ABSTRACT
The membrane-proximal external region (MPER), the V2/glycan site (initially defined by PG9 and PG16 antibodies), and the V3/glycans (initially defined by PGT121–128 antibodies) are targets of broadly neutralizing antibodies and potential targets for anti-HIV-1 antibody-based vaccines. Recent evidence shows that antibodies with moderate neutralization breadth are frequently attainable, with 50% of sera from chronically infected individuals neutralizing ≥50% of a large, diverse set of viruses. Nonetheless, there is little systematic information addressing which specificities are preferentially targeted among such commonly found, moderately broadly neutralizing sera. We explored associations between neutralization breadth and potency and the presence of neutralizing antibodies targeting the MPER, V2/glycan site, and V3/glycans in sera from 177 antiretroviral-naive HIV-1-infected (>1 year) individuals. Recognition of both MPER and V3/glycans was associated with increased breadth and potency. MPER-recognizing sera neutralized 4.62 more panel viruses than MPER-negative sera (95% prediction interval [95% PI], 4.41 to 5.20), and V3/glycan-recognizing sera neutralized 3.24 more panel viruses than V3/glycan-negative sera (95% PI, 3.15 to 3.52). In contrast, V2/glycan site-recognizing sera neutralized only 0.38 more panel viruses (95% PI, 0.20 to 0.45) than V2/glycan site-negative sera and no association between V2/glycan site recognition and breadth or potency was observed. Despite autoreactivity of many neutralizing antibodies recognizing MPER and V3/glycans, antibodies to these sites are major contributors to neutralization breadth and potency in this cohort. It may therefore be appropriate to focus on developing immunogens based upon the MPER and V3/glycans.
IMPORTANCE Previous candidate HIV vaccines have failed either to induce wide-coverage neutralizing antibodies or to substantially protect vaccinees. Therefore, current efforts focus on novel approaches never before successfully used in vaccine design, including modeling epitopes. Candidate immunogen models identified by broadly neutralizing antibodies include the membrane-proximal external region (MPER), V3/glycans, and the V2/glycan site. Autoreactivity and polyreactivity of anti-MPER and anti-V3/glycan antibodies are thought to pose both direct and indirect barriers to achieving neutralization breadth. We found that antibodies to the MPER and the V3/glycans contribute substantially to neutralization breadth and potency. In contrast, antibodies to the V2/glycan site were not associated with neutralization breadth/potency. This suggests that the autoreactivity effect is not critical and that the MPER and the V3/glycans should remain high-priority vaccine candidates. The V2/glycan site result is surprising because broadly neutralizing antibodies to this site have been repeatedly observed. Vaccine design priorities should shift toward the MPER and V3/glycans.
INTRODUCTION
A relatively small number of epitopes that are targets of broadly neutralizing antibodies (Abs) have been identified on the HIV-1 envelope glycoproteins, gp120 and gp41 (1–5). Prominent among them, the membrane-proximal external region (MPER), the V2/glycan site, and the V3/glycans are models for candidate vaccine antigens (1–3). Sophisticated efforts have been made to attach these targets to protein scaffolds in order to create vaccine immunogens to elicit neutralizing antibodies (6), highlighting their importance in vaccine development.
The membrane-proximal external region (MPER) is the target of three broadly neutralizing monoclonal antibodies (MAbs) (7, 8). The MPER appears to be a relatively simple, linear antigen (9) but harbors substantial complexity (10–14). Another set of potent and broadly neutralizing antibodies, PGT121–128 and PGT130–131, bind primarily to glycans at either position 301 or position 332 in the V3 loop (“V3/glycans”) (15). The V2/glycan site is a quaternary epitope (16) that is thought to be stabilized by the presence of the N160 glycan, without forming a direct part of the epitope (17). Antibodies recognizing MPER and the V3/glycans have been reported to be self-reactive (2, 18–20). It has long been suspected that self-reactivity checkpoints may limit the ability of many individuals to produce broadly neutralizing responses to such targets (2, 19, 20).
Little is known about the likelihood that any particular neutralizing anti-HIV antibody will become broadly neutralizing, even though the route of somatic hypermutation to arrive at rare broadly neutralizing antibodies is being elucidated (21, 22). Recent evidence shows that antibodies with this moderate neutralization breadth are frequently attainable (perhaps even in response to a vaccine [23]), more so than the very well-studied and highly broadly neutralizing antibodies found in sera from the top 1% to 2% “elite neutralizers” (24). A total of 50% of sera from chronically infected individuals achieve moderate neutralization breadth, neutralizing ≥50% of a large, diverse set of viruses (23).
There is little systematic information about which specificities are preferentially targeted among moderately broadly neutralizing sera. In this study, we observed that neutralization breadth and potency were significantly positively associated with the presence of MPER-specific neutralization and V3/glycan-specific neutralization but not with anti-V2/glycan site-specific neutralization. These data suggest that many individuals are capable of developing antibody responses of moderate to high neutralization breadth recognizing the MPER and V3/glycans. This may suggest that it would be easier to elicit such antibodies in response to a vaccine.
MATERIALS AND METHODS
Samples.
Blood samples were collected in December 2009 to July 2011 from donors who were >18 years old and HIV-1 infected (>1 year) and were not exposed to antiretroviral therapy (ART), except for ART given for prevention of mother-to-child transmission (>3 months prior). Study participants were recruited from among (i) caregivers of patients at the pediatric HIV clinic at Groote Schuur Hospital and (ii) attendees of the HIV wellness clinic at the Khayelitsha Site B clinic. Both clinics are in Cape Town, South Africa. Written informed consent was received from study participants. This study was approved by the Human Research Ethics Committee, Faculty of Health Sciences of the University of Cape Town. Data were included from another project approved by the Human Research Ethics Committee, Faculty of Health Sciences of the University of Cape Town, and the National Ethics Committee of the Republic of Cameroon.
Pseudovirus constructs.
The envelope constructs for COT6.15, Du151.2, Du156.12, and murine leukemia virus (MLV) envelope were kind gifts from Lynn Morris and Penny Moore, National Institute for Communicable Diseases (NICD), Johannesburg, South Africa. The SG3 HIV-1 genome with an inactivated envelope gene (SG3-Δenv) and envelope constructs (unless specified otherwise) were received via the NIH AIDS Research Reagent Reference Program. The 7312A HIV-2 genomic construct and chimeric versions of it with MPER sequences swapped in from Yu2 (C1 [25]) and consensus C MPER (C1C [26]) were kind gifts from George Shaw, University of Pennsylvania, USA. We generated the chimera displaying 253-11 MPER sequence from C1 by site-directed mutagenesis. CAP45.2.00.G3 N160A and K169E and Du156.12 N160K, K169E, and N332A were kind gifts from Lynn Morris and Penny Moore. QH343.A10.N160A, I169E, and N301A/N332A, Du156.12 N301A/N332A, and CAP45.2 N301A were made from QH343.21M.ENV.A10, Du156.12 N332A, or CAP45.2.00.G3 by site-directed mutagenesis. All constructs made by site-directed mutagenesis were confirmed by sequencing both strands of the open reading frame of the envelope gene.
Neutralization assay.
Neutralization was tested using a standard pseudovirus-based neutralization assay (27). Titers (50% infective doses [ID50]) were calculated using curve fit functions in Prism (GraphPad, La Jolla, CA, USA), except that for the purposes of determining neutralization breadth or potency, many ID50 values were predicted (see below). MLV was used as a negative control; MLV neutralization was low (<20% neutralization), except for two sera with 20% to 30% at a 1/100 dilution.
Pseudovirus panel and assessment of neutralization breadth and potency of sera.
A pseudovirus panel (n = 24) representing the global HIV-1 pandemic was assembled to evaluate the neutralization breadth of sera. The panel was selected based upon neutralization resistance (28–30; R. A. Jacob, unpublished data), subtype, and geographic diversity. Panel viruses are listed and described (see Fig. 2D). All tier (neutralization resistance) designations are according to Seaman and colleagues (28). A neutralization score for each serum was determined by calculating a geometric mean ID50 of all 24 viruses as neutralized by that serum. A neutralization sensitivity score for each virus was determined by calculating a geometric mean ID50 titer of all 177 sera neutralizing that virus. Fold difference in sensitivity of subtypes of panel viruses was determined by calculating the ratio of geometric means of all measurements for viruses of each subtype. The 95% confidence interval (95% CI) of the fold difference for each subtype comparison was calculated from a log linear mixed-regression model.
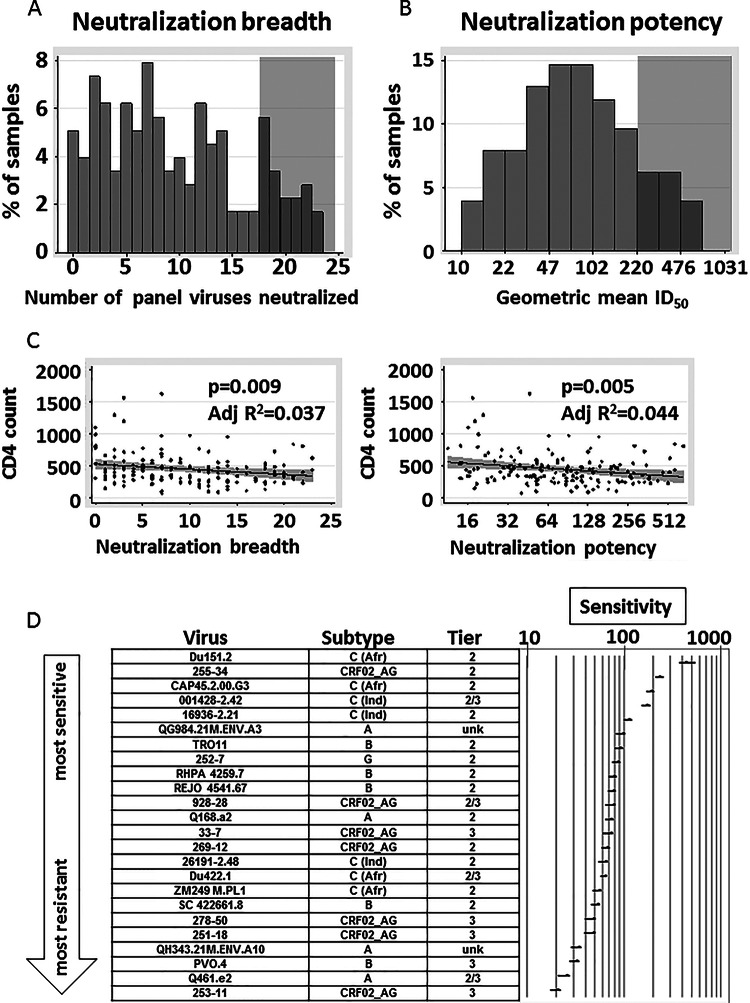
FIG 2.
Neutralization breadth and potency of cohort sera and association with CD4+ T cell count. (A) The distribution of neutralization breadth of the 177 cohort sera is shown by displaying the number of viruses neutralized by each serum. Gray shading indicates at what level samples were scored positive for high neutralization breadth (>3/4 of panel viruses neutralized). (B) The distribution of neutralization potency of the 177 cohort sera is shown by displaying the geometric mean ID50 of each serum neutralizing the 24 panel viruses. Gray shading indicates at what level samples were scored positive for high neutralization potency (geometric mean ID50, >220). (C) Comparison of neutralization breadth and potency to the CD4+ T cell count measured in the same sample. Potency is shown on a log2 scale. Line fits, P values, and adjusted (Adj) R2 values were calculated from a linear regression model. Gray shading represents the 95% CI of the linear regression line. (D) Relative sensitivity ranking of viruses with respect to the 177 cohort sera. Viruses were ranked by the geometric mean ID50 values for all 177 sera neutralizing that virus; 95% prediction intervals (95% PI) from the marginal prediction of a log linear mixed model are depicted. C (Afr), subtype C and derived from an African donor; C (Ind), subtype C and derived from an Indian donor; unk, unknown. Tier designations are from Seaman et al. (28); Tier 2/3, found to be between tiers 2 and tier 3.
Detection of anti-MPER, anti-V2/glycan site, and anti-V3/glycan antibodies.
Chimeric 7312A HIV-2 viruses engrafted with a consensus subtype C MPER (C1C [26]) or a Yu2 MPER (C1 [26, 31]) or the MPER sequence of a CRF02_AG virus, 253-11, were used to detect anti-MPER antibodies. Samples were scored positive for anti-MPER antibodies if they neutralized at least one of three chimeric viruses at an ID50 of >1,000. Anti-MPER-positive sera did not detectably neutralize the 7312A control (data not shown). Dominant anti-V2/glycan antibodies were detected using pseudoviruses with individual mutations at positions 160 and 169. The N160A/K and K/I169E single-amino-acid substitutions abrogate PG9 and PG16 MAb neutralization and have been used to identify anti-V2/glycan antibodies from blood samples (32–35; T. Moyo, unpublished data). Dominant anti-V3/glycan antibodies were detected using pseudoviruses with mutations at position N301 and/or N332. Either N301 or N332 is necessary for the full neutralization activity of anti-V3/glycan MAbs PGT120–131 (15), and N332 is important for neutralization by anti-glycan MAb 2G12 (36). We used three parent (wild-type) viruses: CAP45.2.00.G3 (37), with virus mutants N160A, K169E, and N301A (N at position 332 is not glycosylated in CAP45); Du156.12 (38), with virus mutants N160K, K169E, and N301A/N332A; and QH343.21M.ENV.A10 (29), with virus mutants N160A, I169E, and N301A/N332A. Neutralization mapping was scored positive if a ≥3-fold drop (32) in neutralization ID50 was observed with ≥1 mapping mutant(s) for the site concerned compared to the corresponding wild-type virus.
Comparison of neutralization breadth and potency between groups of sera.
Neutralization breadth and potency were compared for anti-V3/glycan, anti-MPER, and anti-V2/glycan site neutralizing sera by means of the ratios of the geometric mean ID50 titer (potency) and differences (breadth) for the numbers of viruses neutralized at an ID50 of >100, i.e., based on the measured ID50 values (n = 312) and predicted ID50 values (n = 3,936). We calculated the aggregate neutralization breadth and potency for the sera mapped to three different sites and summarized them by means of their ratios and differences compared to sera that did not map to each site or to other groups. Additive linear mixed models were used to model the association of log ID50 and percent neutralization. Bootstrap estimation (39) (1,000 replicates) was used to estimate the confidence intervals of these ratios and differences and to estimate the prediction error associated with the ID50 estimation and model fit.
Depletion of anti-MPER antibodies.
An 11-virus panel was assembled to test sera for their capacity to neutralize HIV-1 viruses by recognition of the MPER. The panel consisted of COT6.15 (subtype C), Du151.2 (C), CAP45.2.00.G3 (C), Du422.1 (C), 001428-2.47 (C), TRO.11 (B), REJO4541.67 (B), RHPA4259.7 (B), 928-28 (CRF02_AG), 269-12 (CRF02_AG), and 253-11 (CRF02_AG). Antibodies were depleted in two rounds of depletion (34, 40) as previously described using a biotinylated MPER peptide (MPR.03 [31, 34, 40, 41]; KKKNEQELLELDKWASLWNWFDITNWLWYIRKKK-biotin-NH2; Peptide Synthetics, Hampshire, United Kingdom). Control depletions were performed as described above using streptavidin-magnetic Dynabeads (Invitrogen, Darmstadt, Germany) and a biotinylated control peptide with a scrambled sequence (KKKNEKSNNDWERLWLEWLYIWLQDWAFTLIKKK-biotin-NH2). A threshold of a ≥2-fold drop (34, 40, 42) in ID50 compared to control peptide depletion was accepted as positive for MPER-mediated neutralization, i.e., as indicating that more than half of the neutralizing activity was directed against MPER. Six serum samples from the cohort and one CRF02_AG-infected plasma sample (30, 43) were tested against at least 7 of the viruses in the 11-virus panel.
RESULTS
Study participants.
The median age of the study participants was 33 years (interquartile range [IQR], 28 to 37 years), with 17 (10%) males and 160 (90%) females, reflecting the general gender imbalance in adults seeking care at our recruiting facilities. The median CD4+ T cell count value was 407 (IQR, 286 to 533). The median known duration of infection was 3.0 years (IQR, 1.7 to 5.9 years). The known duration of infection was determined by the duration of time since the diagnosis of HIV infection or by the earliest CD4+ T cell count documented in clinical records, if possible, or from the study participant's verbal report.
Use of ID50 prediction and its validation.
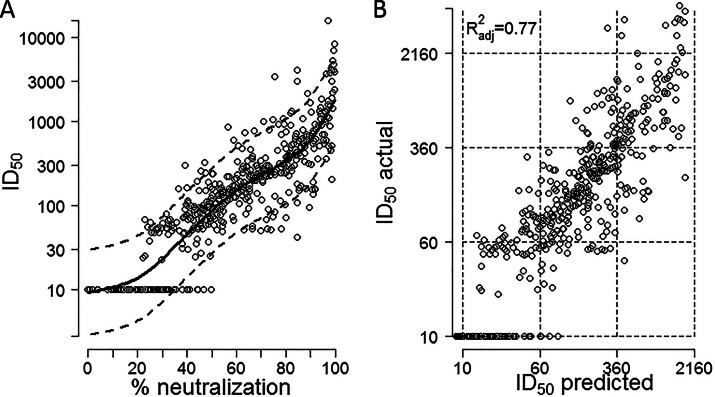
We directly measured (n = 312) or predicted (n = 3,936) the neutralization effect of each of the 177 sera on 24 panel viruses and devised a model to estimate the additional error that was generated by the prediction model (prediction error). To generate the prediction model, the effect of the percentage of neutralization at a 1/100 dilution was modeled both linearly and nonlinearly (spline-based model [44]). We chose the nonlinear model (Fig. 1A) because of its better predictive ability (cross-validation error [cv] [39] = 5.42 versus cv = 5.73 for the linear model). The values for the serum/virus pairs used to generate the model were from this study (290/474) or a previous study (36) (72/474) or other unpublished values (112/474). The model was validated by 10-fold cross-validation, i.e., the data were split into 10 different subsets of the approximately same size; 9 of the subsets were pooled to estimate the model used to predict values for the 10th subset (39). This was repeated 9 times until all data points appeared once in the comparison of predicted versus measured ID50 values (Fig. 1B). The fit was very good (adjusted R2 = 0.7664), and the estimated slope was 0.99 (95% CI, 0.94 to 1.04), close to the expected slope of 1.
FIG 1.
Statistical prediction model of ID50 values from percent neutralization. (A) Prediction function of ID50 determined by percent neutralization at a 1/100 dilution. The dashed lines correspond to ±2 times the residual standard error. This reflects the conditional normal distribution related to the underlying linear model. (B) Testing of the prediction model was performed using a set of 474 virus/serum combinations with measured 1/100 dilution screening values and ID50 values measured by titration. The data were split into 10 different subsets of approximately the same size; 9 of the subsets were pooled to estimate the model which was used to predict values for the 10th subset. This procedure was repeated 10 times so that a predicted value was obtained for each percent neutralization value. The predicted and measured ID50 values are shown for each of the 474 virus/serum combinations.
Measurement of potency/breadth of neutralization and relationship to CD4+ T cell count.
A total of 18% (32/177) of the sera neutralized at least three-fourths of the virus panel (Fig. 2A) and were categorized as broad. A total of 16% (29/177) of the sera had geometric mean ID50 titers of >220 (Fig. 2B), our cutoff for highly potent sera. These frequencies appear similar to previously observed frequencies (24, 31, 32, 34, 45–47), although differences in criteria for neutralization breadth/potency, in the panel viruses used, and in cohort characteristics make precise comparisons difficult.
Neutralization breadth and potency correlated well (Spearman's correlation coefficient, ρ = 0.97, P < 0.0001; data not shown), and each was negatively associated with the CD4+ T cell count (Fig. 2C). The CD4+ T cell count dropped by an average of 8.1 (95% CI, 2.1 to 14.0) for each increase of 1 virus neutralized (adjusted R2 = 0.037, P = 0.009) and by an average of 39.6 (95% CI, 12.4 to 66.7) for each 2-fold increase in geometric mean ID50 (adjusted R2 = 0.044, P = 0.005).
Neutralization sensitivity of panel viruses.
We ranked viruses by neutralization sensitivity using the geometric mean of the ID50 values for all 177 serum samples neutralizing each virus (Fig. 2D). Within-subtype neutralization, i.e., better neutralization of viruses matched to the sera by subtype (8, 28, 48–51), was clearly observed: Four subtype C pseudoviruses were in the most sensitive quartile and none were in the least sensitive quartile when neutralized by the South Africa sera (∼98% subtype C [52]) (Fig. 2D). Subtype C panel viruses were 2.51 times (95% CI, 2.23 to 2.82) more sensitive than subtype A panel viruses, 1.94 times (95% CI, 1.74 to 2.16) more sensitive than subtype B panel viruses, and 1.94 times (95% CI, 1.75 to 2.14) more sensitive than CRF02_AG panel viruses (data not shown). This hierarchy (C→B∼AG→A) is substantially different from that of an almost identical virus panel when neutralized by plasma from CRF02_AG-infected donors; in that case, CRF02_AG viruses were the most sensitive subtype of the viruses (30).
Anti-MPER antibodies within the cohort.
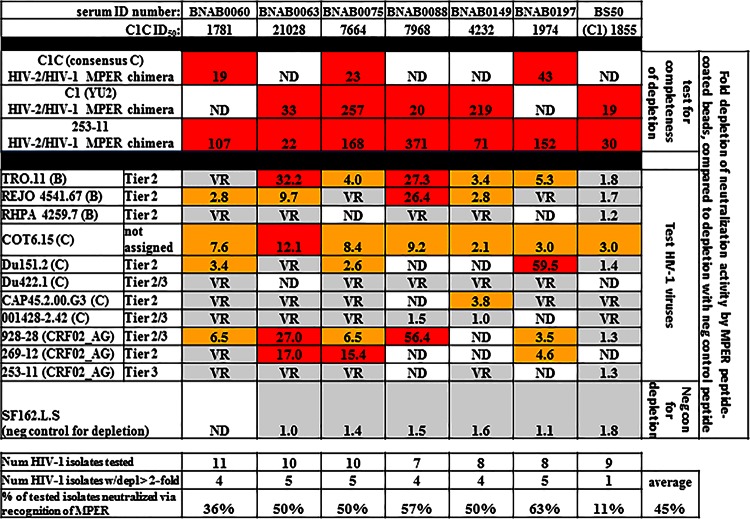
A total of 19% (33/177) of the cohort exhibited significant neutralization activity (ID50 > 1,000) against one or more of the three chimeric construct viruses used to detect anti-MPER neutralizing activity (Fig. 3A and B and Table 1). Previous data demonstrate that high neutralization (ID50 > 1,000) of HIV-2/HIV-1 MPER construct viruses is associated with neutralization of HIV-1 isolates via recognition of MPER (32, 40), while sera with an ID50 of <400 do not neutralize HIV-1 isolates via recognition of MPER (32, 53). In addition, we tested seven samples with neutralization against HIV-2/HIV-1 MPER chimeric viruses ranging from 1,781 to 21,000. Six samples were from among the sera we mapped in this study, and the seventh was a CRF02_AG-infected plasma sample from Cameroon (30, 43). All were tested in MPER peptide depletion assays (34, 40) for dominant (>50%) neutralizing activity in the serum directed against MPER. All seven samples showed dominant neutralizing activity against at least one virus tested. On average, samples neutralized 45% of viruses tested with dominant anti-MPER antibodies, confirming the usefulness of the HIV-2/HIV-1 MPER chimeric viruses for this purpose (Fig. 4).
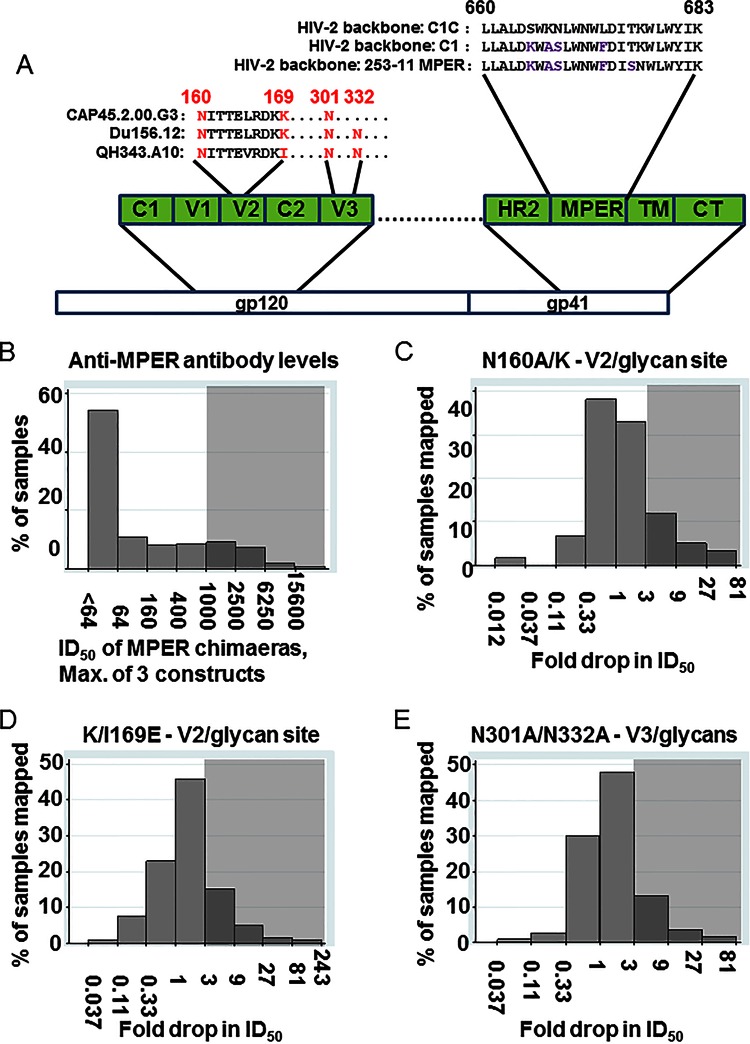
FIG 3.
Mapping of anti-MPER, anti-V2/glycan site, and anti-V3/glycan antibodies in the cohort sera. (A) A depiction of the location of the MPER and the MPER sequences inserted into the HIV-2/HIV-1 MPER chimeric viruses and of the location of the mutations used for mapping the V2/glycan site and V3/glycan epitopes. C, constant region; V, variable loop; HR, heptad repeat; TM, transmembrane domain; CT, cytoplasmic tail. (B) The distribution of anti-MPER ID50 (log scale) is shown, using the highest of the three ID50 values obtained against the three HIV-2/HIV-1 MPER chimeric viruses. Gray shading indicates at what level samples were scored positive for anti-MPER antibodies (ID50 > 1,000). (C, D, and E) The distribution of drops in neutralization due to the introduction of the N160A/K (C), K/I169E (D), or N301A/N332A (E) mutation compared to the unmutated parent virus. Gray shading indicates at what level samples were scored positive for the indicated mapping mutant (≥3-fold drop compared to unmutated parent virus). If mapping of more than one virus was measured, the maximum fold drop is shown, except when the maximum was less than 3 and the minimum was less than 1; minimum fold drop is shown in order to display presumed masking of neutralization epitopes by glycans. Values below 1 indicate an increase in neutralization of the mutant virus compared to the parent.
TABLE 1.
Comparison of the likelihood of an antibody being broadly or potently neutralizing depending upon target recognition of neutralizing antibodies
| Epitope mapping categorya | No. of samples with indicated neutralization potency and mapping category |
Relative risk (95% CI) | P value (χ2)d | No. of samples with indicated neutralization breadth and mapping category |
Relative risk (95% CI) | P value (χ2)d | ||
|---|---|---|---|---|---|---|---|---|
| Less potentb | Potently neutralizingc | Less broade | Broadly neutralizingf | |||||
| Anti-MPER neg | 124 | 20 | 1.00 (reference) | 122 | 22 | 1.00 (reference) | ||
| Anti-MPER pos | 24 | 9 | 1.96 (0.99–3.91) | 0.061 | 23 | 10 | 1.98 (1.04–3.78) | 0.043 |
| Anti-V2/glycan site neg | 63 | 21 | 1.00 (reference) | 62 | 22 | 1.00 (reference) | ||
| Anti-V2/glycan site pos | 29 | 5 | 0.59 (0.24–1.43) | 0.222 | 27 | 7 | 0.79 (0.37–1.67) | 0.522 |
| Anti-V3/glycan neg | 75 | 17 | 1.00 (reference) | 73 | 19 | 1.00 (reference) | ||
| Anti-V3/glycan pos | 12 | 9 | 2.32 (1.21–4.46) | 0.017 | 12 | 9 | 2.08 (1.10–3.92) | 0.033 |
neg, negative; pos, positive.
Geometric mean ID50, <220.
Geometric mean ID50, >220.
Bold values indicate a P value of <0.05.
<18/24 panel viruses neutralized.
≥18/24 panel viruses neutralized.
FIG 4.
Verification of anti-MPER neutralizing antibodies in samples recognizing HIV-2/HIV-1 MPER chimeric viruses. Data represent comparison of ID50-recognizing HIV-2/HIV-1 MPER chimeric target viruses (top; C1C ID50) to tests for dominant anti-MPER neutralizing antibodies measured by bead depletion with anti-MPER coated beads. Fold depletion of neutralizing activity compared to control bead depleted sera is displayed. Depletion of activity against HIV-2/HIV-1 chimeric viruses is displayed to indicate the level of depletion of anti-MPER activity. Tests for depletion of activity against 7 to 11 HIV-1 pseudoviruses are shown, with a >2-fold drop in activity accepted as positive. SF162.L.S was used as a negative control (neg con) for depletion because it is usually recognized by anti-V3 loop neutralizing antibodies (65). The subtype and tier (overall neutralization resistance [28]) of each HIV-1 test virus are indicated. BS50 is a subtype CRF02_AG-infected plasma sample from Cameroon. Neutralization of the Yu2 MPER-swapped chimeric construct (C1) is shown for that plasma sample instead of neutralization of the C1C construct, which contains a consensus C MPER sequence. VR, the virus is resistant to neutralization by the corresponding sample; ND, not determined; num, number; depl, depletion. Color coding: red, >10-fold drop; yellow: 2-to-10-fold drop; gray, <2-fold drop or virus resistant.
Antibodies against the V2/glycan site and V3/glycans within the cohort.
We assessed sera for the presence of dominant neutralizing antibodies recognizing the V2/glycan site or the V3/glycans in any of the pseudoviruses used for mapping, by measuring the drop in neutralization of the mutants with the respective target ablated (mutants depicted in Fig. 3A): N160A/K (Fig. 3C) and K/I169E (Fig. 3D) mutants in V2 and N301A/N332A mutants (Fig. 3E) in V3 (15, 32–36). Because we could assess recognition of V2/glycan and V3/glycan sites only in sera that neutralized Du156.12, CAP45.2, or QH343.A10 parent viruses, we were able to map these epitopes in only a subset of the 177 sera. Mutants were compared to parent viruses CAP45.2.00.G3 (97/177; 95 mapped for V2/glycan site, 90 measured for V3/glycans), Du156.12 (80/171; 80 mapped for V2/glycan site, 77 mapped for V3/glycans), and QH343.21M.ENV.A10 (13/177; 12 measured for V2/glycan site and V3/glycans). In all, 118/177 (66.7%) sera were mapped for the V2/glycan site recognition, and 113/177 (63.8%) were mapped for V3/glycan recognition on ≥1 pseudovirus(es).
Of the tested sera, 29% (34/118) exhibited diminished (≥3-fold drop) neutralization against ≥1 V2 mutant(s) (Table 1). Ten samples (Fig. 3C, values of <0.33) neutralized one N160 mutant substantially better than the corresponding wild type, suggesting that antibody-targeted epitopes shielded by the glycan added at position 160 (54) may be relatively common.
A total of 19% (21/113) of the sera exhibited diminished (≥3-fold drop) neutralization against ≥1 V3 mutant(s) (Table 1). Sera that exhibited increased neutralization for the N301A/N332A mutants appeared less common than those that exhibited increased neutralization for the N160 mutants (Fig. 3D, 2/113 values < 0.33).
Association of anti-MPER antibodies and anti-V3/glycan with neutralization breadth.
We evaluated associations between the presence of neutralizing anti-MPER antibodies and the presence of neutralizing anti-V3/glycan antibodies with neutralization potency and breadth. We used Wilcoxon rank sum analysis to detect differences in distributions among breadths and potencies. In addition, we calculated differences in neutralization breadths and ratios of potencies between groups. We included an estimate of the error arising from our ID50 prediction method that was generated using bootstrapping (95% prediction interval [95%PI]).
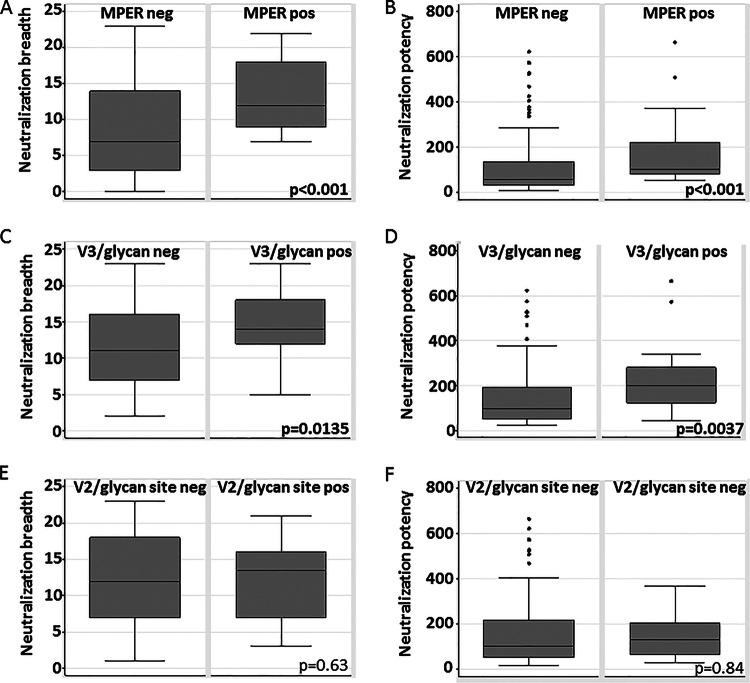
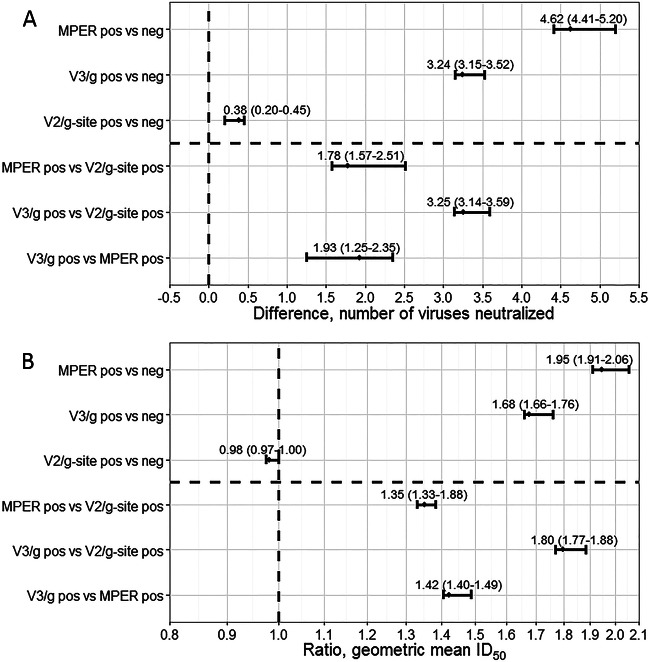
Anti-MPER-positive sera were more broadly (Wilcoxon z = −3.864, P < 0.0001) (Fig. 5A) and potently (z = −3.916, P < 0.001) (Fig. 5B) neutralizing than anti-MPER-negative sera. We also compared the values representing the magnitude of the difference in breadth or fold increase in potency. Anti-MPER-positive sera neutralized 4.62 (95% PI, 4.41 to 5.20) (Fig. 6A) more panel viruses and were 1.95-fold more potent (95% PI, 1.91 to 2.06) (Fig. 6B) than anti-MPER-negative sera. Anti-MPER-positive sera were 1.98 times more likely to be highly broadly neutralizing (Table 1) (95% CI, 1.04 to 3.78, P = 0.043), with a trend toward being more likely to be highly potent (1.96-fold) (95% CI, 0.99 to 3.91, P = 0.061) than anti-MPER-negative sera.
FIG 5.
Differences in neutralization breadth and potency between groups of sera recognizing particular targets. Data represent the results of comparison of the distributions of neutralization breadth scores (A, C, and E) and neutralization potency scores (B, D, and F) based upon detection of functional anti-MPER antibodies (A and B), dominant anti-V3/glycan antibodies (C and D), or dominant anti-V2/glycan site antibodies (E and F). P values were calculated from Wilcoxon rank sum tests. neg, negative; pos, positive.
FIG 6.
Changes in neutralization breadth and potency based upon recognition of MPER, the V2/glycan site, or the V3/glycans. We explored changes in neutralization breadth (A) and potency (B) based upon target mapping of the neutralizing antibodies in each serum. The groups that were compared are indicated on the y axis. The differences in neutralization breadth between the indicated groups are indicated by the differences in the number of viruses neutralized (no difference = 0), and the difference in neutralization potency is indicated by the ratio of aggregate geometric mean ID50 values to those seen with the panel viruses (no difference = 1). Values of 95% prediction intervals (PI) are shown and indicate the bootstrap-based estimate of the error associated with the ID50 prediction algorithm in this data set. g, glycan; g-site, glycan site.
Anti-V3/glycan-positive sera were more broadly (z = −2.470, P = 0.0135) (Fig. 5C) and potently (z = −2.901, P = 0.037) (Fig. 5D) neutralizing than anti-V3/glycan-negative sera, neutralizing 3.24 (95% PI, 3.15 to 3.52) (Fig. 6A) more panel viruses. They were also moderately more potent (1.68-fold; 95% PI, 1.66 to 1.76) (Fig. 6B) than anti-V3/glycan-negative sera. Anti-V3/glycan-positive sera were 2.08 times more likely to be highly broadly neutralizing (Table 1) (95% CI, 1.10 to 3.92, P = 0.033) and 2.32 times more likely to be highly potent (95% CI, 1.21 to 4.46, P = 0.017) than anti-V3/glycan-negative sera.
No association observed between the presence of anti-V2/glycan site antibodies and neutralization breadth or potency.
Anti-V2/glycan site-positive sera were not more broadly (Wilcoxon z = −0.476, P = 0.64) (Fig. 5E) and not more potently (z = −0.208, P = 0.84) (Fig. 5F) neutralizing than anti-V2/glycan-negative sera. They neutralized only 0.38 (95% prediction interval [95% PI], 0.20 to 0.45) (Fig. 6A) more panel viruses and were not more potent (0.98-fold; 95% PI, 0.97 to 1.00) (Fig. 6B) than anti-V2/glycan-negative sera.
The trend for potency and breadth of neutralization was anti-V3/glycan-positive sera > anti-MPER-positive sera > anti-V2/glycan site-positive sera (Fig. 6). Of these comparisons, only the distributions of neutralization breadth for anti-V3/glycans versus anti-V2/glycan site were significantly different (Wilcoxon z = −2.362, P = 0.0182; data not shown), with a strong trend in the same direction for potency (z = −1.941, P = 0.0522; data not shown). We looked for associations between positivity for antibodies to one site and positivity for antibodies to each other site. We were unable to detect any associations (data not shown; all P values > 0.05).
DISCUSSION
In this study, we found neutralization breadth to be strongly positively associated with the presence of neutralizing anti-MPER antibodies and neutralizing anti-V3/glycan antibodies but not with the presence of anti-V2/glycan site-directed antibodies. This is a surprising finding because anti-V2/glycan site antibodies have been observed frequently in broadly neutralizing sera (32, 34, 53) and because anti-MPER and anti-V3/glycan antibodies are often autoreactive (18–20). Self-reactivity has long been proposed to limit responsiveness to targets of broadly neutralizing anti-HIV antibodies (2, 19, 20). It has long been known that many anti-MPER antibodies are self-reactive (20). Recently, of 3 tested anti-V3/glycan MAbs, PGT125 and PGT128 were shown to be polyreactive (18). In contrast, only one of five tested anti-V2/glycan site MAbs was polyreactive (18) and a recent review indicated that, among all broadly neutralizing antibodies responding to this site, only CH103 is polyreactive (2). Our findings suggest that autoreactivity may not be as large a barrier to generating moderately broadly neutralizing and potent anti-HIV-1 neutralizing antibody responses as commonly thought.
Despite these apparent self-reactivity barriers, we show that a substantial proportion of chronically HIV-infected individuals are able to produce at least moderately broadly neutralizing antibody responses to both MPER and the V3/glycans, while such moderately to highly neutralizing antibodies are not enriched among the V2/glycan site-recognizing sera. It has been suggested that moderately broadly neutralizing responses occur frequently and may therefore be more attainable from a vaccine than highly broadly neutralizing responses (23). For example, clusters of related, moderately broadly neutralizing antibodies occur in natural infection and can give high neutralization breadth and potency in aggregate (55). The (presumably more attainable) precursors of broadly neutralizing antibody PGT121 are also, at least sometimes, moderately neutralizing (56).
In this study, 19% (33/177) of the cohort had high titers (ID50 > 1,000) of anti-MPER antibodies (Fig. 3B), which is similar to the proportions of previously studied North American HIV-1 subtype B cohorts (with an ID50 cutoff value of >1,000), 12% (42) and 19% (7), but lower than the prevalence in a sample from one European cohort (57). MPER antibody prevalence in this cohort was higher than that in a blood bank cohort from South Africa (4%) (58), presumably because the blood bank cohort included individuals infected for shorter time periods than our study participants, thus containing lower levels of neutralizing antibodies (32, 35, 47). Our results indicate that prevalence of anti-MPER does not differ substantially by subtype and is often reasonably high.
We found a negative association between neutralization breadth and contemporaneous CD4+ T cell count (Fig. 2C), similarly to a previous study (47). Others found no association with contemporaneous CD4+ T cell count but did find higher neutralization with a greater CD4+ T cell decline (32) and/or with a lower CD4+ T cell count earlier in infection (32, 46). The relationship between CD4+ T cell count and neutralization potency and breadth that we observed in adults appears different from that in children, in whom neutralization potency was lower with depletion of CD4+ T cells (59).
A limitation of our study is that we did not evaluate other targets of broadly neutralizing monoclonal antibodies: the CD4 binding site (CD4bs [60]), the site recognized by MAbs 3BC176 and 3BC315 (61), and the newly identified hinge site at the interface between gp120 and gp41 (5). The hinge region is currently too poorly characterized to allow mutational mapping, and mapping approaches for the 3BC176/315 site are not established. The CD4bs site is not amenable to mapping on the scale necessary for this analysis because the mapping process (62) requires large amounts of serum and recombinant proteins.
Sera with anti-V2/glycan site antibodies (i) are neither more potently neutralizing nor substantially more broadly neutralizing than sera without detectable anti-V2/glycan antibodies and (ii) are less broadly neutralizing than sera with anti-V3/glycan neutralizing sera. There are a series of examples of highly broadly and potently neutralizing V2/glycan site-recognizing MAbs such as PG9 and PG16 (16) and V2/glycan site-recognizing sera (32–34, 53), demonstrating that broadly neutralizing antibodies targeting the V2/glycan site are possible. Higher variability of the V2/glycan site (33) did not prevent the production of very broadly neutralizing and potent antibodies such as PG9 and PG16. It is clear that broadly neutralizing and potent antibodies generally arise after a complex process of somatic hypermutation-mediated evolution of the antibodies (1–3, 63). Reasons for the failure of most serum anti-V2/glycan site antibodies to be broadly neutralizing and potent may include complexity in this evolution process as well as technical difficulties affecting the ability of an immune response to produce a highly broadly neutralizing and potent anti-V2/glycan site antibody response. For example, some part of the structure of these antibodies, perhaps the extended anionic loops (64), may be difficult to fashion effectively by somatic hypermutation of germ line antibodies. If so, our data suggest that a fast progression to high neutralization breadth is not frequently found among the anti-V2/glycan antibodies such as those produced by donor CAP256 (∼5 months [22]). Importantly, such difficulties may also extend to immune responses to an HIV vaccine. Thus, antibodies of moderate neutralization breadth against the MPER or V3/glycans may be easier to induce with a vaccine than those against the V2/glycan site, and these sites might be more amenable models than the V2/glycan site for vaccine design.
ACKNOWLEDGMENTS
We thank the study participants and Dorothy Magwaxaza and Sandra Tshisa for participant recruitment, Marcel Tongo and Eitel Mpoudi Ngole for use of data pertaining to a sample they provided to us, George Shaw for the C1 and C1C HIV-2/HIV-1 chimeras, and Lynn Morris and Penny Moore and colleagues for the env plasmid constructs MLV, COT6.15, Du151.2, and Du156.12 clones, the CAP45 N160A, K169E, and N332A mutants, and the Du156.12 N160K, K169E, and N332A mutants. We also thank F. Gao, M. Li, D. Montefiori, J. Overbaugh, C. Blish, E. M. Long, D. Ellenberger, B. Li, M. Callahan, S. Butera, R. Paranjape, S. Kulkarni, H. Tang, B. H. Hahn, Y. Li, J. F. Salazar-Gonzalez, A. Rice, J. Kappes, and X. Wu for reagents and plasmids we received via the AIDS Research Reference Program.
This study was supported by ICGEB funds. T.M. was supported by studentships from the University of Cape Town and the Poliomyelitis Research Foundation. R.A.J. was supported by studentships from the ICGEB and the Poliomyelitis Research Foundation.
REFERENCES
- 1.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascola JR, Haynes BF. 2013. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev 254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, Moody MA, Alam M, Bonsignori M, Verkoczy L, Ferrari G, Gao F, Tomaras GD, Liao HX, Kelsoe G. 2014. Progress in HIV-1 vaccine development. J Allergy Clin Immunol 134:3–10. doi: 10.1016/j.jaci.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouquet H. 2014. Antibody B cell responses in HIV-1 infection. Trends Immunol 35:549–561. doi: 10.1016/j.it.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou T, Zhu J, Yang Y, Gorman J, Ofek G, Srivatsan S, Druz A, Lees CR, Lu G, Soto C, Stuckey J, Burton DR, Koff WC, Connors M, Kwon PD. 2014. Transplanting supersites of HIV-1 vulnerability. PLoS One 9:e99881. doi: 10.1371/journal.pone.0099881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. 2005. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montero M, Gulzar N, Klaric KA, Donald JE, Lepik C, Wu S, Tsai S, Julien JP, Hessell AJ, Wang S, Lu S, Burton DR, Pai EF, Degrado WF, Scott JK. 2012. Neutralizing epitopes in the membrane-proximal external region of HIV-1 gp41 are influenced by the transmembrane domain and the plasma membrane. J Virol 86:2930–2941. doi: 10.1128/JVI.06349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montero M, van Houten NE, Wang X, Scott JK. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 72:54–84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen X, Dennison SM, Liu P, Gao F, Jaeger F, Montefiori DC, Verkoczy L, Haynes BF, Alam SM, Tomaras GD. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci U S A 107:5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Deng Y, Dey AK, Moore JP, Lu M. 2009. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry 48:2915–2923. doi: 10.1021/bi802303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol 84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Munir Alam S, Gao J, Haynes BF, Kelsoe G. 2015. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol 89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verkoczy L, Diaz M. 2014. Autoreactivity in HIV-1 broadly neutralizing antibodies: implications for their function and induction by vaccination. Curr Opin HIV AIDS 9:224–234. doi: 10.1097/COH.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 21.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, Dekosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, NISC Comparative Sequencing Program, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, Lee FH, Richman DD, Doms RW, Vanham G, Burton DR. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol 81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, Karim SS, Williamson C, Morris L. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol 81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 28.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for neutralizing antibody assessment. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blish CA, Jalalian-Lechak Z, Rainwater S, Nguyen MA, Dogan OC, Overbaugh J. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J Virol 83:7783–7788. doi: 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob RA, Abrahams F, Tongo M, Schomaker M, Roux P, Mpoudi Ngole E, Burgers WA, Dorfman JR. 2012. Refined identification of neutralization-resistant HIV-1 CRF02_AG viruses. J Virol 86:7699–7703. doi: 10.1128/JVI.00804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol 82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, Sibeko S, Mlisana K, Abdool Karim SS, Williamson C, Pinter A, Morris L. 2011. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J Virol 85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. 2011. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol 85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog 7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson C, Morris L, Maughan MF, Ping LH, Dryga SA, Thomas R, Reap EA, Cilliers T, van Harmelen J, Pascual A, Ramjee G, Gray G, Johnston R, Karim SA, Swanstrom R. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res Hum Retroviruses 19:133–144. doi: 10.1089/088922203762688649. [DOI] [PubMed] [Google Scholar]
- 39.Hastie T, Tibshirani R, Friedman J. 2001. The elements of statistical learning: data mining, inference, and prediction. Springer-Verlag, New York, NY. [Google Scholar]
- 40.Gray ES, Madiga MC, Moore PL, Mlisana K, Abdool Karim SS, Binley JM, Shaw GM, Mascola JR, Morris L. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J Virol 83:11265–11274. doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, Donathan M, Bilska M, Gray ES, Abdool Karim SS, Kepler TB, Whitesides J, Montefiori D, Moody MA, Liao HX, Haynes BF. 2011. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One 6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, Migueles SA, Wu X, Phogat A, Shaw GM, Connors M, Hoxie J, Mascola JR, Wyatt R. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol 83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tongo M, Martin DP, Zembe L, Mpoudi-Ngole E, Williamson C, Burgers WA. 2013. Characterization of HIV-1 gag and nef in Cameroon: further evidence of extreme diversity at the origin of the HIV-1 group M epidemic. Virol J 10:29. doi: 10.1186/1743-422X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eilers PH, Marx BD. 1996. Flexible smoothing with B-splines and penalties. Stat Sci 11:89–102. doi: 10.1214/ss/1038425655. [DOI] [Google Scholar]
- 45.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, Schuitemaker H. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J Infect Dis 201:1045–1053. doi: 10.1086/651144. [DOI] [PubMed] [Google Scholar]
- 47.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown BK, Wieczorek L, Sanders-Buell E, Rosa Borges A, Robb ML, Birx DL, Michael NL, McCutchan FE, Polonis VR. 2008. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375:529–538. doi: 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Bures R, Morris L, Williamson C, Ramjee G, Deers M, Fiscus SA, Abdool-Karim S, Montefiori DC. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol 76:2233–2244. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Gils MJ, Edo-Matas D, Schweighardt B, Wrin T, Schuitemaker H. 2010. High prevalence of neutralizing activity against multiple unrelated human immunodeficiency virus type 1 (HIV-1) subtype B variants in sera from HIV-1 subtype B-infected individuals: evidence for subtype-specific rather than strain-specific neutralizing activity. J Gen Virol 91:250–258. doi: 10.1099/vir.0.015693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rademeyer C, Moore PL, Taylor N, Martin DP, Choge IA, Gray ES, Sheppard HW, Gray C, Morris L, Williamson C. 2007. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368:172–181. doi: 10.1016/j.virol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson E, Engelbrecht S. 2009. Molecular characterization of non-subtype C and recombinant HIV-1 viruses from Cape Town, South Africa. Infect Genet Evol 9:840–846. doi: 10.1016/j.meegid.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Karim QA, Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 56.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, Mahan A, Wang S, Kardar M, Yaari G, Walker LM, Simen BB, St John EP, Chan-Hui PY, Swiderek K, Kleinstein SH, Alter G, Seaman MS, Chakraborty AK, Koller D, Wilson IA, Church GM, Burton DR, Poignard P. 2013. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molinos-Albert LM, Carrillo J, Curriu M, Rodriguez de la Concepcion ML, Marfil S, Garcia E, Clotet B, Blanco J. 2014. Anti-MPER antibodies with heterogeneous neutralization capacity are detectable in most untreated HIV-1 infected individuals. Retrovirology 11:44. doi: 10.1186/1742-4690-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83:8925–8937. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agthe M, Nemes E, Jacob RA, Abrahams F, Fainguem N, Ndiang Tetang SM, Cappelli G, Colizzi V, Dorfman JR. 2014. Lower anti-HIV-1 neutralization in HIV-infected children with CD4+ T cell depletion: opposite correlation to that in adults. AIDS 28:1694–1696. doi: 10.1097/QAD.0000000000000281. [DOI] [PubMed] [Google Scholar]
- 60.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. . 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 61.Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, Poignard P, Feizi T, Morris L, Walker BD, Fatkenheuer G, Seaman MS, Stamatatos L, Nussenzweig MC. 2012. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med 209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med 13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol 79:780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]