study possible structural changes of the NPC in IAV-infected cells, we analyzed the spatial distribution of different Nups at the nuclear envelope in the course of IAV infection using MAb 414, an anti-NPC protein antibody that recognizes FXFG sequence repeats conserved in nucleoporins. Immunofluorescence microscopy revealed a readily detectable signal at the nuclear envelope in uninfected cells. The signal changed to a more cytosolic distribution in FPV-infected cells at 6 h and 8 h p.i., that is, at time points when virus-induced caspase activity became detectable (Fig. 4). To analyze this in more detail, MDCK-II cells were infected with IAV for up to 24 h and superresolution microscopy of the NPC was performed using the Nup153-specific antibody QE5. In contrast to mock-infected cells, Nup153-containing nuclear pore structures were found to lose their regular (punctate) nuclear membrane distribution pattern in infected cells (Fig. 3B, panels I and II). Also, we observed large aggregates of Nup153-containing structures that partly localized to the nucleo- and cytoplasm (Fig. 3B, panels III and IV). This effect was further investigated on a subcellular level using immunogold labeling and transmission electron microscopy (TEM). MDCK-II cells were infected with strain FPV at a high MOI (= 15) to ensure infection of all cells. Figure 5A shows a representative image of a nuclear pore detected by TEM analysis, whereas Fig. 5B and C show representative immunogold labeling in uninfected and infected cells, respectively. Determination of relative frequencies of distances between individual immunogold signals and the nuclear lamina (Fig. 5D and data not shown) revealed that, in uninfected and untreated cells, Nup153 was predominantly located in the nucleus below the inner nuclear membrane, consistent with the known localization of nuclear baskets of which Nup153 is an integral part (43, 44). In contrast, a redistribution of immunogold signals was detected in IAV-infected cells (Fig. 5C and D). Here, relatively dispersed signals with reduced density were detected at or near the nuclear envelope, whereas an increased amount of Nup153 was observed in the periphery or deeper in the nucleus (mock-infected and untreated versus infected and untreated cells, P ≤ 0.002). These effects were significantly reduced in the presence of CI (infected and untreated versus infected and CI-treated cells, P ≤ 0.004). No significant difference of Nup153 localization was seen between uninfected and untreated and uninfected and CI-treated cells. In line with the data presented above (Fig. 3), the TEM study suggests that IAV infection triggers both degradation and relocalization of Nup153.
FIG 4.
Nup153 distribution in IV-infected cells. FPV-infected MDCK-II cells (MOI = 1) treated with or without caspase 3/7 inhibitor (CI) were stained for DNA (blue) and for viral NP (green) and with an anti-NPC protein antibody, Mab414, that recognizes FXFG repeats conserved in various nucleoporins (red). Spatial distribution of different Nups at the nuclear envelope was analyzed at 4, 6, and 8 h p.i. by confocal laser scanning microscopy. Partial or (nearly) complete delocalization of Nup in infected cells is indicated by arrowheads.
FIG 5.
Transmission electron microscopy and immunogold labeling. (A) Ultrastructural image of a nuclear pore (white arrowhead) in MDCK-II cells. The nuclear envelope is formed by the outer nuclear membrane (onm) and the inner nuclear membrane (inm). Cyt, cytoplasm; Nuc, nucleoplasm; Mit, mitochondrion; ne, nuclear envelope. (B and C) Typical TEM images after immunogold labeling for Nup153 in noninfected (B) and FPV-infected (C) MDCK-II cells. Circles highlight labeled areas, and arrows indicate the position of the nuclear lamina just above the strongly contrasted heterochromatin. The nuclear ne is seen as an area with reduced electron density, whereas the inm and onm are not visible due to the omission of osmium treatment (see protocol). (D) Spatial distribution of distances between Nup153 signals and the nuclear lamina. Bars represent the central 50% (interquartile range) of ranked data of each group. Black lines indicate median values. Significance levels are indicated (n.s., no significant difference; *, P ≤ 0.004; **, P ≤ 0.002). In each test group, immunogold data were collected from two experiments and 10 different cell profiles, except in test group Mock + CI, for which data were collected from two experiments and 6 cell profiles.
Influenza virus-induced NPC degradation causes enlargement of nuclear pores.
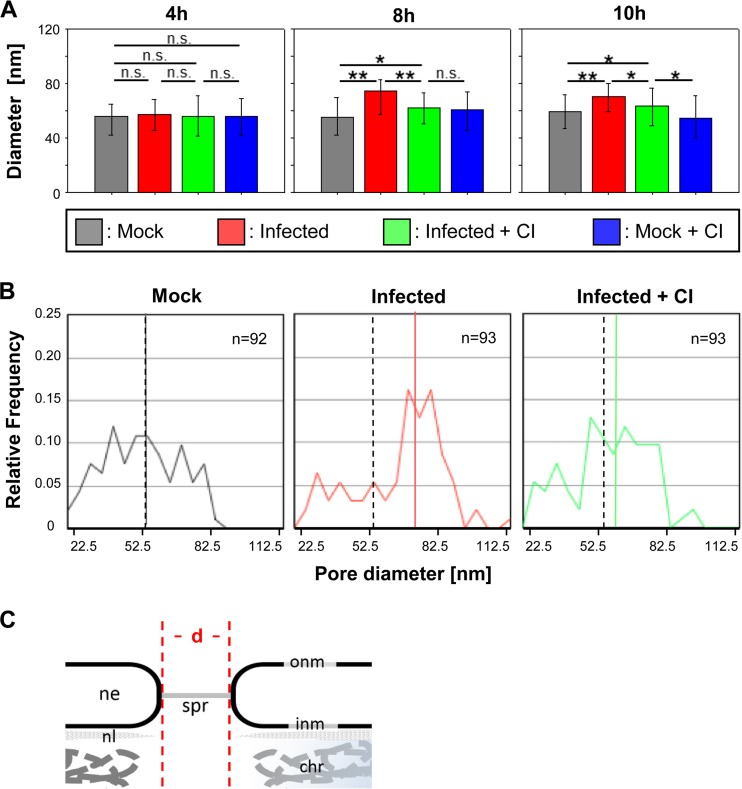
Previous studies suggest that caspase activation initiates a coordinated disruption of the nuclear lamina which, in IV-infected cells may increase the passive diffusion limit of nuclear pores and thereby facilitate migration of viral RNPs to the cytoplasm in late phases of the viral life cycle (11, 16). The data presented above confirm and extend these hypotheses. More specifically, they suggest that IAV-induced apoptotic caspase activities are critically involved in changes of the NPC architecture that, possibly, cause an increase in nuclear pore sizes. To address the latter possibility more directly, we performed another set of TEM studies. MDCK-II cells were infected with strain FPV (MOI = 15), images were taken at specific time points (Fig. 6A), and diameters of NPC lumenal spoke rings were measured. Figure 5A gives an example of a typical TEM micrograph used to determine pore diameters in this study. Nuclear pore diameters were found to be significantly increased at late time points p.i. (8 to 10 h) (Fig. 6) (cf. Fig. 1A). The increase of nuclear pore sizes in IV-infected cells was blocked by CI, again confirming the critical role of IAV-induced caspase activation in this process. The data also show that the enlargement of nuclear pores is a relatively late event, commencing at about 8 h p.i., whereas no significant changes were seen at 4 h p.i. (Fig. 6A). In the absence of CI, nuclear pore diameters in infected cells were found to be increased by nearly 20 nm compared to mock-infected cells (P ≤ 0.001). The increase of nuclear pore size was less profound in infected and CI-treated cells compared to infected and untreated cells (8 nm, P ≤ 0.001). Consistent results were obtained when median values and distributions of pore diameters were analyzed (Fig. 6B). Analysis of nuclear pore sizes in mock-infected cells in the presence or absence of CI revealed no significant differences (P = 0.194). Nuclear pore diameters determined in uninfected and untreated cells and infected and CI-treated cells revealed moderate differences (6 nm, P = 0.036), while a much more profound increase in nuclear pore size was determined for infected and untreated cells (20 nm, P < 0.001). At 10 h p.i., similar results were obtained and no further increase of nuclear pore diameters was seen in infected cells (Fig. 6A). These data are in perfect agreement with the observations described above and reveal a quite impressive enlargement of nuclear pores by up to 37% in IAV-infected cells at late time points (≥8 h p.i.).
FIG 6.
Increase of nuclear pore diameters in IV-infected cells. (A) Diameters of nuclear pores (median values) in MDCK-II cells under different experimental conditions (mock treatment, FPV infection, FPV infection plus caspase 3/7 inhibition [CI], and mock treatment plus CI) and at different time points p.i. Error bars indicate interquartile ranges. Significance levels are indicated (n.s., no significant difference; *, P ≤ 0.05; **, P ≤ 0.001). (B) Distribution of nuclear pore diameters in FPV-infected MDCK-II cells at 8 h p.i. after mock treatment (left), after infection (middle), and after infection plus CI treatment (right). The colored lines represent the median value for each sample. As a reference, the median value of the control (mock-treated) group is also indicated for the two other groups by a dashed line. (C) Panel showing the main structural elements of the NPC seen in the electron microscopy (em) images and indication of how the diameters (d) of the pores were measured. chr, chromatin; ne, nuclear envelope; onm and inm, outer and inner nuclear membranes; spr, spoke ring; nl, nuclear lamina.
Influenza virus-induced enlargement of nuclear pores leads to increased diffusion limits.
IV RNPs are thought to have an approximate molecular mass of up to 5.6 MDa and dimensions of about 30 to 120 nm by 10 to 15 nm (45). We therefore speculated that, despite their large molecular mass, viral RNPs might pass NPCs if (i) they are oriented along their longitudinal axis and (ii) the diffusion limits of NPCs are sufficiently increased. We sought to obtain additional evidence for increased nuclear pore sizes in IAV-infected cells and investigated possible consequences for the transport of large protein complexes across the nuclear membrane. MDCK-II cells were transfected with p3xEGFP-NLS (3xGFP-NLS), a plasmid that encodes a 75-kDa green fluorescent protein (GFP) trimer carrying a nuclear localization signal (NLS). The protein was actively imported and accumulated in the nucleus. In contrast, a 125-kDa GFP pentamer (5xGFP, encoded by pEGFP-GFP5x) accumulated in the cytoplasm and, because of its large size, was prevented from entering the nucleus by free diffusion. At 36 h p.t., cells expressing the appropriate GFP construct were infected with strain FPV (or mock infected) and cultivated in the presence or absence of CI. At 10 h p.i., the cells were fixed, stained for viral NP and cellular DNA, and analyzed by confocal laser scanning microscopy.
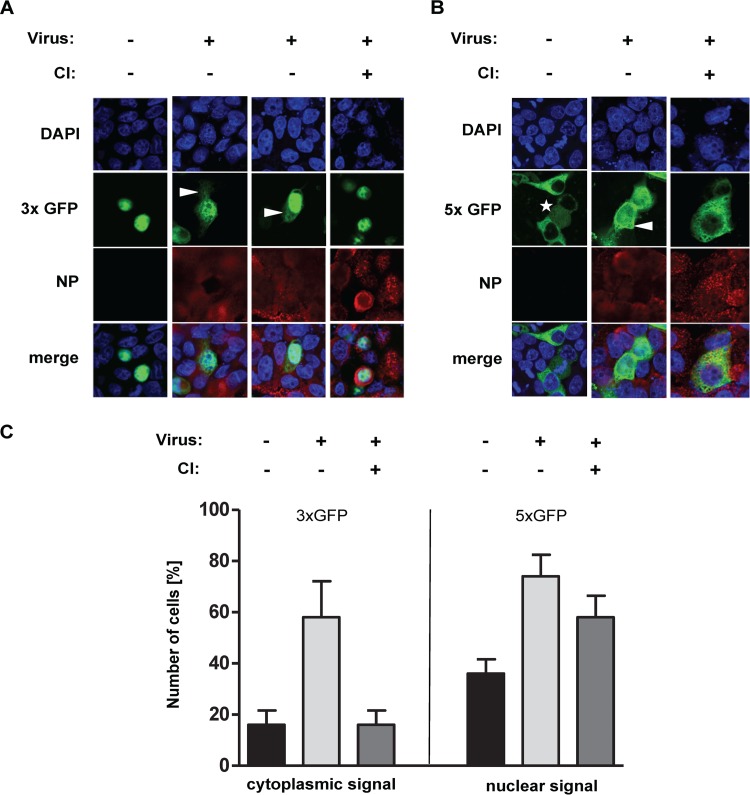
As expected, the 3xGFP-NLS protein accumulated in the nucleus of transfected cells (Fig. 7A). In contrast, in IAV-infected cells, the protein was also observed in the cytoplasm (10 h p.i.), indicating significant diffusion of 3xGFP-NLS from the nucleus to the cytoplasm and insufficient compensation by appropriate reimport mechanisms. In line with this hypothesis, we observed that 5xGFP, which, in mock-infected cells, could be detected only in the cytoplasm, became detectable in the nucleus of IAV-infected cells, suggesting dramatic changes in the free diffusion limits of NPCs in these cells (Fig. 7B). The massive redistribution of either marker protein was abolished in the presence of CI. To quantify these effects, the proportion of cells with detectable GFP expression in the nucleus or cytoplasm was determined in 2 independent samples (Fig. 7C). The quantitative analyses were consistent with the data shown in Fig. 7A and B.
FIG 7.
Intracellular localization of multimeric GFP constructs in IV-infected cells treated with or without caspase 3/7 inhibitor (CI). (A and B) MDCK-II cells were transfected with a plasmid that codes either for a GFP trimer that carries a nuclear localization signal (3xGFP) and accumulates in the nucleus or for a GFP pentamer that lacks a nuclear localization signal (5xGFP) and accumulates in the cytoplasm. At 36 h posttransfection, cells were superinfected with FPV (MOI = 5) and treated with or without CI. The cells were fixed at 10 h p.i., and the intracellular GFP localization was analyzed. Intracellular GFP (green) and NP (red) localization was detected by confocal laser scanning microscopy. DNA was stained with DAPI (blue). Star, noninfected, dividing cell with chromosomal condensation. (C) A total of 50 cells in 2 independent samples identified as transfected and infected, with or without CI treatment, were analyzed by confocal laser scanning microscopy for intracellular GFP localization. For 3xGFP, the proportions of cells (indicated in percentages) displaying a cytoplasmic GFP signal are shown. For 5xGFP, the proportions of cells (indicated in percentages) showing a nuclear GFP signal are shown.
An accumulation of 5xGFP in both the nucleus and cytoplasm was also seen in noninfected cells undergoing mitosis, the latter involving chromosomal condensation (Fig. 7B, asterisk) and disintegration of the nuclear membrane. Similarly, increased diffusion limit was detected in cells in which apoptotic caspase activity was induced by actinomycin D (data not shown). Taken together, our results suggest that the enlargement of nuclear pores caused by virus-induced caspase activities leads to an increase of the NPC diffusion limit of at least 40 kDa, resulting in a diffusion limit of >125 kDa.
Increasing role of passive RNP export mechanisms later in infection.
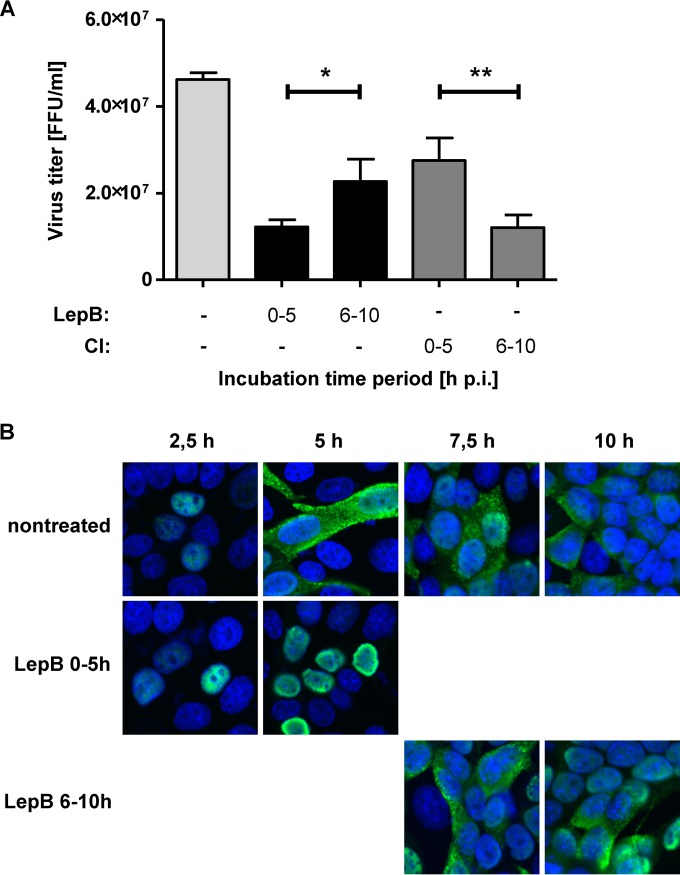
The data presented above suggested that the previously described nucleocytoplasmic RNP transport pathway involving Crm1 (26) may be complemented by passive transport mechanisms at late time points p.i. To further investigate the specific roles of active versus passive mechanisms involved in IAV RNP transport, we determined the kinetics of vRNP export in the presence of LepB, a specific inhibitor of Crm1. LepB was added to virus-infected cells (MOI = 1) from 0 to 5 h p.i. or from 6 to 10 h p.i. At 10 h p.i., virus titers were determined and compared to those seen with infected but nontreated cells. As shown in Fig. 8A, the production of infectious virus was strongly reduced (ca. 70%) when LepB was added to the culture medium at 0 to 5 h p.i. Virus titers were reduced less profoundly (ca. 50%) when LepB was added at a later time point (6 to 10 h p.i.). Inhibition of caspase 3/7 activity 6 to 10 h p.i. resulted in an approximately 70% reduction of virus titers, while inhibition of caspase activities at earlier time points (0 to 5 h p.i.) caused significantly (ca. 40%) less virus titer reduction. The data support the idea that, at later time points, active nuclear transport functions may become less critical to infectious particle formation.
FIG 8.
Relevance of Crm1-dependent transport pathways for nuclear RNP export at different time points in the viral replication cycle. (A) MDCK-II cells were infected with FPV (MOI = 1), and inhibitors of Crm1 (Leptomycin B [LepB], black bars) or caspase 3/7 (CI, gray bars) were added for different time periods p.i. as indicated. Virus titers in cell culture supernatants collected at 10 h p.i. were determined. Experiments were done in triplicate, and standard deviations are indicated. (*, P = 0.029; **, P = 0.004). FFU, focus-forming units. (B) MDCK-II cells were infected with FPV (MOI = 0.5). LepB was included from 0 to 5 h or from 6 to 10 h p.i. as indicated. Intracellular RNP localization was detected by confocal laser scanning microscopy using a NP-specific MAb (green) at the indicated time points p.i. DNA was stained with DAPI (blue).
In a final set of experiments, we investigated the intracellular localization of viral RNPs in infected or mock-infected cells cultured in the presence or absence of LepB. The cells were analyzed at different time points p.i. by immunofluorescence microscopy using an NP-specific MAb. As shown in Fig. 8B, viral RNPs were retained in the nucleus when LepB was included in the culture medium from 0 to 5 h p.i., confirming the previously established important role of Crm1 in nucleocytoplasmic RNP export. In contrast, if the inhibitor was included later in infection (6 to 10 h p.i.), viral RNPs were no longer retained in the nucleus, suggesting that, in the course of IAV infection, the role of Crm1 in nuclear export of viral RNPs was diminishing or even that it was becoming dispensable over time.
DISCUSSION
IAV replication occurs in the host cell nucleus and, therefore, requires bidirectional RNP transport processes across the nuclear membrane to (i) import primary RNPs into the nucleus and (ii) export newly produced RNPs across the nuclear membrane to the cell membrane, where viral assembly takes place (2, 4). The subunits of the viral RdRp and the NP contain NLS (4) and interact with the nuclear import machinery (46). This ensures that newly translated PB1, PB2, PA, and NP are efficiently imported into the nucleus, where viral genome replication and transcription take place. The viral proteins M1, NEP/NS2, and NP are required for the nuclear export of newly formed RNPs. The proteins tightly interact with each other and contain nuclear export signals that are recognized by Crm1 (reviewed in reference 47). The available evidence for effective inhibition of RNP egress by caspase 3/7 inhibitors suggests that Crm1-mediated mechanisms may be complemented by other (caspase-dependent) mechanisms which, however, have not been characterized in detail. Previously, we were able to show that IAV-induced NF-κB-dependent TRAIL expression and FasL expression as well as IAV-induced caspase activity are important for efficient nuclear RNP export (23, 24). Based on these observations, we proposed that IAV infection may result in NF-κB-controlled TRAIL and FasL expression that triggers apoptosis by auto- and paracrine mechanisms and activates cellular caspases (11). We further hypothesized that these caspase activities might affect NPC functions to facilitate nuclear RNP export.
Evidence to support this hypothesis was previously obtained for avian IAV strain FPV (23). We now extended these studies to human A-, B-, and C-type IVs (Fig. 1A) and obtained convincing evidence to suggest that caspase activation is a general requirement for efficient nuclear RNP export across different IV genera propagated in a wide range of human and avian permanent cell lines as well as primary cells (Fig. 2).
To further analyze the downstream effects of virus-induced caspase activation on nuclear RNP export, we focused on Nup153, a known caspase substrate and an important component of the NPC (16, 48). We were able to show that IAV-triggered caspase activities caused a progressive degradation of this nuclear pore component (Fig. 3A). The time courses of caspase activation and Nup153 degradation were shown to follow similar kinetics (Fig. 1B), and Nup153 degradation could be blocked by using CI, further supporting the idea of the critical role of caspase 3/7 in this process. Indirect immunofluorescence and superresolution microscopy data revealed an irregular distribution of NPC components in IV-infected cells and relocation of Nup153 or, more likely, specific degradation products derived from this protein (Fig. 3B). The antibodies used in this study do not allow us to distinguish between signals derived from intact protein and those derived from proteolytically cleaved protein. Our data consistently support the idea that caspase activation at late time points p.i. causes Nup153 degradation which, in turn, affects the structural and functional integrity of NPCs (Fig. 3A and 5). As expected for a major component of the nuclear basket, Nup153 was found to be located at the nuclear surface of the NPC next to the inner nuclear membrane in both uninfected and untreated cells (17, 21, 49). In contrast, the intracellular localization of the Nup153 signal changed dramatically in IAV-infected cells, consistent with our immunofluorescence and superresolution microscopy data. The observed effects could be blocked by CI, yet again confirming the key role of virus-induced caspase activities in Nup153 degradation and relocation. Electron microscopy provided evidence that the diameters of NPC luminal spoke rings, which form the central channel between the inner and outer nuclear membranes (17) and thus control the transport of large cargo through the nuclear pore, are significantly increased in IAV-infected cells (Fig. 6A and B). NPC disintegration (Fig. 3B and 5) could be blocked by using CI and was particularly evident at late time points p.i. when (i) caspase 3 was activated (Fig. 1B), (ii) Nup153 was degraded (Fig. 3A), and (iii) NPC spoke rings were widened (Fig. 6). The combined data lead us to propose a model in which IV-induced caspase 3 activity promotes nuclear RNP export by initiating a process that eventually results in enlarged and/or dysfunctional nuclear pores.
In previous studies, many other viruses were found to support their own replication or suppress antiviral host responses by disrupting nuclear import pathways or inhibiting mRNA export through interactions of viral with cellular proteins (reviewed in reference 50). This also includes the degradation of Nups. For members of the family Picornaviridae, it has been shown that nuclear components, such as Nup153, may be cleaved by the viral 2Apro protein (51). In other cases, classical nuclear import pathways were found to be blocked, resulting in cytoplasmic retention (and accumulation) of nuclear cargo; also, increased diffusion limits by mechanisms that do not depend on caspase activation have been described to be induced in picornavirus-infected cells (51–53). In contrast to these picornavirus studies, our data suggest that IVs use cellular (rather than viral) protease-mediated mechanisms to modulate nuclear pore functions. It remains to be studied whether (and to what extent) these mechanisms also affect the export of cellular mRNAs and, as a result, expression of antiviral proteins.
IV RNPs have molecular masses of up to 5.6 MDa, depending on the size of the respective genome segments (ranging from 890 to 2,341 nucleotides [nt]). Nevertheless, due to their rod-like structure with a size of about 10 to 15 nm by 30 to 120 nm (45, 54–56), it is reasonable to speculate that (i) the observed increase in the NPC diameter by up to 20 nm, (ii) the increase of passive diffusion limits at later stages of infection, and (iii) the specific vRNP shape might allow IV RNPs to diffuse passively through the NPC along their longitudinal axis.
Taken together, the data lead us to hypothesize that IVs (largely) employ active Crm1-dependent mechanisms for nucleocytoplasmic RNP transport early in infection whereas passive transport mechanisms are suggested to play a more prominent role at later time points p.i. when NPCs increasingly lose their structural integrity and specific functions (25, 31, 32). The study data advance our understanding of the multiple roles of caspase activities in viral infections and suggest that these enzymes (and probably other molecules involved in cellular apoptotic pathways) might be promising targets for future antiviral drugs.
ACKNOWLEDGMENTS
We thank S. Ludwig (Muenster, Germany) for providing the mouse anti-IAV NP antibody for titration, R. Webster (Memphis, TN, USA) for providing the goat anti-IAV NP antibody (for SRM), M. Marschall (Erlangen, Germany) for providing the mouse anti-ICV NP antibody, P. V. Litsky (Moscow, Russia) for providing the 3xGFP-NLS plasmid, and Y. Lazebnik (Cold Spring Harbor, NY, USA) for providing the pEGFP-GFP5x plasmid. In addition, we thank E. Erdogan, M. Riedl, S. Agel, and A. Moebus for excellent technical assistance. We also thank V. Schubert (Gatersleben, Germany) and M. Mieth (Berlin, Germany [SFB-TR84, Z1a]) for their help with superresolution microscopy.
This work was supported in part by grants from the DFG (SFB-TR 84 “Innate immunity of the lung” fellowship to J.D., SFB-TR 84 “Innate immunity of the lung” project B2 to S.H., S. P., and T. W., and project Z1a to A.H.), the German Centre for Infection Research (DZIF), partner site Giessen, Germany (short-term fellowship to D.M. and a grant within the TTU-Emerging Infections to S.P. and J.Z.), and the BMBF-founded FluResearchNet—Molecular Signatures Determining Pathogenicity and Species Transmission of Influenza A Viruses (01 KI 07136 to S.P.) and a fellowship of the German-Egyptian Research Long-Term Scholarship “GERLS” program cofunded by the Egyptian government and the German Academic Exchange Service (DAAD) (to A.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Wilschut JC, McElhaney JE, Palache AM. 2006. Rapid reference to influenza, 2nd ed Mosby, Orlando, FL. [Google Scholar]
- 2.Wright PF, Neumann G, Kawaoka Y. 2013. Orthomyxoviruses, p 1186–1243. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Short KR, Kroeze EJ, Fouchier RA, Kuiken T. 2014. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis 14:57–69. doi: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 4.Shaw ML, Palese P. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Herold S, Ludwig S, Pleschka S, Wolff T. 17 February 2012, posting date Apoptosis signaling in influenza virus propagation, innate host defense, and lung injury. J Leukoc Biol doi: 10.1189/jlb.1011530. [DOI] [PubMed] [Google Scholar]
- 6.Kerr JF, Wyllie AH, Currie AR. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia-Sastre A, Munz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Yang Y, Zhou X, Yang Z, Liu X, Cao Z, Song H, He Y, Huang P. 2011. The NS1 protein of influenza A virus interacts with heat shock protein Hsp90 in human alveolar basal epithelial cells: implication for virus-induced apoptosis. Virol J 8:181. doi: 10.1186/1743-422X-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, Wutzler P, Zell R. 2011. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol 200:69–75. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- 10.Schultz-Cherry S, Dybdahl-Sissoko N, Neumann G, Kawaoka Y, Hinshaw VS. 2001. Influenza virus ns1 protein induces apoptosis in cultured cells. J Virol 75:7875–7881. doi: 10.1128/JVI.75.17.7875-7881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig S, Pleschka S, Planz O, Wolff T. 2006. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell Microbiol 8:375–386. doi: 10.1111/j.1462-5822.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 12.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, Seeger W, Lohmeyer J. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med 205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Högner K, Wolff T, Pleschka S, Plog S, Gruber AD, Kalinke U, Walmrath HD, Bodner J, Gattenlohner S, Lewe-Schlosser P, Matrosovich M, Seeger W, Lohmeyer J, Herold S. 2013. Macrophage-expressed IFN-beta contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog 9:e1003188. doi: 10.1371/journal.ppat.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walczak H, Haas TL. 2008. Biochemical analysis of the native TRAIL death-inducing signaling complex. Methods Mol Biol 414:221–239. doi: 10.1007/978-1-59745-339-4_16. [DOI] [PubMed] [Google Scholar]
- 15.Falschlehner C, Schaefer U, Walczak H. 2009. Following TRAIL's path in the immune system. Immunology 127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer A, Liashkovich I, Oberleithner H, Ludwig S, Mazur I, Shahin V. 2008. Apoptosis leads to a degradation of vital components of active nuclear transport and a dissociation of the nuclear lamina. Proc Natl Acad Sci U S A 105:11236–11241. doi: 10.1073/pnas.0801967105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 18.Panté N. 2004. Nuclear pore complex structure: unplugged and dynamic pores. Dev Cell 7:780–781. doi: 10.1016/j.devcel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Tran EJ, King MC, Corbett AH. 2014. Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim Biophys Acta 1843:2784–2795. doi: 10.1016/j.bbamcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams RL, Wente SR. 2013. Uncovering nuclear pore complexity with innovation. Cell 152:1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Goryaynov A, Sarma A, Yang W. 2012. Self-regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci U S A 109:7326–7331. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suntharalingam M, Wente SR. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4:775–789. doi: 10.1016/S1534-5807(03)00162-X. [DOI] [PubMed] [Google Scholar]
- 23.Wurzer WJ, Planz O, Ehrhardt C, Giner M, Silberzahn T, Pleschka S, Ludwig S. 2003. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J 22:2717–2728. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurzer WJ, Ehrhardt C, Pleschka S, Berberich-Siebelt F, Wolff T, Walczak H, Planz O, Ludwig S. 2004. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J Biol Chem 279:30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen KT, Holloway MP, Altura RA. 2012. The CRM1 nuclear export protein in normal development and disease. Int J Biochem Mol Biol 3:137–151. [PMC free article] [PubMed] [Google Scholar]
- 26.Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol 75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattaj IW, Englmeier L. 1998. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 29.Ding Q, Zhao L, Guo H, Zheng AC. 2010. The nucleocytoplasmic transport of viral proteins. Virol Sin 25:79–85. doi: 10.1007/s12250-010-3099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mor A, White MA, Fontoura BM. 2014. Nuclear trafficking in health and disease. Curr Opin Cell Biol 28:28–35. doi: 10.1016/j.ceb.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakielny S, Shaikh S, Burke B, Dreyfuss G. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer U, Janicke RU, Schulze-Osthoff K. 2003. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ 10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma W, Brenner D, Wang Z, Dauber B, Ehrhardt C, Hogner K, Herold S, Ludwig S, Wolff T, Yu K, Richt JA, Planz O, Pleschka S. 2010. The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV. J Virol 84:2122–2133. doi: 10.1128/JVI.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herold S, Tabar TS, Janssen H, Hoegner K, Cabanski M, Lewe-Schlosser P, Albrecht J, Driever F, Vadasz I, Seeger W, Steinmueller M, Lohmeyer J. 2011. Exudate macrophages attenuate lung injury by the release of IL-1 receptor antagonist in gram-negative pneumonia. Am J Respir Crit Care Med 183:1380–1390. doi: 10.1164/rccm.201009-1431OC. [DOI] [PubMed] [Google Scholar]
- 35.Panté N, Jarmolowski A, Izaurralde E, Sauder U, Baschong W, Mattaj IW. 1997. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA 3:498–513. [PMC free article] [PubMed] [Google Scholar]
- 36.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aris JP, Blobel G. 1989. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J Cell Biol 108:2059–2067. doi: 10.1083/jcb.108.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrler G, Klenk HD. 1987. The surface receptor is a major determinant of the cell tropism of influenza C virus. Virology 159:102–108. doi: 10.1016/0042-6822(87)90352-7. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. 1998. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem 273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 40.Schrader N, Koerner C, Koessmeier K, Bangert JA, Wittinghofer A, Stoll R, Vetter IR. 2008. The crystal structure of the Ran-Nup153ZnF2 complex: a general Ran docking site at the nuclear pore complex. Structure 16:1116–1125. doi: 10.1016/j.str.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Ball JR, Ullman KS. 2005. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 114:319–330. doi: 10.1007/s00412-005-0019-3. [DOI] [PubMed] [Google Scholar]
- 42.Kihlmark M, Rustum C, Eriksson C, Beckman M, Iverfeldt K, Hallberg E. 2004. Correlation between nucleocytoplasmic transport and caspase-3-dependent dismantling of nuclear pores during apoptosis. Exp Cell Res 293:346–356. doi: 10.1016/j.yexcr.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 43.Panté N, Aebi U. 1994. Toward the molecular details of the nuclear pore complex. J Struct Biol 113:179–189. doi: 10.1006/jsbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 44.Panté N. 2007. Contribution of electron microscopy to the study of the nuclear pore complex structure, composition and function, p 144–153. In Méndez-Vilas A, Diaz J (ed), Modern research and educational topics in microscopy, vol 1 Formatex Research Center, Badajoz, Spain. [Google Scholar]
- 45.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 46.Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutchinson EC, Fodor E. 2013. Transport of the influenza virus genome from nucleus to nucleus. Viruses 5:2424–2446. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrando-May E, Cordes V, Biller-Ckovric I, Mirkovic J, Gorlich D, Nicotera P. 2001. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ 8:495–505. doi: 10.1038/sj.cdd.4400837. [DOI] [PubMed] [Google Scholar]
- 49.Cordes VC, Reidenbach S, Kohler A, Stuurman N, van Driel R, Franke WW. 1993. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol 123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM. 2014. Viral subversion of nucleocytoplasmic trafficking. Traffic 15:127–140. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gustin KE, Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park N, Katikaneni P, Skern T, Gustin KE. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belov GA, Evstafieva AG, Rubtsov YP, Mikitas OV, Vartapetian AB, Agol VI. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275:244–248. doi: 10.1006/viro.2000.0427. [DOI] [PubMed] [Google Scholar]
- 54.Boulo S, Akarsu H, Ruigrok RW, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res 124:12–21. doi: 10.1016/j.virusres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. 2009. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J 28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Compans RW, Content J, Duesberg PH. 1972. Structure of the ribonucleoprotein of influenza virus. J Virol 10:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]