ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) evades host defenses through tight suppression of autophagy by targeting each step of its signal transduction: by viral Bcl-2 (vBcl-2) in vesicle nucleation, by viral FLIP (vFLIP) in vesicle elongation, and by K7 in vesicle maturation. By exploring the roles of KSHV autophagy-modulating genes, we found, surprisingly, that vBcl-2 is essential for KSHV lytic replication, whereas vFLIP and K7 are dispensable. Knocking out vBcl-2 from the KSHV genome resulted in decreased lytic gene expression at the mRNA and protein levels, a lower viral DNA copy number, and, consequently, a dramatic reduction in the amount of progeny infectious viruses, as also described in the accompanying article (A. Gelgor, I. Kalt, S. Bergson, K. F. Brulois, J. U. Jung, and R. Sarid, J Virol 89:5298–5307, 2015). More importantly, the antiapoptotic and antiautophagic functions of vBcl-2 were not required for KSHV lytic replication. Using a comprehensive mutagenesis analysis, we identified that glutamic acid 14 (E14) of vBcl-2 is critical for KSHV lytic replication. Mutating E14 to alanine totally blocked KSHV lytic replication but showed little or no effect on the antiapoptotic and antiautophagic functions of vBcl-2. Our study indicates that vBcl-2 harbors at least three important and genetically separable functions to modulate both cellular signaling and the virus life cycle.
IMPORTANCE The present study shows for the first time that vBcl-2 is essential for KSHV lytic replication. Removal of the vBcl-2 gene results in a lower level of KSHV lytic gene expression, impaired viral DNA replication, and consequently, a dramatic reduction in the level of progeny production. More importantly, the role of vBcl-2 in KSHV lytic replication is genetically separated from its antiapoptotic and antiautophagic functions, suggesting that the KSHV Bcl-2 carries a novel function in viral lytic replication.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV; also referred to as human herpesvirus 8 [HHV-8]) belongs to the gammaherpesvirus family, which includes Epstein-Barr virus (EBV), herpesvirus saimiri (HSV), and murine gammaherpesvirus 68 (MHV-68) (1). KSHV infection is associated with Kaposi's sarcoma (KS), the most common cancer in HIV-infected patients (1). KSHV is also linked to the development of several other lymphoproliferative malignancies, including primary effusion lymphoma (PEL) and a subset of multicentric Castleman's disease (2). Similar to other herpesviruses, the life cycle of KSHV consists of latent and lytic replication phases (3). Following acute infection, KSHV establishes latency in the immunocompetent hosts, where KSHV maintains its genome as an episome and expresses only a limited number of viral proteins or viral mRNAs. Thus, KSHV latency is an effective strategy for evading host immune detection (3). In KS lesions, most of the tumor cells are latently infected by KSHV, indicating that viral latency and latent products are likely essential for the development of KS tumors (3, 4). In contrast, latent KSHV can be reactivated into lytic replication by specific stimulations. During lytic replication, KSHV expresses a full panel of viral genes in a cascade fashion, beginning with immediate early genes, followed by early genes and then late genes (5). Successful completion of this lytic replication leads to the release of progeny viruses and, ultimately, cell death. Despite the destruction of cells, lytic replication is also believed to play a critical role in KSHV tumorigenesis (3, 5).
Programmed cell death (PCD) is a major component of host innate immunity against pathogen infection. Besides the well-characterized apoptosis (in which the cell kills itself), autophagy (in which the cell eats itself) is an emerging PCD pathway that is a highly regulated homeostatic process where worn-out proteins, malfunctioning organelles, and invading pathogens are swept up and degraded by tiny vacuum cleaners, called autophagosomes. Thus, autophagy is an important innate safeguard mechanism for protecting the organism against unwanted guests like pathogens to keep it healthy. Specifically, autophagy combats and defends infected cells by enhancing the degradation of intracellular pathogens (6, 7). On the other hand, to avoid the host's autophagy-mediated immune responses, herpesviruses have evolved elaborate mechanisms to block different aspects of the autophagy pathway for their persistent infection (8–10). To overcome cellular autophagy, KSHV has evolved several viral gene products to modulate different steps of autophagy signaling (8, 9, 11, 12). For instance, viral Bcl-2 (vBcl-2) interacts with the Beclin-1 complex to downregulate autophagy at the vesicle nucleation step (11), viral FLIP (vFLIP) suppresses autophagy at the vesicle elongation step by preventing the Atg3 E2 enzyme from binding and processing light chain 3 (LC3) (12), and K7 interacts with autophagy inhibitor Rubicon to impair the autophagosome maturation step (9). These pieces of evidence further underscore the importance of autophagy as the host's critical immune control.
The KSHV genome encodes the vBcl-2 (open reading frame 16 [ORF16]) protein, which shares sequence, structural, and functional homology with cellular Bcl-2 family members (13). The Bcl-2 family proteins, which are widely known to be positive or negative regulators of apoptosis, are characterized as containing up to four conserved stretches of amino acids, known as Bcl-2 homology (BH) domains (2, 14, 15). For example, members of the proapoptotic Bcl-2 family, including Bim, Bad, PUMA, and Noxa, sense prodeath signals to activate the downstream proapoptotic members Bax and Bak (16, 17), consequently causing mitochondrial dysfunction and the release of cytochrome c (18, 19). The antiapoptotic members, such as Bcl-2 and Bcl-XL, prevent apoptosis by interacting with the BH3 domain of the proapoptotic members (20, 21). In addition to being regulators of apoptosis, the Bcl-2 family proteins have emerged as critical regulators of autophagy. Cellular Bcl-2 interacts with and negatively regulates the autophagy-promoting activity of Beclin-1 (22, 23), while BH3-only protein Bad plays an autophagy-inducing function by disrupting the interaction of Bcl-2 with Beclin-1 (24). Compared to cellular Bcl-2, the herpesvirus-encoded Bcl-2 homologs Bcl-2 of KSHV and M11 of MHV-68 show a much tighter affinity of binding to Bak and Beclin-1, resulting in much stronger inhibitory activity on apoptosis and autophagy, respectively (11). Mutations on the central hydrophobic cleft of herpesvirus genome-encoded Bcl-2 (84WGR86 of KSHV Bcl-2 or 85SGR87 of MHV-68 M11) disrupt the structure of the BH3-peptide binding groove and block the interaction with Bak or Beclin-1, resulting in the release of the inhibition of apoptosis or autophagy (11). Furthermore, a mutant MHV-68 strain lacking the antiautophagic activity of vBcl-2 demonstrates an impaired ability to maintain chronic infections in mice, whereas a mutant virus lacking the antiapoptotic activity of vBcl-2 establishes chronic infections as efficiently as the wild-type virus but displays a compromised ability for ex vivo reactivation (11). Thus, while the MHV-68 M11 protein is not required for lytic replication in vitro, its antagonism of autophagy confers persistent infections (11, 25–28).
For successful infection and propagation, viruses use and modulate cellular signaling machineries, including the autophagy pathway (8–10). While KSHV is well equipped to overcome cellular autophagy, all of these conclusions were obtained from individual gene studies with overexpression cell lines in the absence of true KSHV infection. In this study, we utilized a clone of the new bacterial artificial chromosome (BAC) 16 (BAC16) KSHV genetic modification system to knock out vBcl-2, vFLIP, and K7 individually to study their roles in KSHV lytic replication (29). To our surprise, unlike vFLIP and K7, vBcl-2 is essential for KSHV lytic replication. We show that knockout of vBcl-2 dramatically reduces KSHV lytic gene expression, viral DNA replication, and progeny virus production (see also the accompanying article [30]). Most importantly, we found that this novel function of vBcl-2 for lytic replication does not depend on its antiapoptotic and antiautophagic functions. These results further suggest that KSHV Bcl-2 carries a novel function in viral lytic replication that is not shared with the MHV-68 M11 protein.
MATERIALS AND METHODS
Cell culture and viruses.
HEK-293T and SLK cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin (Gibco-BRL). iSLK cells were cultured in the presence of 1 μg/ml puromycin and 250 μg/ml G418. BAC16 and its derivatives were introduced into iSLK cells via transfection with the Fugene HD reagent (Roche), and transfected iSLK cells were selected with 200 μg/ml hygromycin (Invitrogen). Wild-type (WT) KSHV and its mutant derivatives were all produced from iSLK-BAC16 or iSLK-BAC16 mutant cell lines upon doxycycline (1 μg/ml) induction for 3 to 4 days. SLK stable cell lines were established using lentivirus vector infection with selection with 200 μg/ml hygromycin.
Plasmid construction.
The vBcl-2-coding sequence was amplified from KSHV BAC16 genomic DNA and cloned into the pCDH-CMV-MSC-ef1-hygromycin vector (System Biosciences) carrying an N-terminal hemagglutinin (HA) tag (pCDH-HA-vBcl-2). Deletions and mutations in the vBcl-2 gene were generated using a QuikChange site-directed mutagenesis kit (Stratagene). MHV-68 M11 was subcloned into the pCDH-CMV-MSC-ef1-hygromycin vector carrying an N-terminal V5 tag.
Immunoblotting.
Cell lysates were collected in 1% NP-40 buffer and quantified by the Bradford protein assay (Thermo Scientific). Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Bio-Rad) by semidry transfer at 25 V for 30 min. All membranes were blocked in 5% milk in phosphate-buffered saline–Tween 20 and probed overnight with the antibodies indicated below in 5% bovine serum albumin at 4°C. Primary antibodies included mouse Flag (Sigma), mouse V5 (Invitrogen), mouse HA (Covance), rat KSHV latency-associated nuclear antigen (LANA; Advanced Biotechnologies), rabbit KSHV ORF8/gB (GeneTex), rabbit KSHV interferon regulatory factor 3 (IRF3; Novus Biologicals), mouse LC3 (Cosmo Bio), and actin (Santa Cruz). Antibodies for mouse KSHV ORF65, rabbit KSHV K8.1, rabbit KSHV K3, rabbit KSHV K8, and rabbit KSHV ORF50/RTA were described previously (9, 31, 32). Appropriate horseradish peroxidase-conjugated secondary antibodies were incubated on membranes in 5% milk, and bands were developed with the enhanced chemiluminescence reagent (Thermo Scientific) and imaged on a Fuji LAS-4000 imager.
TEM.
Cells were fixed at 48 h postinduction (by doxycycline and butyrate treatment), and the samples were prepared as previously described (33). The prepared samples were subjected to transmission electron microscopy (TEM) at the USC/Norris Cell & Tissue Imaging Core within the Vision Research Center.
RNA extraction and qRT-PCR.
Total RNA was isolated from cells with an RNeasy minikit (Qiagen) and treated with RNase-free DNase per the manufacturer's protocol. cDNA was reverse transcribed from 2 μg of total RNA using an iScript cDNA synthesis kit (Bio-Rad), and quantitative reverse transcription-PCR (qRT-PCR) was performed with the iQ SYBR green Supermix (Bio-Rad). The primer sequences used for qRT-PCR are listed in Table 1.
TABLE 1.
Primer sequences for qRT-PCR
| Primer name | Sequence (5′–3′) |
|---|---|
| ORF11-Fwd | GGCACCCATACAGCTTCTACGA |
| ORF11-Rv | CGTTTACTACTGCACACTGCA |
| LANA-Fwd | GAGTCTGGTGACGACTTGGAG |
| LANA-Rv | AGGAAGGCCAGACTCTTCAAC |
| ORF36-Fwd | ATTGCCAACGACCTGATGCA |
| ORF36-Rv | ACTCCAGTCCAGCTGCAGCA |
| ORF25-Fwd | ACAGTTTATGGCACGCATAGTG |
| ORF25-Rv | GGTTCTCTGAATCTCGTCGTGT |
| ORF64-Fwd | CTTCCTCGAGGGCATCATATAC |
| ORF64-Rv | TATACGGTGATGGACTTGATGG |
| RTA-Fwd | TTGCCAAGTTTGTACAACTGCT |
| RTA-Rv | ACCTTGCAAAGACCATTCAGAT |
| ORF48-Fwd | CCACATCTTCATAGAGCACAT |
| ORF48-Rv | ATTGCATCACCAGGGTATCCA |
| ORF57-Fwd | AGGGATATCACCGCTCTCATAAGA |
| ORF57-Rv | CTGCGGTTTCTCGACGGCAACTCA |
| 18S-Fwd | TTCGAACGTCTGCCCTATCAA |
| 18S-Rv | GATGTGGTAGCCGTTTCTCAGG |
| HS1-Fwd | TTCCTATTTGCCAAGGCAGT |
| HS1-Rv | CTCTTCAGCCATCCCAAGAC |
Viral mutagenesis.
Mutagenesis of the KSHV genome in BAC16 was performed as described previously (9, 29). Briefly, PCR amplification was used to generate a linear DNA fragment containing a kanamycin resistance expression cassette, an I-SceI restriction enzyme site, and flanking sequences derived from KSHV genomic DNA, each of which includes a roughly 40-bp copy of a duplication. The fragment was then electroporated into GS1783 cells harboring BAC16 and transiently expressing gam, bet, and exo. The proteins encoded by these three genes are required for homologous recombination of a linear DNA fragment with a target sequence and can be expressed in a temperature-inducible manner from the bacteriophage lambda Red operon engineered within the endogenous GS1783 chromosome. Integration of the Kanr/I-SceI cassette was verified by PCR and restriction enzyme digestion of the purified BAC16 DNA. The GS1783 strain is also equipped with an arabinose-inducible gene encoding the I-SceI enzyme. Upon treatment with 1% l-arabinose, the integrated Kanr/I-SceI cassette is cleaved, resulting in a transiently linearized BAC16. A second bacteriophage lambda Red-mediated recombination between the duplicated sequences results in recircularization of the BAC DNA and the scarless loss of the Kanr/I-SceI cassette. Kanamycin-sensitive clones were screened via replica plating. The genomic sequences of target regions were amplified by PCR and subsequently sequenced. The primers used to generate BAC16-vBcl-2-HA, the BAC16-vBcl-2 knockout (KO), BAC16-vFLIP-Flag, and the BAC16-vFLIP KO are listed in Table 2.
TABLE 2.
Primer sequences for KSHV BAC16 mutagenesis
| Primer name | Sequence (5′–3′) |
|---|---|
| vBcl-2 KO-Fwd | GCCCTGGTGACCGTCCACAATGGACGAGGACGTTTTGCCTTAAGGGAGAGGTGTTGGCCAAGGATGACGACGATAAGTAGGG |
| vBcl-2 KO-Rv | GGCCATGAATATCCCTTCAATGGCCAACACCTCTCCCTTAAGGCAAAACGTCCTCGTCCAAACCAATTAACCAATTCTGATTAG |
| vBcl-2-HA-Fwd | ATTGGCCACTATATTGGCAGCGGTCGCGATGAGCAGGAGATACCCTTACGATGTCCCTGATTACGCAGGGAGCTACCCATTATGCCAAGTACGCCCCCTATTGAC |
| vBcl-2-HA-Rv | GCAACCCTCTACTCTTCCGGGGACCTCGAATTACGCGTTAGGAGCCGGCGTAGTCGGGCACGTC |
| vFLIP KO-Fwd | CATCCGTGCCCAGTTTCCGCGCCACCTCACAGAGAACCTCACTAGTAAGTGGCCATGGTGCCGGAGGATGACGACGATAAGTAGGG |
| vFLIP KO-Rv | GGTGCCTTCACATATACAAGCCGGCACCATGGCCACTTACTAGTGAGGTTCTCTGTGAGGTGGCAACCAATTAACCAATTCTGATTAG |
| vFLIP-Flag-Fwd | ACATTCTACGGACCAAAAATTAGCAACAGCTTGTTATCTACTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAGGGATAACAGGGTAATCGATTTATTC |
| vFLIP-Flag-Rv | TTCCGGCCCATCACACTCCCAACACTATCGCCATACACCAGACTACAAAGACCATGACGG |
Autophagy analyses.
SLK stable cells expressing the WT or mutant forms of vBcl-2 were treated with 2 μM rapamycin (Sigma) in DMEM containing 1% fetal bovine serum for 2 to 4 h. LC3 mobility shift was detected by immunoblotting. Quantitation of punctate green fluorescent protein (GFP)-LC3 was performed by confocal microscopy as previously described (34, 35).
Apoptosis analyses.
SLK cells stably expressing the WT or mutant forms of vBcl-2 were seeded at 106 cells per well of 6-well plates and grown for 24 h. The cells were then treated with fresh medium containing 5 ng/ml tumor necrosis factor alpha (TNF-α) plus 5 μg/ml cycloheximide (CHX) for 12 h. For the analysis of apoptotic cells, the samples were prepared using a Deadend fluorometric terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) system kit (Promega) per the manufacturer's protocol. For the caspase-3 activation assay, cleaved caspase-3 was measured from cell lysates using a cleaved caspase-3 (Asp175) sandwich enzyme-linked immunosorbent assay (ELISA) kit (Cell Signaling).
Statistical analysis.
All data were analyzed using a 2-tailed Student's t test with data from a minimum of 3 experiments. P values of less than 0.05 were considered significant.
RESULTS
Generation of knockout or tagged mutations of autophagy-modulating genes in the KSHV genome.
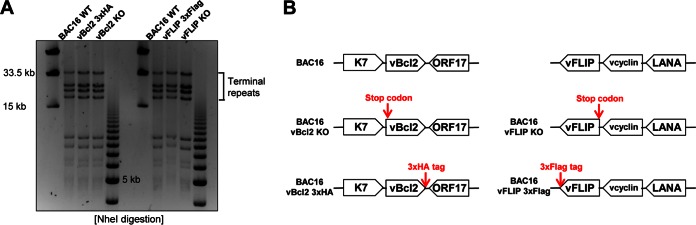
To study the roles of the antiautophagy-related genes of KSHV in the context of virus, we introduced the stop codon or C-terminal tagging for vBcl-2 or vFLIP into BAC16, a highly infectious BAC clone derived from KSHV.219 (29, 36), using two-step bacteriophage lambda Red-mediated seamless recombination (37, 38). Putative mutant BAC clones were analyzed for the integrity of terminal repeats by digestion with the restriction enzyme NheI (Fig. 1A). Sequencing of the genomic region further confirmed that all mutants had the desired sequences (Fig. 1B). The BAC16-K7-KO and BAC16-K7-Flag constructs were described previously (9).
FIG 1.
Generation of individual knockout or tagged mutants of vFLIP or vBcl-2 in the BAC16 genetic system. BAC16-vFLIP-Flag, BAC16-vFLIP-KO, BAC16-vBcl-2-HA, and BAC16-vBcl-2-KO constructs were generated by two-step bacteriophage lambda Red-mediated recombination. (A) BAC16 DNAs isolated from overnight cultures were digested by NheI and subjected to gel electrophoresis. (B) The sequence of each BAC16 mutant clone was confirmed by DNA sequencing. vcyclin, viral cyclin.
vBcl-2 is essential for KSHV production.
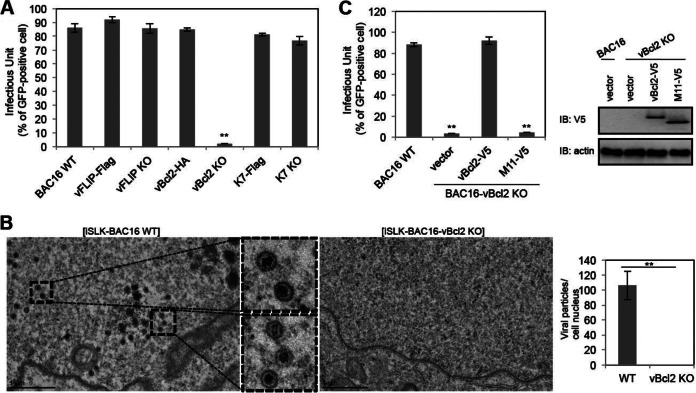
In an effort to investigate the role of autophagy-modulating genes in KSHV lytic replication, vBcl-2-KO, vBcl-2-HA, vFLIP-KO, vFLIP-Flag, K7-KO, K7-Flag, and wild-type BAC16 DNAs were transfected into iSLK cells, which express KSHV RTA under the control of a doxycycline-inducible promoter (39). After hygromycin selection of BAC16 DNA-transfected cells, comparable levels of GFP expression and viral DNA were found in iSLK cells harboring the different KSHV BACs (data not shown). Upon doxycycline and butyrate treatment, RTA is expressed to initiate the lytic replication program, which results in the expression of lytic genes and, ultimately, the release of infectious viral particles (39). The culture supernatants containing progeny viruses were collected from each individual iSLK-BAC16 cell line at 3 days postinduction, and these supernatants were directly used to infect SLK cells. GFP expression was examined at 1 day postinfection, and the results indicated that knocking out vFLIP or K7, as well as tagging vBcl-2, vFLIP, or K7 in the C terminus, did not affect the production of KSHV progeny upon induction of RTA expression; however, knocking out vBcl-2 blocked the production of KSHV progeny (Fig. 2A). On the basis of transmission electron microscopy (TEM), numerous premature viral particles accumulated in wild-type iSLK-BAC16 cells at 2 days postinduction, but almost no premature viral particles existed in vBcl-2-knockout cells (Fig. 2B). To further confirm the role of vBcl-2 in KSHV lytic replication, we utilized a lentiviral vector to express vBcl-2 and its MHV-68 homolog, M11, in iSLK-BAC16-vBcl-2-KO cells and induced lytic replication. Expression of vBcl-2, but not that of MHV-68 M11, in iSLK-BAC16-vBcl-2-KO cells restored virus production upon induction of RTA expression (Fig. 2C). Collectively, these results showed that vBcl-2 plays a critical role in KSHV progeny production.
FIG 2.
vBcl-2 is essential for KSHV production in iSLK cells. (A) iSLK-BAC16 (wild type), iSLK-BAC16-vFLIP-Flag, iSLK-BAC16-vFLIP-KO, iSLK-BAC16-vBcl-2-HA, iSLK-BAC16-vBcl-2-KO, iSLK-BAC16-K7-Flag, and iSLK-BAC16-K7-KO cells were induced by doxycycline (1 μg/ml) and sodium butyrate (0.3 mM) for 3 days, and the supernatants were harvested and used for infection of SLK cells. At 24 h postinfection, cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. (B) iSLK-BAC16 (wild type) and iSLK-BAC16-vBcl-2-KO cells were induced by doxycycline and sodium butyrate for 2 days, and cells were harvested and processed for transmission electron microscopy analysis. The number of viral particles in each cell was quantified. (C) iSLK-BAC16-vBcl-2-KO cells were complemented with empty vector, V5-tagged Bcl-2 of KSHV, or V5-tagged M11 of MHV-68. At 36 h postcomplementation, these cells were induced by doxycycline and sodium butyrate for 3 days, and the supernatants were harvested and used for infection of SLK cells. At 24 h postinfection, cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. The expression of complemented vBcl-2 or M11 was analyzed by immunoblotting (IB) with anti-V5 antibody. **, P < 0.002.
Effect of vBcl-2 knockout on viral gene expression.
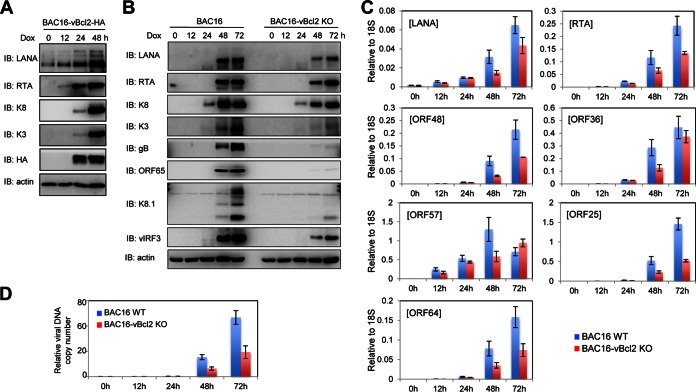
vBcl-2 is a lytic gene, and its expression starts 24 h after stimulation of lytic replication (Fig. 3A). To pinpoint exactly how vBcl-2 affects KSHV production, we examined the lytic cycle in the iSLK-BAC16 and iSLK-BAC16-vBcl-2-KO cell lines. We analyzed by Western blotting the expression levels of proteins encoded by some viral lytic genes at 0, 12, 24, 48, and 72 h postinduction and found that the immediate early gene RTA, early genes K3, K8, and vIRF3, and late genes ORF65, gB, and K8.1 accumulated in cells during the lytic cycle in wild-type iSLK-BAC16 cells; however, vBcl-2 knockout weakened the expression of RTA, K3, K8, and vIRF3 and almost eliminated the expression of the late genes ORF65, gB, and K8.1 (Fig. 3B). To assess the impact of vBcl-2 knockout on KSHV gene expression at the mRNA level, we analyzed the transcription of several KSHV ORFs at 0, 12, 24, 48, and 72 h postinduction by quantitative reverse transcription-PCR (qRT-PCR). Consistent with the levels of viral proteins, the results of qRT-PCR indicated that the mRNA levels of latent genes, immediate early genes, early genes, and late genes were reduced in iSLK-BAC16-vBcl-2-KO cells (Fig. 3C). These data demonstrate that knockout of vBcl-2 has a broad impact on viral gene expression upon lytic reactivation.
FIG 3.
vBcl-2 is essential for KSHV lytic gene expression in iSLK cells. (A) iSLK-BAC16-vBcl-2-HA cells were induced by doxycycline (Dox) and sodium butyrate, and whole-cell lysates (WCLs) were collected at 0, 12, 24, and 48 h postinduction and subjected to immunoblotting with the indicated antibodies. (B to D) iSLK-BAC16 (wild type) and iSLK-BAC16-vBcl-2-KO cells were induced by doxycycline and sodium butyrate, and cells were collected at 0, 12, 24, 48, and 72 h postinduction. The whole-cell lysates were subjected to immunoblotting with the indicated antibodies (B); total RNA or DNA was extracted and subjected to qRT-PCR with the indicated primers to examine viral gene expression (C) or viral genome copy number (D), respectively.
The intracellular viral genome copy number was also examined by qRT-PCR at 0, 12, 24, 48, and 72 h postinduction. KSHV DNA started to replicate at about 48 h postinduction, and its copy number dramatically increased at 72 h postinduction. However, the loss of vBcl-2 resulted in a 2- to 3-fold decrease in the viral DNA copy number at 48 and 72 h postinduction (Fig. 3D). Since viral DNA replication is required for the effective transcription and translation of KSHV late genes, the defect in late gene expression was much more significant than the defect in immediate early and early gene expression in vBcl-2-knockout cell lines during the lytic cycle (Fig. 3B). These results demonstrate that the loss of vBcl-2 dramatically impairs lytic gene expression and viral DNA replication, which resulted in reduced progeny virus production during the lytic cycle.
The essential role of vBcl-2 for lytic replication does not depend on its antiapoptotic or antiautophagic activities.
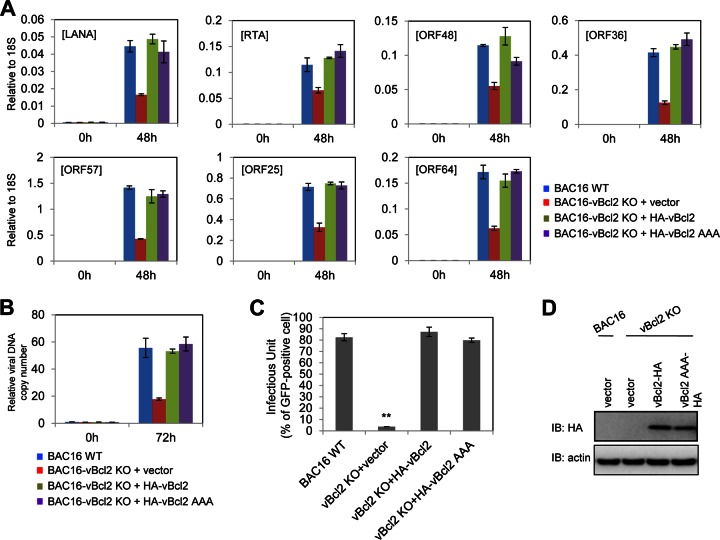
Since Bcl-2 of KSHV and its homologs in other herpesviruses demonstrated antiapoptotic and antiautophagic activities by interaction with Bak and Beclin-1, respectively (11), we investigated whether these two critical functions of vBcl-2 are involved in KSHV lytic replication by using the antiapoptosis- and antiautophagy-defective vBcl-2 AAA mutant (in which the amino acids in the 84WGR86 sequence were changed to alanine). We introduced the vBcl-2 AAA mutant, wild-type vBcl-2, and the vector control into iSLK-BAC16-vBcl-2-KO cells by use of a lentivirus vector, induced reactivation, and monitored lytic gene expression, viral DNA copy number, as well as progeny virus production. As shown in Fig. 4, both the vBcl-2 AAA mutant and wild-type vBcl-2 restored the induction of lytic genes (Fig. 4A), viral DNA copy number (Fig. 4B), and the production of progeny infectious viruses (Fig. 4C) to similar levels. These results indicate that vBcl-2 plays an important role in KSHV lytic replication independently of its antiapoptotic and antiautophagic activities and further explain why its close homolog, MHV-68 M11, cannot restore lytic replication in iSLK-BAC16-vBcl-2-KO cells, suggesting a novel additional function of vBcl-2 in the KSHV lytic life cycle.
FIG 4.
The essential role of vBcl-2 for KSHV lytic replication does not depend on its antiapoptotic and antiautophagic functions. iSLK-BAC16-vBcl-2-KO cells were complemented with empty vector, V5-tagged vBcl-2 (wild type), or V5-tagged vBcl-2 AAA. At 36 h postcomplementation, these cells were induced by doxycycline and sodium butyrate, and cells were collected at 0, 48, and 72 h postinduction. Total RNA or DNA was extracted and subjected to qRT-PCR with the indicated primers to examine viral gene expression (A) or viral genome copy number (B), respectively. The supernatant collected from complemented iSLK cells at 3 days postinduction was used for SLK cell infection. (C) At 24 h postinfection, SLK cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. (D) The expression of complemented vBcl-2 or its mutant was analyzed by immunoblotting with anti-V5 antibody.
Identification of the functional regions of vBcl-2 for KSHV lytic replication.
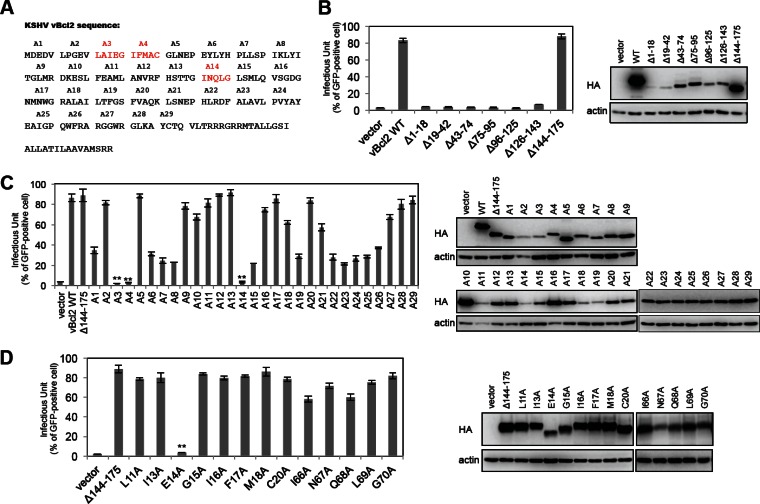
To further investigate how vBcl-2 regulates KSHV lytic replication, we decided to map the functional region of vBcl-2 necessary for lytic replication. KSHV Bcl-2 is a small protein with 175 amino acids (aa) and shares a similar structure with cellular Bcl-2 and MHV-68 M11 (Fig. 5A) (11, 40). Based on the structure, we constructed a series of internal deletion mutants on the pCDH-HA-vBcl-2 lentiviral vector and individually reconstituted these mutants, along with wild-type vBcl-2 and the empty vector, in iSLK-BAC16-vBcl-2-KO cells. Following RTA induction, expression of only the vBcl-2 mutant lacking aa 144 to 175 (the vBcl-2 Δ144-175 mutant) restored KSHV production as well as wild-type vBcl-2 did (Fig. 5B), indicating that the C-terminal region consisting of aa 144 to 175 is not required for KSHV lytic replication. Since KSHV Bcl-2 is relatively small and therefore mutations consisting of 20- to 30-aa internal deletions may dramatically change the structure and disrupt the function of vBcl-2, we decided to perform alanine scanning through the region from aa 1 to 143. We generated 29 mutants (named A1 to A29) of pCDH-HA-vBcl-2 with mutations from aa 1 to 143, and each mutant contained 4 to 5 amino acids mutated to alanines (Fig. 5A). Following the reconstitution of each mutant and lytic cycle induction in iSLK-BAC16-vBcl-2-KO cells, the production of progeny infectious viruses was measured. As shown in Fig. 5C, mutations in A3, A4, and A14 almost eliminated KSHV production, mutations in A1, A6 to A8, A15, A19, and A22 to A26 impaired KSHV production, and the rest of the mutations showed no effect on KSHV production (Fig. 5C). We focused on the mutated regions in the A3, A4, and A14 mutants and further generated a series of mutants with a single alanine mutation. As shown in Fig. 5D, a change of glutamic acid 14 (E14) to alanine in vBcl-2 aa 1 to 143 completely abolished KSHV production, and other single mutations in mutants A3, A4, and A14 showed little or no effect (Fig. 5D). These results suggest that the E14 residue of KSHV Bcl-2 is critical for KSHV lytic replication and progeny infectious virus production.
FIG 5.
Identification of the functional regions of vBcl-2 for KSHV lytic replication. (A) The 175-amino-acid sequence of KSHV Bcl-2. (B) iSLK-BAC16-vBcl-2-KO cells were complemented with empty vector, HA-tagged vBcl-2 (wild type), or its internal deletion mutants (the Δ1-18, Δ19-42, Δ43-74, Δ75-95, Δ96-125, Δ126-143, and Δ144-175 mutants). At 36 h postcomplementation, these cells were induced by doxycycline and sodium butyrate for 3 days, and the supernatants were harvested and used for infection of SLK cells. At 24 h postinfection, cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. The expression of complemented vBcl-2 or its mutants was analyzed by immunoblotting with anti-HA antibody. (C) iSLK-BAC16-vBcl-2-KO cells were complemented with empty vector, HA-tagged vBcl-2 (wild type), or its alanine scanning mutants (mutants A1 to A29). At 36 h postcomplementation, these cells were induced by doxycycline and sodium butyrate for 3 days, and the supernatants were harvested and used for infection of SLK cells. At 24 h postinfection, cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. The expression of complemented vBcl-2 or its mutants was analyzed by immunoblotting with anti-HA antibody. (D) iSLK-BAC16-vBcl-2-KO cells were complemented with empty vector, HA-tagged vBcl-2 Δ143-175 (wild type), or its mutants with a single mutation to alanine (the L11A, I13A, E14A, G15A, I16A, F17A, M18A, C20A, I66A, N67A, Q68A, L69A, and G70A mutants). At 36 h postcomplementation, these cells were induced by doxycycline and sodium butyrate for 3 days, and the supernatants were harvested and used for infection of SLK cells. At 24 h postinfection, cells were collected and the infectious units were quantified by fluorescence-activated cell sorter analysis. The expression of complemented vBcl-2 or its mutants was analyzed by immunoblotting with anti-HA antibody. The size of vBcl-2 Δ143-175 E14A is about 16 kDa.
The essential role of vBcl-2 in lytic replication is genetically separated from its antiapoptotic or antiautophagic activity.
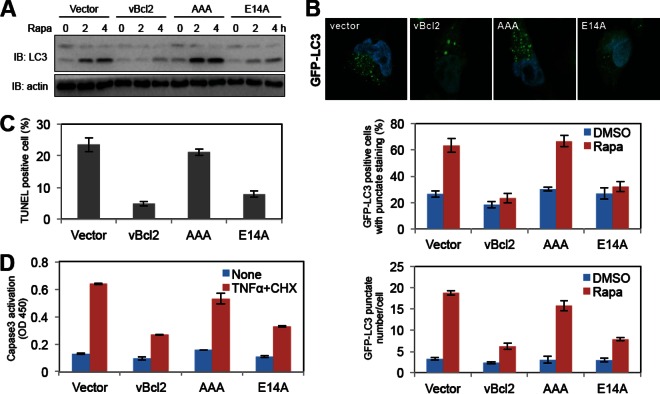
Since KSHV Bcl-2 contains at least three functions, including antiapoptosis, antiautophagy, and lytic replication functions, and these functions rely on different regions of vBcl-2 (84WGR86 for antiapoptosis and antiautophagy and E14 for lytic replication) (11), we determined whether these functions were genetically separable. We generated SLK stable cell lines individually expressing wild-type vBcl-2, vBcl-2 AAA, and vBcl-2 E14A and treated these cells to induce autophagy (by rapamycin treatment) or apoptosis (by TNF-α and cycloheximide treatment). Upon autophagy induction, soluble microtubule-associated protein light chain 3 (LC3-I) is converted to a lapidated form (LC3-II), forming punctate cytoplasmic structures (6). As shown in Fig. 6A and B, rapamycin treatment increased LC3-II levels, the percentage of LC3 punctate-positive cells, and the numbers of LC3 punctated regions per cell in vector-containing or vBcl-2 AAA SLK cells. However, expression of wild-type vBcl-2 as well as the vBcl-2 E14A mutant blocked these autophagy processes upon stimulation, suggesting that E14 is not required for vBcl-2 antiautophagic activity (Fig. 6A and B).
FIG 6.
The essential function of vBcl-2 for KSHV lytic replication is genetically separable from its antiapoptotic and antiautophagic functions. (A) SLK vector, SLK-vBcl-2 (wild type), SLK-vBcl-2 E14A, and SLK-vBcl-2 AAA stable cells were treated with rapamycin (Rapa; 2 μM). At 4 h posttreatment, cells were harvested and whole-cell lysates were subjected to immunoblotting with LC3 and actin antibodies. (B) SLK-GFP-LC3 vector, SLK-GFP-LC3-vBcl-2 (wild type), SLK-GFP-LC3-vBcl-2 E14A, and SLK-GFP-LC3-vBcl-2 AAA stable cells were treated with rapamycin (2 μM). At 4 h posttreatment, cells were fixed and stained with DAPI (4′,6-diamidino-2-phenylindole) and subjected to confocal microscopy. The number of GFP-LC3 punctate regions per cell and the percentage of GFP-LC3-punctate-positive cells were quantified. (C and D) SLK vector, SLK-vBcl-2 (wild type), SLK-vBcl-2 E14A, and SLK-vBcl-2 AAA stable cells were treated with TNF-α (5 ng/ml) and CHX (5 μg/ml) for 12 h. (C) After treatment, the cells were collected and subjected to TUNEL staining, and the apoptotic cells were quantified. (D) Whole-cell lysates of the treated cells were subjected to ELISA analysis for caspase-3 cleavage. DMSO, dimethyl sulfoxide; OD 450, optical density at 450 nm.
To compare the abilities of wild-type vBcl-2 and its mutants to confer apoptosis resistance, SLK stable cells were treated with TNF-α and cycloheximide for 12 h and subjected to apoptotic assays. Quantification of the apoptotic cells via TUNEL staining revealed that wild-type vBcl-2 and E14A vBcl-2 have similar antiapoptotic activities. In contrast, mutations to 84WGR86 abolished this inhibitory activity (Fig. 6C). Unlike the vBcl-2 AAA mutant, the vBcl-2 E14A mutant had activity inhibiting caspase-3 activation similar to that of wild-type vBcl-2 upon apoptosis stimulation, as shown by the detection of cleaved caspase-3 with a commercial ELISA kit (Fig. 6D). These data show that vBcl-2 controls cellular apoptosis and autophagy through the 84WGR86 region, consistent with the findings of previous studies (11), but controls KSHV lytic replication through the E14 residue, and these functions are genetically separable. Overall, our results identify the novel essential function of vBcl-2 for KSHV lytic replication, which relies on glutamic acid 14 but not the antiapoptotic or antiautophagic activity of vBcl-2.
DISCUSSION
Here we utilized the new KSHV BAC16 genome modification system and the iSLK virus production cell line to study the role of viral autophagy-modulating genes on KSHV lytic replication, and we provide evidence that KSHV Bcl-2 is essential for KSHV lytic replication. By inserting an HA tag in the C terminus of vBcl-2, we detected the endogenous expression of the vBcl-2 protein during lytic replication for the first time. We also found that mutation of glutamic acid 14 of vBcl-2 abolished KSHV lytic replication, whereas mutations of the 84WGR86 region did not. Our study indicates for the first time that vBcl-2 contains an additional function besides antiapoptotic and antiautophagic functions which is directly essential for the KSHV lytic cycle.
Herpesviruses have evolved multiple strategies to overcome or modulate cellular signaling pathways, ultimately leading to the establishment of persistent infection and tumorigenesis (8). KSHV dedicates a large portion of its genome to functions that sabotage almost every aspect of host immunity, including autophagy (8, 9, 11, 12). Bcl-2 of KSHV targets Beclin-1 to downregulate the nucleation step of autophagy (11), FLIP of KSHV blocks the elongation step of autophagy by preventing the interaction between Atg3 and LC3 (12), and K7 of KSHV is associated with Rubicon to impair the autophagosome maturation step (9). Bcl-2 homologs in other herpesviruses exhibit similar antiautophagic activity. For instance, Epstein-Barr virus (EBV) encodes two Bcl-2 homologs, BHRF1 and BALF1, which demonstrate antiapoptotic activity and are required for transforming primary resting B lymphocytes (41). MHV-68 encodes one Bcl-2 homolog, M11, which demonstrates antiapoptotic as well as antiautophagic activity (11, 25–28). In vivo studies suggested that MHV-68 lacking M11 exhibits an impaired ability to establish latency and compromised reactivation from latency; however, M11 is not required for MHV-68 lytic replication in vitro (11, 25–28). Compared to these virus genome-encoded homologs, vBcl-2 is critical for KSHV lytic replication, besides its antiapoptotic and antiautophagic activities. Although Bcl-2 of KSHV and M11 of MHV-68 have similar crystal structures, they have a low degree of sequence identity (11, 40). KSHV Bcl-2 and MHV-68 M11 bind to Bak and Beclin-1 with different binding affinities (11, 40), suggesting that KSHV Bcl-2 may have additional cellular or viral interaction partners compared to those of MHV-68 M11. The different positions with respect to the viral genome may cause the differences in function between KSHV Bcl-2 and MHV-68 M11. The gene for KSHV Bcl-2 is a lytic gene and is located right after the origin of lytic replication (29); however, the gene for MHV-68 M11 is located within the latency locus (42). The positional difference between KSHV Bcl-2 and MHV-68 M11 suggests that during evolution KSHV Bcl-2 may have tried to gain some additional functions for effective lytic replication.
Since viral genomes are relative small, viruses always try to compact more genetic information into their limited genome. Many viral proteins, such as NS1 of influenza virus (43), VP35 of Ebola Zaire virus (44), and ICP0 of herpes simplex virus 1 (45), perform multiple functions. Therefore, it is not surprising that KSHV Bcl-2 harbors several functions which are not only required for modulating cellular cell death signaling but also essential for viral lytic replication. By a series of mutagenesis analyses, we identified the crucial amino acid of vBcl-2 (E14) for KSHV lytic replication and found that it is genetically separable from its antiapoptotic and antiautophagic functions. The E14 residue of KSHV Bcl-2 is located in the first α helix (the α1 helix) (40), which has a low degree of sequence conservation with the α1 helix sequence of MHV-68 M11 (11). The α1 helix forms a BH4 domain but is not involved in the formation of the central hydrophobic BH3-peptide binding groove, which structurally explains why the vBcl-2 E14A mutant still showed antiapoptotic and antiautophagic activities similar to those of wild-type vBcl-2. The BH4 domain appears to be responsible for protein-protein interaction and may be sensitive to the change of amino acid polarity. In fact, the E14A mutation changes its side chain charge and polarity, which may affect the migration rate of this mutant in SDS-PAGE compared to that of wild-type vBcl-2. However, the mutation of E14 does not considerably affect the overall structure and function of KSHV Bcl-2, since the vBcl-2 E14A mutant was still capable of blocking apoptosis and autophagy. On the other hand, the vBcl-2 AAA mutant can still support KSHV lytic replication. Although the detailed mechanism by which vBcl-2 supports KSHV lytic replication is still elusive, we propose that vBcl-2 might interact with some novel cellular or viral binding partners through its α1 helix peptide, and this interaction might be essential for the tight regulation of KSHV lytic gene expression, viral DNA replication, or viral particle assembly. In summary, we have identified a critical additional function of vBcl-2 which is required for KSHV lytic replication. The detailed mechanism is under further investigation, and it will not be surprising if further exploration uncovers additional novel functions of this small viral protein.
ACKNOWLEDGMENTS
This work was partly supported by NIH grants CA82057, CA31363, CA115284, CA180779, HL110609, DE023926, AI105909, AI073099, and AI116585; the Hastings Foundation; and the Fletcher Jones Foundation (to J.U.J.); by NIH grants CA140964, CA161436, and ACS RSG-11-121-01-CCG (to C.L.); and by the GRL Program (K20815000001) of the National Research Foundation of Korea (to B.-H.O. and J.U.J.).
We thank Rina Amatya for manuscript preparation.
REFERENCES
- 1.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gross A, McDonnell JM, Korsmeyer SJ. 1999. BCL-2 family members and the mitochondria in apoptosis. Genes Dev 13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 3.Ye F, Lei X, Gao SJ. 2011. Mechanisms of Kaposi's sarcoma-associated herpesvirus latency and reactivation. Adv Virol 2011:193860. doi: 10.1155/2011/193860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest 120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol 73:2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ. 2005. The molecular machinery of autophagy: unanswered questions. J Cell Sci 118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodgers MA, Bowman JW, Liang Q, Jung JU. 2014. Regulation where autophagy intersects the inflammasome. Antioxid Redox Signal 20:495–506. doi: 10.1089/ars.2013.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang C, Lee JS, Jung JU. 2008. Immune evasion in Kaposi's sarcoma-associated herpes virus associated oncogenesis. Semin Cancer Biol 18:423–436. doi: 10.1016/j.semcancer.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Q, Chang B, Brulois KF, Castro K, Min CK, Rodgers MA, Shi M, Ge J, Feng P, Oh BH, Jung JU. 2013. Kaposi's sarcoma-associated herpesvirus K7 modulates Rubicon-mediated inhibition of autophagosome maturation. J Virol 87:12499–12503. doi: 10.1128/JVI.01898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Q, Fu B, Wu F, Li X, Yuan Y, Zhu F. 2012. ORF45 of Kaposi's sarcoma-associated herpesvirus inhibits phosphorylation of interferon regulatory factor 7 by IKKepsilon and TBK1 as an alternative substrate. J Virol 86:10162–10172. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, Kowalik TF, Sun R, Jung JU, Liang C. 2009. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog 5:e1000609. doi: 10.1371/journal.ppat.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, Jung JU. 2009. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med 3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 14.Skommer J, Wlodkowic D, Deptala A. 2007. Larger than life: mitochondria and the Bcl-2 family. Leuk Res 31:277–286. doi: 10.1016/j.leukres.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 15.Walensky LD. 2006. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ 13:1339–1350. doi: 10.1038/sj.cdd.4401992. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. 2006. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 17.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC. 2007. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 18.Antignani A, Youle RJ. 2006. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol 18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Green DR, Kroemer G. 2004. The pathophysiology of mitochondrial cell death. Science 305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 20.Adams JM, Cory S. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci 26:61–66. doi: 10.1016/S0968-0004(00)01740-0. [DOI] [PubMed] [Google Scholar]
- 21.Opferman JT, Korsmeyer SJ. 2003. Apoptosis in the development and maintenance of the immune system. Nat Immunol 4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- 22.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 72:8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. 2007. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lima BD, May JS, Marques S, Simas JP, Stevenson PG. 2005. Murine gammaherpesvirus 68 bcl-2 homologue contributes to latency establishment in vivo. J Gen Virol 86:31–40. doi: 10.1099/vir.0.80480-0. [DOI] [PubMed] [Google Scholar]
- 26.Gangappa S, van Dyk LF, Jewett TJ, Speck SH, Virgin HW IV. 2002. Identification of the in vivo role of a viral bcl-2. J Exp Med 195:931–940. doi: 10.1084/jem.20011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herskowitz JH, Jacoby MA, Speck SH. 2005. The murine gammaherpesvirus 68 M2 gene is required for efficient reactivation from latently infected B cells. J Virol 79:2261–2273. doi: 10.1128/JVI.79.4.2261-2273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loh J, Huang Q, Petros AM, Nettesheim D, van Dyk LF, Labrada L, Speck SH, Levine B, Olejniczak ET, Virgin HW IV. 2005. A surface groove essential for viral Bcl-2 function during chronic infection in vivo. PLoS Pathog 1:e10. doi: 10.1371/journal.ppat.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelgor A, Kalt I, Bergson S, Brulois KF, Jung JU, Sarid R. 2015. Viral Bcl-2 encoded by the Kaposi's sarcoma-associated herpesvirus is vital for virus reactivation. J Virol 89:5298–5307. doi: 10.1128/JVI.00098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciufo DM, Cannon JS, Poole LJ, Wu FY, Murray P, Ambinder RF, Hayward GS. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J Virol 75:5614–5626. doi: 10.1128/JVI.75.12.5614-5626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, MacKey J, Czajak SC, Desrosiers RC, Lackner AA, Jung JU. 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J Virol 73:1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, Jung JU. 2013. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, Jung JU. 2014. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Q, Seo GJ, Choi YJ, Ge J, Rodgers MA, Shi M, Jung JU. 2014. Autophagy side of MB21D1/cGAS DNA sensor. Autophagy 10:1146–1147. doi: 10.4161/auto.28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vieira J, O'Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 37.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 38.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 39.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. 2008. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog 4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu TT, Liao HI, Tong L, Leang RS, Smith G, Sun R. 2011. Construction and characterization of an infectious murine gammaherpesivrus-68 bacterial artificial chromosome. J Biomed Biotechnol 2011:926258. doi: 10.1155/2011/926258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marc D. 2014. Influenza virus non-structural protein NS1: interferon antagonism and beyond. J Gen Virol 95:2594–2611. doi: 10.1099/vir.0.069542-0. [DOI] [PubMed] [Google Scholar]
- 44.Audet J, Kobinger GP. 2015. Immune evasion in ebolavirus infections. Viral Immunol 28:10–18. doi: 10.1089/vim.2014.0066. [DOI] [PubMed] [Google Scholar]
- 45.Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol 78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]