Abstract
Silica impregnated polymer monolithic columns may provide a simple method for lysing and extracting DNA from bacteria inside of microfluidic chips. Here we use Escherichia coli as a test organism for a point of care thermoplastic microfluidic module designed to take in a urine sample, mix it with lysis buffer, and perform a hybrid chemical/mechanical lysis and solid phase extraction of nucleic acids from the sample. To demonstrate proof-of-concept, we doped human hematuric urine samples with E. coli at concentrations ranging from 101–105 colony-forming units/mL (CFU/mL) to simulate patient samples. We then performed on-chip lysis and DNA extraction. The bacterial DNA was amplified using real-time PCR demonstrating lysis and isolation down to 101 CFU/mL. Results were comparable to a commercial kit at higher concen trations and performed better at recovering DNA at lower concentrations.
Keywords: Urine, Gram-negative, Bacteria, Lysis, PCR, Urinary tract infection, Microfluidic, Thermoplastic, Cyclic polyolefins
1 Introduction
In order to ensure easy and inexpensive point-of-care diagnostics, little or no sample preprocessing at the bench should be required. In this paper, we present a microfluidic sample preparation module and demonstrate the lysis, extraction and purification of DNA from pathogenic microorganisms infecting human urine. We have simulated a urinary tract infection (UTI) by spiking human urine samples with human whole blood and Escherichia coli, the organism responsible for the majority of UTIs.
Symptomatic UTIs are the most frequent bacterial infections encountered in primary care practice. Forty to 50% of all adult women have had a UTI(Franz and Horl 1999) and one quarter of women with UTIs will experience a recurrence within 6 months, primarily due to re-infection (Foxman et al. 2000). E. coli is the pathogen in 85% of episodes of community acquired acute uncomplicated cystitis(Hooton et al. 2004) and emerging drug resistance is beginning to complicate empiric treatment strategies (Brown et al. 2002). Based on clinical evaluation, it is reasonable to offer empiric treatment for UTI when the probability of uncomplicated infection is high. However, additional diagnostic testing is warranted when the diagnosis is unclear or if the symptoms are recurrent.
Dipstick tests are the current gold standard. These tests detect leukocyte esterase, which indicates the presence of white blood cells and nitrite, which can indicate the presence of bacteria. Urine cultures are not typically obtained in uncomplicated inflammation of the bladder, largely because cultures are most often irrelevant due to long turn around times (24 h). However, the need to obtain urine cultures prior to initiation of antibiotic treatment becoming more important given the increasing prevalence of antibiotic-resistant uropathogens(Gupta et al. 2001). A molecular diagnostic would be capable of providing information that the current dipstick tests cannot, including strain information to identify antibiotic resistant infections. Further, molecular diagnostics based on urine samples have many potential applications outside of diagnosing UTIs. Urine nucleic acid tests have been proposed for use in cancer diagnostics, bacterial vaginosis and other sexually transmitted diseases(Gaydos et al. 2004; Lindan et al. 2005).
Other on-chip lysis techniques reported in the literature have included a nanoscale barb design which uses pressure driven flow with sharp nanostructures(Di Carlo et al. 2003), ultrasound(Belgrader et al. 1999), electrical lysis (Wang et al. 2007)and induced local changes in pH(Di Carlo et al. 2005). Solid phase extraction (SPE) methods based on the work of Boom (Boom et al. 1990) for DNA extraction have been successfully miniaturized and incorporated in microfluidic chips. Here, we refer to the microfluidic SPE columns as μSPE. Sol-gel/silica bead mixtures have been shown to have good extraction efficiencies and reproducibility in micro-fluidic systems(Breadmore et al. 2002, 2003; Easley et al. 2006; Wolfe et al. 2002). However, the sol-gel process involves high temperatures or long processing times. Others have demonstrated extraction using silica bead packed columns in glass channels(Chen et al. 2007; Poeckh et al. 2008; Tian et al. 2000). These structures require frits.
There has been a longstanding interest in the detection of bacteria in urine using rapid techniques capable of multiplexing. A method using flow cyotmetry was published by Van Dilla et al. in 1983 (Van Dilla et al. 1983). More recently, Liao and coworkers have demonstrated a compact biosensor using an rRNA-based method for multiplexed analysis that does not require amplification(Liao et al. 2006; Liao et al. 2007).
We have described a method of immobilizing silica particles in a porous polymer monolith to form a microscale solid-phase extraction system inside of a thermoplastic chip (Bhattacharyya and Klapperich 2006). The plastic chips can be injection molded or compression molded. The monolithic columns are formed by light initiated polymerization through the sealed chips. The devices are meant for one time use and are not cleaned or regenerated. Further, these devices can be run completely by a hand-held syringe, making them interesting for global health applications.
Here, we used simulated human UTI samples (urine samples doped with whole blood and E. coli) to evaluate the ability of the lysis and extraction module to perform lysis of E. coli and extract the bacterial DNA on-chip. A polymer monolith with small pores for mechanically assisted lysis is combined in series with a silica bead packed solid-phase extraction (μSPE) monolith to isolate the liberated DNA from the sample. Here, the samples are forced through the small pores at high pressure (ca. 150 psi) in the presence of a high salt buffer. The porous structure is also filled with silica particles that are trapped in the polymer. These particles bind the nucleic acids in the sample. The channels are then washed, and finally the isolated nucleic acids are eluted in water. Lysis efficiency was measured using a fluorescence assay. The overall ability of the module to extract DNA from the microorganisms was assessed using real time PCR.
2 Experimental
2.1 Materials
Cyclic polyolefin (Zeonex 690R) was obtained as a gift from Zeon Chemicals Inc. (Louisville, KY). Butyl methacrylate (99%, BuMA), ethylene glycol dimethacrylate (98%, EGDMA), methyl methacrylate (99%, MMA), 1-dodecanol (98%), cyclohexanol (99%), benzophenone (99%), and 2,2-dimethoxy-2-phenylacetophenone (99%, DMPAP), were purchased from Sigma-Aldrich (St. Louis, MO). A Qiagen DNeasy Blood & Tissue Kit, proteinase K and guanidine thiocyanate (GuSCN) containing lysis buffer (buffer RLT) was purchased from Qiagen Inc. (Catalog #69504, Valencia, CA). Luria Broth was purchased from DIFCO (Franklin Lakes, NJ). SYBR®Green PCR master mix was purchased from Applied Biosystems (Foster City, CA). Excel 3 cc disposable syringes were purchased from Fisher Scientific (Fairlawn, NJ). Silica microspheres (0.7 μm) were purchased from Polysciences, Inc. (Warrington, PA). Polyetheretherke-tone (PEEK) capillaries of 360 μm-i.d. and NanoPort assemblies for device-based fluidic connections were purchased from Upchurch Scientific (Oak Harbor, WA). Human urine and blood samples were purchased from Innovative Research (Novi, Michigan). Human urine, catalog #IR991-B03, and human whole blood (heparin anticoagulant), catalog #IPLA-WB1.
2.2 Microchip fabrication
The microchannels were formed by hot embossing with a nickel-cobalt electroplated mold (NiCoForm, Inc., Rochester, NY) from a silicon master. The channels were 2 cm in length, 400 μm wide and 100 µm deep. The silicon master was fabricated by spinning a negative resist (NR5-8000, Futurrex, Franklin, NJ) to a thickness of 12 μm onto the wafers. After pre-baking the wafers for 1 min. at 150°C, the pattern was transferred through a mask by proximity contact lithography. This step was followed by post-exposure baking, developing with RD6 resist developer (Futurrex, Franklin, NJ) and hard-baking the wafers for 2 min. at 100°C. The exposed areas of the wafer were then etched on the STS DRIE (STS, Newport, UK). Hotembossing was performed with a hot press (Heated Press 4386, Carver, Wabash, IN) at 176°C (30° above the glass transition temperature (Tg) of Zeonex 690R) at a pressure of 500 psi for 4 min. Overall plastic chip size during fabrication was 3 in. in diameter. After embossing, the master and the substrate were removed from the hot press and allowed to cool at room temperature for 30 s on an aluminum plate and were manually separated. 1.5 mm wells were drilled at the ends of the hot-embossed channels. The imprinted channels were sealed with another Zeonex wafer of the same dimensions by thermally bonding at the Tg (136°C) for 4 min at 500 psi. Nanoports (Upchurch Scientific, Oak Harbor, WA) were epoxied to the chip at the location of the wells to provide secure attachment of PEEK tubing from a syringe pump.
2.3 Monolith formation and characterization
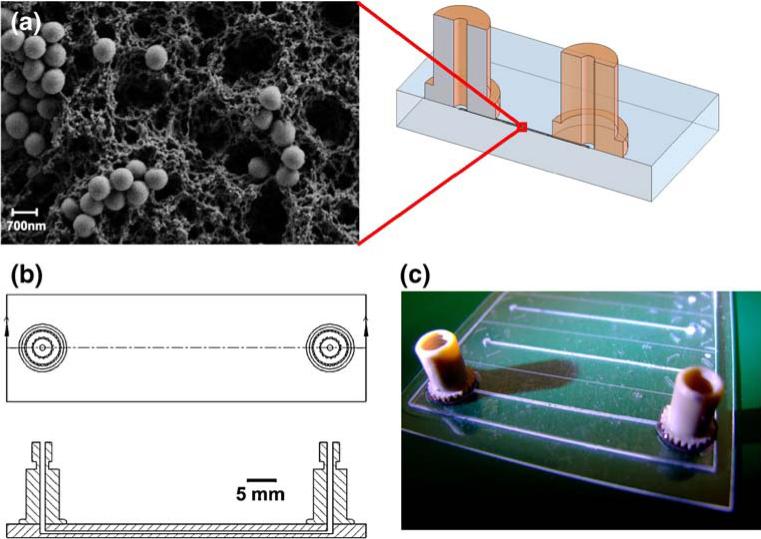
The hot-embossed channels were surface grafted prior to the formation of the polymer monolith columns as previously described (Bhattacharyya and Klapperich 2006; Rohr et al. 2001; Stachowiak et al. 2003; Tan et al. 2003). The grafting step allowed for covalent attachment of the lysis monolith to the inside of the channels. After surface-modification, the monolith was formed using in situ photopolymerization through the sealed channel (Bhattacharyya and Klapperich 2006). The pre-polymer solution for the monolith consisted of BuMA (15 wt.%), EGDMA (10 wt.%), 1-dodecanol (52.5 wt.%), cyclohexanol (22.5 wt.%). This solution was well mixed and 1.13% w/v of DMPAP and the 700 nm silica particles were added to this solution. Each channel was filled with the same amount (3.6 μL) of the monlith/silica solution. Each 3.6 μL aliquot of pre-polymer solution contained 360 μg of the silica particles. The modified channels were filled with the monolith solution and UV irradiated at wavelength of 365 nm in a UV oven (CL-1000 UV Crosslinker, UVP, Inc., Upland, CA) at 120 mJ/cm2 for 1.8 min and then were washed with 50 μL of methanol. An SEM image of a typical solid-phase extraction column, schematics, and a picture of an μSPE in a microchannel are shown in Fig. 1.
Fig. 1.
(a) SEM Image of a lysis/solid-phase extraction monolith taken at ×10,000 magnification. The spheres in the image are the silica particles, (b) CAD drawing of chip components, and (c) a photograph of an array of monolith filled channels. Only the front most channel has nanoports in place
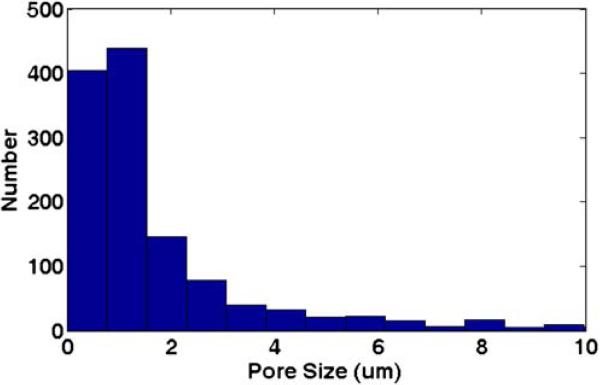
The pore size distribution of the polymer monoliths was obtained from analyzing scanning electron microscope (SEM) images of the monolith using ImageJ (http://rsbweb.nih.gov/ij/). Representative channels were sectioned and images were taken of the monolith in the channel. In total, 38 scale-calibrated images of different locations in two representative monolith columns were batch processed using the built-in „Analyze Particles” function of ImageJ. Specifically, the size limit was set such that only pores greater than 500 nm were counted. The options “Show outlines”, “Exclude on Edges” and “Include Holes” were selected. The output files showed outlines around each pore, and the area measurement results were saved and processed using MATLAB (The Mathworks, Natick, MA) to produce a pore size distribution histogram (Fig. 2).
Fig. 2.
Pore size distribution determined using image analysis of scanning electron micrographs of representative monoliths
2.4 Bacterial strains and culture
Liquid cultures of either K-12 Escherichia coli or DH5-alpha Escherichia coli (GenBank Accession#: U57608) with GFP (green-fluorescent protein) plasmids were grown in 3 mL of Luria Bertani (LB) bacterial growth media at 37°C for 14– 16 h. We chose to use this second strain of Escherichia coli because we have validated specific PCR primers for the transfected GFP gene. Concentrations were confirmed using OD measurements and viable plate counting.
2.5 Simulated UTI samples
The E. coli were grown to stationary phase in liquid culture and OD readings were taken at 600 nm with a biophotometer (Eppendorf BioPhotometer, Eppendorf Scientific, Inc., Westbury, NY) with densities falling within 0.20– 0.22 A, corresponding to a count of approximately 105 CFU/mL. To simulate human UTI samples, the E. coli were resuspended in a solution of human urine with 1% whole blood. While a UTI is commonly defined as a bacterial count of ≥ 105 CFU/mL, it has been suggested that for symptomatic patients and those with lower tract UTIs, a bacterial count of 102 CFU/mL would be a more accurate standard (Bent et al. 2002; Franz and Horl 1999). Furthermore, a lower threshold may be more appropriate due to the often higher fluid intake (and output) of someone sick with a UTI (Franz and Horl 1999), so we performed experiments using a range of E. coli concentrations.
2.6 On-chip lysis and fluorescence quantification of products
Simulated UTI samples with 105 CFU/mL K-12 E. coli were introduced into chips in a 1:1 mixture (100 µL total) of 0.85% NaCl and 0.8 mg/mL proteinase K solution at 300 μL/h. The NaCl solution was used to mitigate any chemical lysis that the high salt lysis and extraction buffer might introduce. In other words, we wanted to test whether the lysis and extraction columns could lyse the bacteria without the chemical assist. Various flow rates between 100 and 450 μL/h were tried, and no flow rate dependence on lysis was seen in this range. The pressure drop across the channel was measured for several of the samples using an inline transducer. Samples were run over the column, and then removed from the device. The collected sample was gently filtered using a 0.2-μm filter to remove any intact cells or cell debris that might contribute to the fluorescence signal. The resulting solution was then ethanol precipitated (again as a cleaning step to ensure that we were measuring only fluorescence from liberated nucleic acids) and resus-pended in clean water. The amount of DNA was determined using a Quant-IT PicoGreen Assay (Invitrogen, Carlsbad, CA). The positive controls were 100 μL samples extracted using a Qiagen Blood and Cell Culture DNA kit (Qiagen, Valencia, CA). The negative controls were samples mixed with the appropriate buffer and left to sit on the bench for the duration of the experiment. This experiment was repeated for simulated UTI samples introduced into the chip in a 1:1 mixture with the lysis and extraction buffer, GuSCN containing 0.8 mg/mL proteinase K and 0.01% SDS.
2.7 Lysis and extraction of bacteria
Simulated UTI samples (101–105 CFU/mL DH5-alpha Escherichia coli) were introduced into the combined lysis and μSPE chip in a 1:1 mixture of GuSCN containing 0.8 mg/mL proteinase K and 0.01% SDS (100 μL total volume). Samples were flowed through the chips at a pump setting 450 µL/h. Once a sample had been passed through the column, the column was washed once with cold 70% ethanol and the isolated DNA was eluted in 70 μl of clean water. Two fractions of 70 μl were eluted from each chip and are labeled F1 and F2. These three steps took a combined 30–40 min to complete. Controls were the Qiagen kit (positive) and empty microchannels (negative). We also ran water as a PCR negative control.
2.8 Quantitative PCR
Eluted DH5-alpha Escherichia coli samples were amplified using real-time PCR (Applied Biosystems 7300, Foster City, CA) with a SYBR®Green assay and compared to the Qiagen kit isolated samples. The primers for the green fluorescent protein (GFP) plasmid were 5′-atgcccgaaggc tacgtcca-3′and 3′-caggaccatgtgatcgcgct-5′. Twenty-five microliter assays were run in 96 well plates. Plates were preheated and incubated a 94°C for 3 min. The PCR program was thirty cycles of 94°C for 30 s, 57°C for 1 min followed by 72°C for 1 min. All samples were run in multiple wells to minimize the effects of pipetting errors. Results were confirmed using polyacrylamide gel electrophoresis (data not shown).
3 Results and discussion
Characterization of the average pore size of the monoliths used for this study was performed using image analysis as described. The data are presented in a histogram in Fig. 2. The histogram shows over 800 counts of size below 1.5 μm. The final average size over all images was 2.56 μm ± 1.00. E. coli vary in size but are most often about 1 μm in diameter and 2 μm in length. So, the tortuous nature of the small open pore system is likely mechanically shearing the bacteria. This effect may be enhanced by the presence of a chaotropic agent.
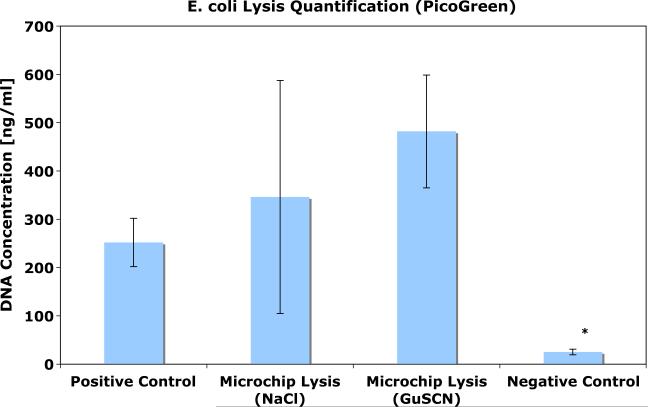
The fluorescence quantification results are presented in Fig. 3. Comparable amounts of DNA were recovered from samples run through the lysis monolith and cleaned up off-chip in comparison to the positive control. The negative control (bench sample) showed a small amount of lysis, but much less than both the microcolumn and the positive control. The results were not significantly altered by the addition of the high salt buffer, indicating that a substantial amount of the lysis in the microcolumn is due to mechanical effects.
Fig. 3.
Summary of lysis quantification experiments. DNA concentration was measured after samples were run over a column in the presence of both low and high salt buffer solutions. The positive control is the Qiagen kit. The negative control is a sample mixed with NaCl buffer and left on the bench for the duration of the experiment. The experimental samples and the negative controls were gently filtered (0.2 μm filter) and ethanol precipitated to remove any fluorescent cell debris prior to fluorescent quantification. *p<0.001
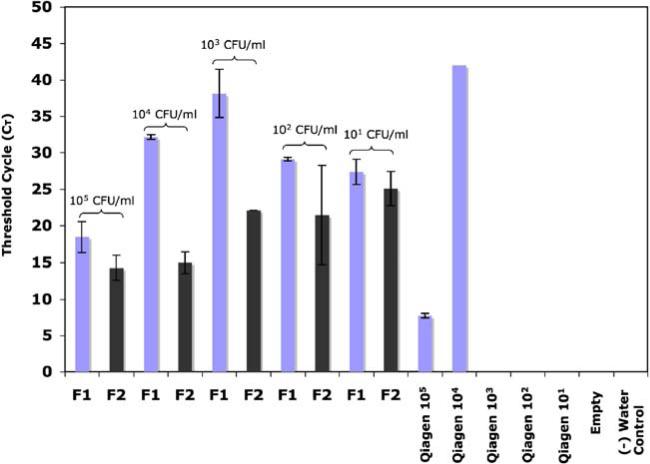
The real time PCR results are presented in Fig. 4. The CT is the cycle at which the fluorescence generated by the amplification of the target sequence crosses a threshold value defined by the user. All amplification below this threshold is considered noise, or a negative result. In general, the lower the CT value, the more initial template was present.
Fig. 4.
Summary of amplification threshold values for E. coli and whole blood spiked urine samples. A lower threshold value indicates more DNA was present in the extracted sample. F1 refers to the first 70 μl sample collected after elution and F2 refers to the second. Elutions that never amplified are not included in this data. 22/27 (81%) of the microchannels produced amplifiable results
A higher value means less of the gene was present.
We compared the CT values of the DNA recovered with the μSPE column to the positive control for bacteria concentrations from 105 CFU/mL to 101 CFU/mL. Figure 4 shows a summary of the data. The Qiagen kit showed a low CT value only for the highest concentration of input sample (105 CFU/mL). The μSPE columns (n=3 were run at each concentration) performed comparably or better than the Qiagen kit at all of the other concentrations. The Qiagen kit was unable to isolate PCRable DNA at the three lowest concentrations. The empty channel and no template controls did not amplify.
Recall that two 70 μL fractions, F1 and F2 were recovered from each channel. Not all of the sample fractions collected from the microchannels amplified. Those fractions that did not amplify (CT=Ø) are not included in the plot. Of the 27 fractions collected, 22 of them produced amplifiable DNA (81%). At 105 CFU/mL, all of the collected fractions amplified. At 104 CFU/mL, 5/6 fractions amplified. At 103 CFU/mL, 4/6 fractions amplified. At 102 CFU/mL, 6/6 fractions amplified and at 101 CFU/mL 4/6 amplified. For the 104 CFU/mL and 103 CFU/mL cases, the non-amplifying fractions were always the first fraction; the second fractions amplified in all cases. For the 101 CFU/mL one of the channels (both fractions) never amplified, while the other two channels had amplifiable DNA in both the first and second fractions. In every case, the second fraction had a lower CT value (Fig. 4), indicating the presence of PCR inhibitors in the first elution fraction. Since ethanol can be a PCR inhibitor, it is possible that better elution in the first fraction might be achieved if the microcolumns are dried prior to elution. In addition, we need to optimize the channel cleaning procedures, as we suspect that unreacted pre-polymer solution may be present and may inhibit the downstream PCR.
At the lowest concentration, 101 CFU/mL, one would expect that roughly 2/3 (61%) of 50 μL samples taken from the test samples would not contain a bacterium due to sampling effects. In this set of experiments we saw 2/3 fractions amplifying, but n=3 is a relatively small sample size. Microfluidic assay developers need to remain mindful of the Poisson effect when dealing with dilute solutions, however; since in many biological fluids, the presence of the causative agent is low, and the sampling volumes are necessarily small(Stenman and Orpana 2001).
We saw comparable results to the Qiagen kit at the highest concentration and the microchannels performed better at recovering amplifiable DNA than the control at lower concentrations. At higher concentrations of bacteria, we may be overloading and saturating the μSPE column, but this is unlikely due to the small amount of nucleic acids present. However, saturation can be complicated by other factors(Hara et al. 2005), and incomplete elution may be a problem(Arroyo et al. 2005). A rough calculation based on literature values showing that the capacity of these silica particles is on the order of 10–30 ng/mg of DNA(Tian et al. 2000), indicates that each channel should be capable of binding roughly 4 ng of DNA. For the highest concentration case here, 105 CFU/mL, if all of the organisms present in a 50 μL sample (5,000) were completely lysed, then roughly one ng of DNA should be present (taking into account the additional plasmid DNA). It is unlikely that the channels are saturated at these concentrations and sample sizes, leaving incomplete elution and/or the presence of PCR inhibitors as potential complicating factors. At lower concentrations we may be capturing and eluting more of the captured DNA present than the commercial kit because of the small volume and closed nature of the system. For small volume samples, it can be easy to leave behind a significant sample volume in a Qiagen tube.
In determining the effectiveness of the μSPE column for isolation at lower concentrations we were able to successfully amplify target down to 101 CFU/mL. If each organism contained only one plasmid, this number would indicate one live bacteria in a sample of 100 μl. The DH5-alpha Escherichia coli nominally contain 500 plasmids per organism as received, and this number can decrease significantly over long term culture (Koenig 2003). So, we cannot use this data to back calculate the limit of detection in terms of copy number of genomic DNA. The PicoGreen assay results (Fig. 2) suggest that we are able to lyse bacteria and liberate nucleic acids at lower concentrations, since those experiments were performed with a strain that does not contain a transfected plasmid. It is also important to note that CFU counts are based on the number of living organisms in a sample and are approximate and less reliable at low concentrations. In addition, we are able to detect dead organisms that are not capable of generating a colony on an agar plate, so our results may actually capture more starting material than we are able to accurately count.
Additionally, while the samples here were eluted in 70 μL of water, we have demonstrated that the isolated DNA can be eluted in smaller volumes. When aliquots of 5 μL are sequentially used to elute standard samples of human genomic DNA in buffer from the extraction columns, the bulk of the sample is recovered in the first 10 μL, suggesting that smaller elution volumes can be used to obtain even more concentrated samples, while shortening the overall time for sample preparation.
4 Conclusions
We have fabricated a microscale module that can be used with a pressure system to perform lysis and extract DNA from bacteria infected human urine samples in the presence of human whole blood contamination. We have established proof-of-concept for a sample preparation microfluidic device that uses shear and frictional forces coupled with a high salt buffer to achieve these goals. We were able to successfully isolate bacterial DNA from simulated UTI samples with a range of concentrations, 105–101 CFU/mL. The integrated sample preparation channel processes a 100 μl sample with one wash in less than 40 min. Reducing the elution volume used here and increasing the channel volumes will lead to more time savings. With further design development, this system will be suitable for integration with in-line amplification and detection technologies in thermoplastic platforms.
Acknowledgments
We thank the Wallace H. Coulter Foundation for funding this research and H. Muayad for help with image analysis.
Contributor Information
M. Dominika Kulinski, Department of Mechanical Engineering, Boston University, 44 Cummington St., Boston, MA, USA.
Madhumita Mahalanabis, Department of Biomedical Engineering, Boston University, 44 Cummington St., Boston, MA, USA.
Sara Gillers, Boston University Medical Center, 720 Harrison Ave., Boston 02118 MA, USA.
Jane Y. Zhang, Department of Biomedical Engineering, Boston University, 44 Cummington St., Boston, MA, USA
Satish Singh, Boston University Medical Center, 720 Harrison Ave., Boston 02118 MA, USA.
Catherine M. Klapperich, Department of Mechanical Engineering, Boston University, 44 Cummington St., Boston, MA, USA Department of Biomedical Engineering, Boston University, 44 Cummington St., Boston, MA, USA.
References
- Arroyo E, Wheeler EK, Shediac R, Hindson B, Nasarabadi S, Vrankovich G, Bell P, Bailey C, Sheppod T, Christian AT. Flow through pcr module of biobriefcase. Smart Medical and Biomedical Sensor Technology III, Proceedings of the SPIE, Vol. 6007. Proceedings of the SPIE. 2005 [Google Scholar]
- Belgrader P, Hansford D, Kovacs GT, Venkateswaran K, Mariella Jr R, Milanovich F, Nasarabadi S, Okuzumi M, Pourahmadi F, Northrup MA. A minisonicator to rapidly disrupt bacterial spores for DNA analysis. Anal. Chem. 1999;71(19):4232–4236. doi: 10.1021/ac990347o. doi:10.1021/ac990347o. [DOI] [PubMed] [Google Scholar]
- Bent S, Nallamothu BK, Simel DL, Fihn SD, Saint S. Does this woman have an acute uncomplicated urinary tract infection? JAMA. 2002;287(20):2701–2710. doi: 10.1001/jama.287.20.2701. doi:10.1001/jama.287.20.2701. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Klapperich CM. Thermoplastic microfluidic device for on-chip purification of nucleic acids for disposable diagnostics. Anal. Chem. 2006;78(3):788–792. doi: 10.1021/ac051449j. doi:10.1021/ac051449j. [DOI] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breadmore MC, Shrinivasan S, Wolfe KA, Power ME, Ferrance JP, Hosticka B, Norris PM, Landers JP. Towards a microchip-based chromatographic platform. Part 1: Evaluation of sol-gel phases for capillary electrochromatography. Electrophoresis. 2002;23(20):3487–3495. doi: 10.1002/1522-2683(200210)23:20<3487::AID-ELPS3487>3.0.CO;2-5. doi:10.1002/1522-2683(200210) 23:20<3487::AID-ELPS3487>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Breadmore MC, Wolfe KA, Arcibal IG, Leung WK, Dickson D, Giordano BC, Power ME, Ferrance JP, Feldman SH, Norris PM, Landers JP. Microchip-based purification of DNA from biological samples. Anal. Chem. 2003;75(8):1880–1886. doi: 10.1021/ac0204855. doi:10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- Brown PD, Freeman A, Foxman B. Prevalence and predictors of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli isolates in Michigan. Clin. Infect. Dis. 2002;34(8):1061–1066. doi: 10.1086/339491. doi:10.1086/339491. [DOI] [PubMed] [Google Scholar]
- Chen XW, Xu ZR, Qu BY, Wu YF, Zhou J, Zhang HD, Fang J, Wang JH. DNA purification on a lab-on-valve system incorporating a renewable microcolumn with in situ monitoring by laser-induced fluorescence. Anal. Bioanal. Chem. 2007;388(1):157–163. doi: 10.1007/s00216-007-1196-0. doi:10.1007/s00216-007-1196-0. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Jeong KH, Lee LP. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3(4):287–291. doi: 10.1039/b305162e. doi:10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Ionescu-Zanetti C, Zhang Y, Hung P, Lee LP. On-chip cell lysis by local hydroxide generation. Lab Chip. 2005;5(2):171–178. doi: 10.1039/b413139h. doi:10.1039/b413139h. [DOI] [PubMed] [Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc. Natl. Acad. Sci. U S A. 2006;103(51):19272–19277. doi: 10.1073/pnas.0604663103. doi:10.1073/ pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B, Gillespie B, Koopman J, Zhang L, Palin K, Tallman P, Marsh JV, Spear S, Sobel JD, Marty MJ, Marrs CF. Risk factors for second urinary tract infection among college women Am. J. Epidemiol. 2000;151(12):1194–1205. doi: 10.1093/oxfordjournals.aje.a010170. [DOI] [PubMed] [Google Scholar]
- Franz M, Horl WH. Common errors in diagnosis and management of urinary tract infection. I: Pathophysiology and diagnostic techniques. Nephrol. Dial. Transplant. 1999;14(11):2746–2753. doi: 10.1093/ndt/14.11.2746. doi:10.1093/ndt/14.11.2746. [DOI] [PubMed] [Google Scholar]
- Gaydos CA, Theodore M, Dalesio N, Wood BJ, Quinn TC. Comparison of three nucleic acid amplification tests for detection of chlamydia trachomatis in urine specimens. J. Clin. Microbiol. 2004;42(7):3041–3045. doi: 10.1128/JCM.42.7.3041-3045.2004. doi:10.1128/JCM.42.7.3041-3045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Hooton TM, Stamm WE. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 2001;135(1):41–50. doi: 10.7326/0003-4819-135-1-200107030-00012. [DOI] [PubMed] [Google Scholar]
- Hara C, C., N., Wheeler E, Sorensen K, Arroyo E, Vrankovich G, Christian A. Small sample whole-genome amplification. Smart Medical and Biomedical Sensor Technology III. Proceedings of the SPIE. 2005 [Google Scholar]
- Hooton TM, Besser R, Foxman B, Fritsche TR, Nicolle LE. Acute uncomplicated cystitis in an era of increasing antibiotic resistance: a proposed approach to empirical therapy. Clin. Infect. Dis. 2004;39(1):75–80. doi: 10.1086/422145. doi:10.1086/422145. [DOI] [PubMed] [Google Scholar]
- Koenig GL. Viability of and plasmid retention in frozen recombinant Escherichia coli over time: a ten-year prospective study. Appl. Environ. Microbiol. 2003;69(11):6605–6609. doi: 10.1128/AEM.69.11.6605-6609.2003. doi:10.1128/AEM.69.11.6605-6609.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Mastali M, Gau V, Suchard MA, Moller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, McCabe ER, Churchill BM, Haake DA. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J. Clin. Microbiol. 2006;44(2):561–570. doi: 10.1128/JCM.44.2.561-570.2006. doi:10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Mastali M, Li Y, Gau V, Suchard MA, Babbitt J, Gornbein J, Landaw EM, McCabe ER, Churchill BM, Haake DA. Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection. J. Mol. Diagn. 2007;9(2):158–168. doi: 10.2353/jmoldx.2007.060052. doi:10.2353/jmoldx.2007.060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindan C, Mathur M, Kumta S, Jerajani H, Gogate A, Schachter J, Moncada J. Utility of pooled urine specimens for detection of chlamydia trachomatis and neisseria gonorrhoeae in men attending public sexually transmitted infection clinics in mumbai, india, by pcr. J. Clin. Microbiol. 2005;43(4):1674–1677. doi: 10.1128/JCM.43.4.1674-1677.2005. doi:10.1128/JCM.43.4.1674-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeckh T, Lopez S, Fuller AO, Solomon MJ, Larson RG. Adsorption and elution characteristics of nucleic acids on silica surfaces and their use in designing a miniaturized purification unit. Anal. Biochem. 2008;373(2):253–262. doi: 10.1016/j.ab.2007.10.026. doi:10.1016/j. ab.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr T, Yu C, Davey MH, Svec F, Frechet JM. Porous polymer monoliths: Simple and efficient mixers prepared by direct polymerization in the channels of microfluidic chips. Electrophoresis. 2001;22(18):3959–3967. doi: 10.1002/1522-2683(200110)22:18<3959::AID-ELPS3959>3.0.CO;2-5. doi:10.1002/1522-2683(200110) 22:18<3959::AID-ELPS3959>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Stachowiak TB, Rohr T, Hilder EF, Peterson DS, Yi M, Svec F, Frechet JM. Fabrication of porous polymer monoliths covalently attached to the walls of channels in plastic microdevices. Electrophoresis. 2003;24(21):3689–3693. doi: 10.1002/elps.200305536. doi:10.1002/ elps.200305536. [DOI] [PubMed] [Google Scholar]
- Stenman J, Orpana A. Accuracy in amplification. Nat. Biotechnol. 2001;19(11):1011–1012. doi: 10.1038/nbt1101-1011b. doi:10.1038/nbt1101-1011b. [DOI] [PubMed] [Google Scholar]
- Tan A, Benetton S, Henion JD. Chip-based solid-phase extraction pretreatment for direct electrospray mass spectrometry analysis using an array of monolithic columns in a polymeric substrate. Anal. Chem. 2003;75(20):5504–5511. doi: 10.1021/ac030196w. doi:10.1021/ac030196w. [DOI] [PubMed] [Google Scholar]
- Tian H, Huhmer AF, Landers JP. Evaluation of silica resins for direct and efficient extraction of DNA from complex biological matrices in a miniaturized format. Anal. Biochem. 2000;283(2):175–191. doi: 10.1006/abio.2000.4577. doi:10.1006/abio.2000.4577. [DOI] [PubMed] [Google Scholar]
- Van Dilla MA, Langlois RG, Pinkel D, Yajko D, Hadley WK. Bacterial characterization by flow cytometry. Science. 1983;220(4597):620–622. doi: 10.1126/science.6188215. doi:10.1126/science.6188215. [DOI] [PubMed] [Google Scholar]
- Wang HY, Banada PP, Bhunia AK, Lu C. Rapid electrical lysis of bacterial cells in a microfluidic device Methods. Mol. Biol. 2007;385:23–35. doi: 10.1007/978-1-59745-426-1_3. doi:10.1007/978-1-59745-426-1_3. [DOI] [PubMed] [Google Scholar]
- Wolfe KA, Breadmore MC, Ferrance JP, Power ME, Conroy JF, Norris PM, Landers JP. Toward a microchip-based solid-phase extraction method for isolation of nucleic acids. Electrophoresis. 2002;23(5):727–733. doi: 10.1002/1522-2683(200203)23:5<727::AID-ELPS727>3.0.CO;2-O. doi:10.1002/1522-2683(200203) 23:5<727::AID-ELPS727>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]