Abstract
Herpesviruses, including human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), and Kaposi’s sarcoma-associated herpesvirus, establish latency by modulating or mimicking antiapoptotic Bcl-2 proteins to promote survival of carrier cells. BH3 profiling, which assesses the contribution of Bcl-2 proteins towards cellular survival, was able to globally determine the level of dependence on individual cellular and viral Bcl-2 proteins within latently infected cells. Moreover, BH3 profiling predicted the sensitivity of infected cells to small-molecule inhibitors of Bcl-2 proteins.
TEXT
Human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (KSHV) belong to the Herpesviridae family of viruses that are able to establish lifelong latent infections. Within infected hosts, HCMV establishes latency within the myeloid compartment (1–4), while EBV and KSHV establish latency within B cells (5–9). Although infections by these viruses are generally asymptomatic in immunocompetent individuals, a multitude of illnesses can arise from the persistent nature of latency. HCMV is a major cause of posttransplantation illness and death in hematopoietic-cell and solid-organ transplant recipients (10–12). Reactivation from latently infected myeloid cells, which are the predominant infiltrating cell type found in the infected organs of these patients (13), can lead to overt inflammation-mediated multiorgan failure (14, 15). EBV is the etiologic agent in the development of various B-cell cancers, such as Hodgkin's lymphoma, non-Hodgkin's lymphoma, and Burkitt's lymphoma (16). KSHV is associated with B-cell lymphoproliferative diseases and cancers, including primary effusion lymphoma, multicentric Castleman's disease, and Kaposi's sarcoma (17). Thus, despite the generally benign nature of herpesvirus infections, the ability of these viruses to establish lifelong infections is not without disease consequence in a significant proportion of infected individuals.
To initiate and maintain latency, herpesviruses must sustain the survival of carrier cells with a minimal complement of viral proteins, which is necessary for immune evasion. One strategy utilized by herpesviruses is to stimulate cell survival via the modulation of cellular apoptotic machinery (18), specifically through the enhanced expression and/or activation of the antiapoptotic B-cell lymphoma 2 (Bcl-2) family of proteins, including Bcl-2, myeloid cell leukemia 1 (Mcl-1), and B-cell lymphoma extra large (Bcl-xL). HCMV is known to upregulate the expression of Mcl-1 and Bcl-2 in monocytes and CD34+ bone marrow myeloid progenitor cells (19–21), as well as Mcl-1 in the THP-1 monocytic cell line (20). The upregulation of Bcl-2 family members in latently infected myeloid cells was shown to be responsible for establishing a prosurvival state in the absence of lytic proteins (19–21). EBV has been reported to induce survival of B cells via increased expression of Mcl-1 (22, 23), Bcl-2 (24), and Bcl-xL (25). KSHV also upregulates Bcl-2 (26) and Mcl-1 (27) to promote survival of infected B cells. Despite studies showing the individual roles that Bcl-2 members play in the survival of cells latently infected with herpesviruses, a global picture of how each antiapoptotic Bcl-2 protein interplays with other Bcl-2 members to maintain survival, i.e., whether one or multiple Bcl-2 proteins play a predominant role over others to maintain the viability of latently infected cells, is still unclear. In addition, both EBV and KSHV encode viral homologs of prosurvival Bcl-2 proteins that also potently inhibit mitochondrion-mediated apoptosis; however, the contribution of these viral Bcl-2 homologs toward cell survival during latency is uncertain, as their expression during latency appears to be dependent on cell type and virus strain (28, 29).
Similar to latently infected cells, cancer cells often express multiple prosurvival Bcl-2 proteins simultaneously, yet display dependence on or “addiction” to only a specific subset of Bcl-2 proteins (30, 31). The Bcl-2 protein(s) that a cancer cell is dependent on can be “diagnosed” using a technique called BH3 profiling (30). BH3 profiling is a functional assay that provides information about cellular dependence on individual antiapoptotic proteins. Consequently, it can be used for personalized medicine, allowing for the design of effective chemotherapy treatment regimens involving small-molecule inhibitors of Bcl-2 proteins (32). Given that both cancer cells and latently infected cells modulate antiapoptotic Bcl-2 proteins for survival, we asked if BH3 profiling can be utilized as a comprehensive approach to functionally identify the subset of Bcl-2 proteins which latently infected cells predominantly rely on for survival.
BH3 profiling reveals distinct patterns of dependence on Bcl-2 proteins in the survival of persistently infected cells.
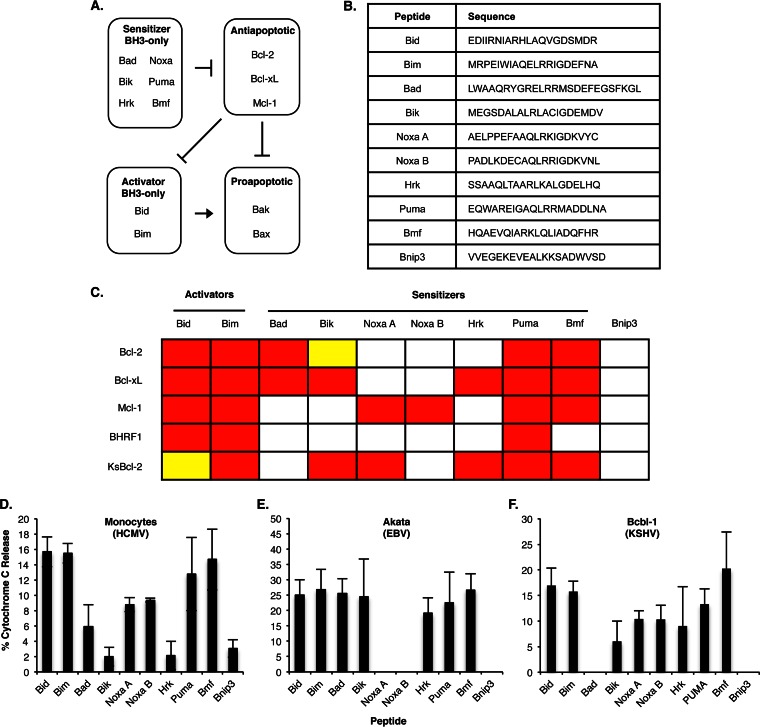
Antiapoptotic Bcl-2 proteins, including Bcl-2, Bcl-xL, and Mcl-1, regulate apoptosis by inhibiting proapoptotic effectors Bax and Bak (30), which, upon activation, undergo allosteric modifications leading to oligomerization within the outer mitochondrial membrane, allowing for the release of cytochrome c and apoptosis (Fig. 1A). Antiapoptotic Bcl-2 proteins bind and sequester activator BH3-only proteins (aBH3) such as Bid and Bim, which activate Bax and Bak (33). Repression of aBH3 proteins by antiapoptotic Bcl-2 proteins can be relieved by competitive inhibition with sensitizer BH3-only proteins (sBH3) such as Bad, Bik, Noxa, Hrk, Puma, and Bmf. Alternatively, antiapoptotic Bcl-2 proteins can directly bind and block oligomerization of Bax or Bak (33). Similar to aBH3 proteins, Bax and Bak can be freed from antiapoptotic Bcl-2 proteins by competitive inhibition with sBH3 proteins. BH3 profiling is based on the principle that sBH3 proteins bind to antiapoptotic Bcl-2 proteins with different selectivities and affinities (Fig. 1C). The strength and selectivity of protein-protein interactions among Bcl-2 proteins directly correlate with the ability of sBH3 proteins to antagonize Bcl-2 proteins from blocking cytochrome c release (30–32, 34, 35). To determine if BH3 profiling can decipher the level of dependence on individual Bcl-2 proteins for the survival of persistently infected cells, we performed BH3 profiling on HCMV-infected monocytes, Akata cells (a B-cell line latently infected with EBV), and body cavity-based lymphoma 1 (Bcbl-1) cells (a B-cell line latently infected with KSHV). Procedural details are similar to those described by Ryan et al. (36). Briefly, infected cells were lysed in mitochondrion isolation buffer (250 mM sucrose, 10 mM Tris-HCl [pH 7.4], 0.1 mM EGTA) and passed once through a 27-gauge needle or a Dounce homogenizer. After samples were centrifuged at 600 × g for 10 min, the resulting supernatant was centrifuged at 10,000 × g for 10 min to obtain mitochondria. Mitochondria were then resuspended in experimental buffer (125 mM KCl, 10 mM Tris-MOPS [morpholinepropanesulfonic acid] [pH 7.4], 5 mM glutamate, 2.5 mM malate, 1 mM KPO4, and 10 μM EGTA-Tris [pH 7.4]) to a concentration of 0.3 to 0.5 mg/ml protein and exposed to BH3 domain peptides (Fig. 1B) at 100 μM for 40 min at room temperature. Following treatment with BH3 peptides, mitochondria were separated and cytochrome c concentrations in the pellet and supernatant fractions were measured by enzyme-linked immunosorbent assay (ELISA).
FIG 1.
BH3 profiling reveals distinct patterns of dependence on Bcl-2 proteins in cells latently infected with herpesviruses. (A) Model depicting the control of mitochondrial depolarization by the Bcl-2 family of proteins. (B) BH3 peptide sequences used for BH3 profiling analysis (30). (C) Interactions between BH3 proteins and antiapoptotic Bcl-2 proteins (30, 34, 35). Red indicates high-affinity binding, yellow indicates intermediate-affinity binding, and white indicates low-affinity binding. (Adapted from reference 63 with permission of the publisher.) (D to F) BH3 profiling results for HCMV-infected monocytes (D), EBV-infected Akata cells (E), and KSHV-infected Bcbl-1 cells (F). The assays were performed with 100 μM BH3-only peptides. Shown are the means of the results of 3 to 6 independent experiments; error bars show standard deviation.
As expected, aBH3 peptides (Bid and Bim) that directly bind and activate proapoptotic proteins Bax and Bak induced cytochrome c release in all three persistently infected cell types (Fig. 1D to F), indicating that survival was not due to a loss of Bax and Bak, but rather to the increased activities of antiapoptotic Bcl-2 proteins. In accordance, treatment with sBH3 peptides Puma and Bmf, which bind indiscriminately to all Bcl-2 proteins, induced cytochrome c release from mitochondria of latently infected cells. Next, we examined the effects of sBH3 peptides that exhibit different binding specificities to individual antiapoptotic Bcl-2 proteins. It has been reported that BH3 peptides from Bad, Noxa, and Hrk can be used as functional probes for Bcl-2, Mcl-1, and Bcl-xL, respectively (30). Mitochondria from HCMV-infected monocytes released cytochrome c upon treatment with Noxa A and Noxa B, as well as with Bad, suggesting that HCMV-infected monocytes are dependent on Mcl-1 and Bcl-2 for survival (Fig. 1D). Bik and Hrk did not induce more cytochrome c release than the negative peptide control Bnip3, a human BH3-only protein that does not bind antiapoptotic Bcl-2 proteins nor activate Bax or Bak, indicating that Bcl-xL is not involved in regulating mitochondrial permeability within HCMV-infected monocytes. Moreover, of these selective sBH3 peptides, Noxa A and Noxa B induced the greatest amount of cytochrome c release, indicating that Mcl-1 may play a more predominant role in blocking the death of HCMV-infected cells, a result consistent with our previous findings that HCMV-induced Mcl-1 inhibited apoptosis of infected monocytes (19). It should also be pointed out that the basal release of cytochrome c from mitochondria isolated from uninfected monocytes was ≥90% (data not shown), which is in accordance with the biological programming of monocytes to rapidly undergo apoptosis upon entry into circulation (37). These data suggest that HCMV specifically induces the upregulation of antiapoptotic Bcl-2 proteins in order to drive the survival of infected monocytes. In EBV-infected (Akata) cells, Bad, Bik, and Hrk peptides induced similar levels of cytochrome c release, which is indicative of Bcl-xL-mediated survival (Fig. 1E). Finally, cells latently infected with KSHV (Bcbl-1) were sensitive to Noxa A and Noxa B peptides, indicating that Mcl-1 mediates cell survival. In addition, Bcbl-1 also showed a distinct BH3 profiling signature of sensitivity to Bik and Hrk, but not to Bad, a pattern not associated with any cellular Bcl-2 proteins (Fig. 1F). Flanagan and Letai previously demonstrated that purified mitochondria treated with recombinant Kaposi’s sarcoma virus Bcl-2 (KsBcl-2) were sensitized to cytochrome c release upon treatment with Bik, Noxa A, Hrk, Puma, and Bmf peptides but not with Bad (34). Indeed, our results from mitochondria isolated from latently infected B cells exhibited a similar BH3 profiling, indicating a role for KsBcl-2, as well as cellular Mcl-1, during latency. Although KsBcl-2 is a lytic gene (38, 39), low-level expression during latency has been observed (18, 40–42), underscoring the necessity of teasing out the specific Bcl-2 proteins responsible for the survival of latently infected cells. Taken together, these data demonstrate potential for the use of BH3 profiling to globally assess the levels to which individual cellular and viral Bcl-2 proteins exert their activities in the presence of other Bcl-2 family members.
Antiapoptotic Bcl-2 protein expression levels weakly correspond to BH3 profiling analysis.
Previous studies demonstrated that herpesviruses upregulate a multitude of prosurvival Bcl-2 proteins in latently infected cells to mediate survival. Since BH3 profiling of latently infected cells identified specific Bcl-2 proteins which played a predominant role in preventing mitochondrion depolarization, we asked if there was a correlation between Bcl-2 protein levels in virally infected cells and BH3 profiling analysis.
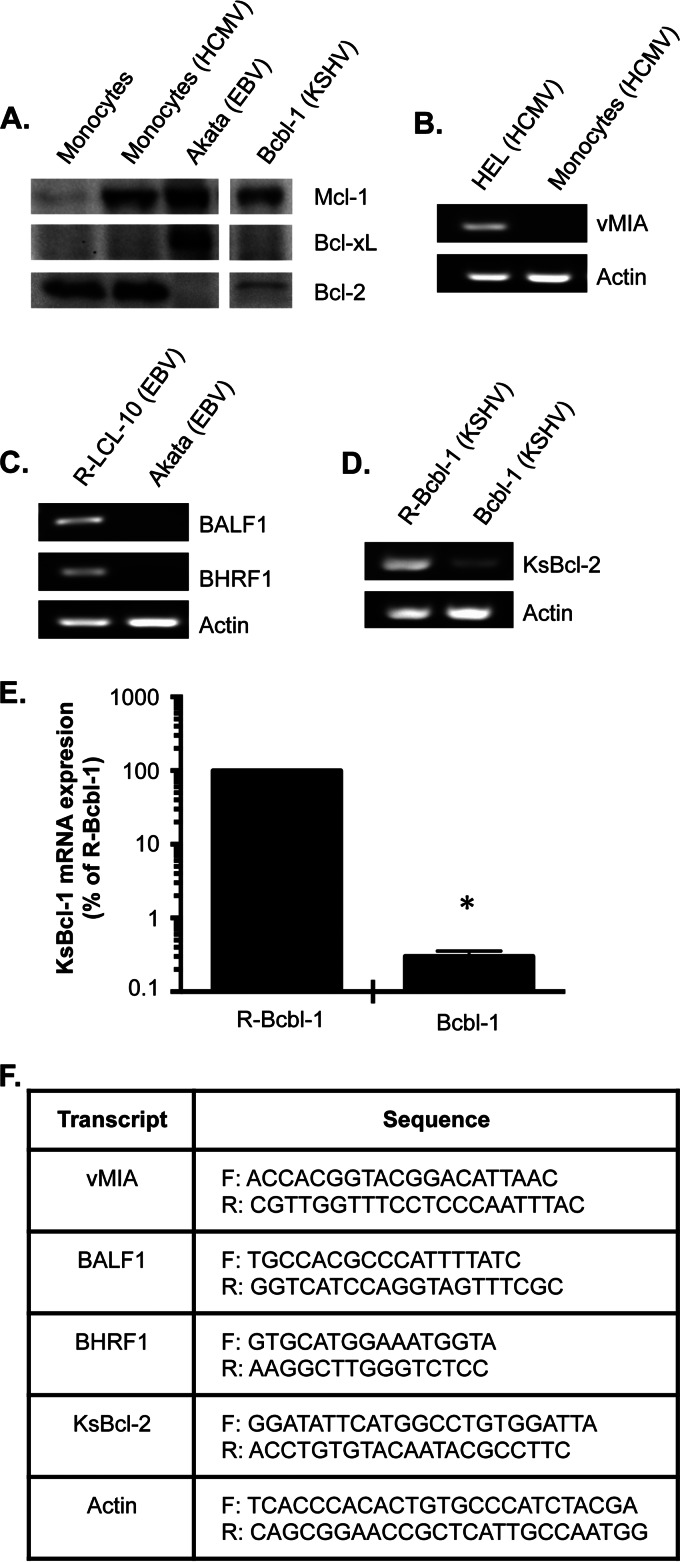
Unlike EBV and KSHV, HCMV does not encode a viral homolog of Bcl-2 (43, 44). Instead, HCMV encodes vMIA, a potent viral mitochondrial inhibitor of apoptosis expressed during lytic infection (45–47) which is not synthesized in persistently infected monocytes (Fig. 2B) (19), indicating that cellular antiapoptotic Bcl-2 proteins play a central role in preventing depolarization of mitochondria in HCMV-infected monocytes. We showed a robust induction of Mcl-1 expression in monocytes following HCMV infection (Fig. 2A), which leads to the establishment of an antipoptotic state (19, 20, 48) and is consistent with our previous findings and BH3 profiling analysis (Fig. 1D). BH3 profiling of HCMV-infected cells also predicted Bcl-2 to be required for survival. Accordingly, Bcl-2 was highly expressed in HCMV-infected cells (Fig. 2A), suggesting a correlation between protein expression and BH3 profiling analysis. However, the relatively higher expression levels of Bcl-2 compared to those of Mcl-1 would seem to hint at Bcl-2 being the principal factor mediating the block in mitochondrion depolarization. To the contrary, BH3 profiling indicated Mcl-1 to be the key determinant of cytochrome c release following infection. In support of this finding, Bcl-2 expression was similar in uninfected and infected monocytes (Fig. 2A), suggesting Bcl-2 to be a general survival factor of monocytes and not specifically for infected cells. These data highlight the ability of BH3 profiling to more stringently decipher subtle differences between the activities of Bcl-2 proteins within a given latently infected cell type.
FIG 2.
Antiapoptotic Bcl-2 protein expression levels do not correspond with BH3 profiling analyses of latently infected cells. (A) Equal amounts of total protein from lysates from HCMV-infected monocytes and EBV- and KSHV-infected B cells were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Blots were probed with an anti-Bcl-2, an anti-Bcl-xL, or an anti-Mcl-1 antibody. (B to E) RNA was isolated from lytic HCMV-infected human embryonic lung fibroblasts (HEL) (B), HCMV-infected peripheral blood monocytes (B), LCL-10 (B cells latently infected with EBV) reactivated with tetradecanoyl phorbol acetate (TPA) and sodium butyrate to induce EBV lytic replication (R-LCL-10) (C), Akata cells (B cells latently infected with EBV) (C), Bcbl-1 cells (B cells latently infected with KSHV) (D and E), or Bcbl-1 cells reactivated with valproic acid (R-Bcbl-1 cells) (D and E). (B to E) Semiquantitative (B to D) or quantitative (E) PCR analyses were performed. Results shown in panels B, C, and D are representative of 3 independent experiments. (E) Shown are means of results of 3 independent experiments; error bars show standard deviation. The asterisk (*) represents a P value of ≤0.001. (F) Primer sets used to perform PCR analysis. F, forward primer; R, reverse primer.
We next examined the expression levels of the EBV Bcl-2 homologs BamHI fragment H rightward open reading frame 1 (BHRF1) and BamHI fragment A leftward reading frame 1 (BALF1), since their expression has been observed during latency despite both proteins being expressed from lytic genes (49–53). We found that neither BHRF1 nor BALF1 are expressed in Akata cells, but are expressed in a reactivated EBV-transformed autologous B lymphoblastoid cell line (LCL), LCL 10 (R-LCL-10) (Fig. 2C), emphasizing a critical role of cellular antiapoptotic Bcl-2 proteins in promoting the survival of cells latently infected with EBV. BH3 profiling predicted a reliance on Bcl-xL for survival of EBV-infected cells, which we found to be highly expressed (Fig. 2A). Surprisingly, Akata cells also expressed high levels of Mcl-1, which was not shown by BH3 profiling analysis to be involved in mediating survival. A possible explanation is that Mcl-1 may be localized to the cytoplasm and thus unable to inhibit mitochondrial depolarization (33). Alternatively, Mcl-1 molecules may be completely saturated with proapoptotic sensitizers, thus preventing Mcl-1 from inhibiting proapoptotic effectors or activator BH3-only proteins. Nonetheless, our data indicate that antiapoptotic protein levels do not necessarily correlate with activity and that a global functional approach such as BH3 profiling is required to elucidate the individual contribution of each Bcl-2 protein in mediating survival of infected cells.
In contrast to BH3 profiling of HCMV- and EBV-infected cells, BH3 profiling of KSHV-infected cells predicted a requirement for the viral Bcl-2 homolog KsBcl-2 in preventing mitochondrial depolarization (Fig. 1F). Despite KsBcl-2 being a lytic protein (29, 38), we found that cells latently infected with KSHV expressed lower levels of KsBcl-2 than Bcbl-1 cells undergoing viral reactivation (Fig. 2D and E), which is consistent with other studies (29). Although other studies have demonstrated that viral Bcl-2 homologs and cellular Bcl-2 proteins have similar binding affinities to BH3-only proteins (34, 35), our data provide the proof-of-principle that BH3 profiling can concurrently reveal dependence on both cellular and viral Bcl-2 proteins from mitochondria directly isolated from latently infected cells. BH3 profiling also showed a survival addiction of KSHV-infected cells to Mcl-1, which we found to be highly expressed (Fig. 2A). However, we also observed low levels of cellular Bcl-2 in KSHV-infected cells, although it was not predicted by BH3 profiling to be involved in maintaining the viability of Bcbl-1 cells. These data again illustrate the weak correlation between protein expression levels and activity.
Overall, we observed that levels of antiapoptotic Bcl-2 proteins loosely correlate with BH3 profiling predictions, thus exemplifying why measuring the levels of antiapoptotic proteins alone is insufficient to determine how latently infected cells utilize Bcl-2 proteins to overcome apoptosis. A functional global approach such as BH3 profiling is necessary to gain a comprehensive overview of the activities of individual Bcl-2 proteins in the context of those of other family members within latently infected cells, together with their expression levels. We also showed for the first time that BH3 profiling is a viable tool to predict addiction to both cellular Bcl-2 proteins and viral Bcl-2 homologs, highlighting the potential use of BH3 profiling to prognosticate sensitivities of latently infected cells to small-molecule Bcl-2 inhibitors. To validate this prediction, we next used small-molecule Bcl-2 inhibitors, which selectively bind and target different Bcl-2 proteins, to confirm the mechanism by which herpesviruses inhibit apoptosis in latently infected cells and to concurrently assess the validity of BH3 profiling technology in predicting sensitivity to potential therapeutics.
BH3 profiling predicts sensitivities of latently infected cells to small-molecule Bcl-2 inhibitors.
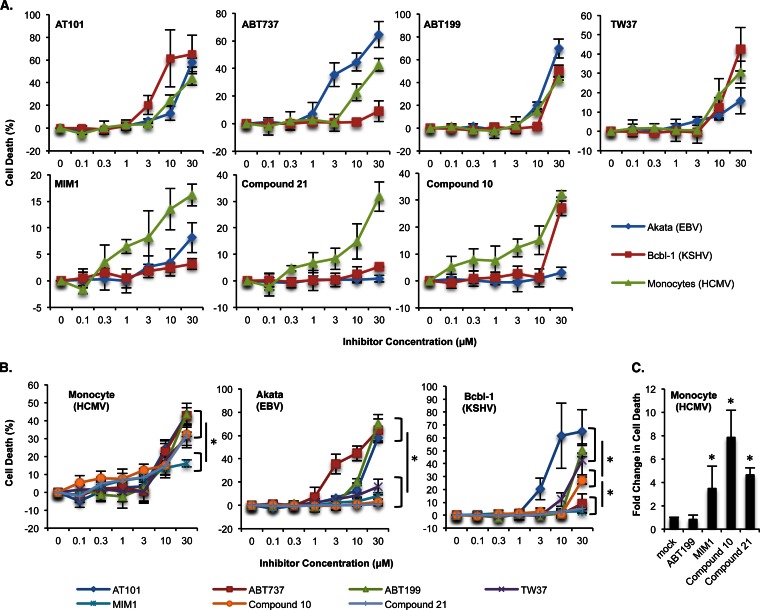
Monocytes are programed to rapidly undergo apoptosis upon entry into the circulatory system from the bone marrow, and we found that, in the absence of HCMV infection, ∼60% to 90% of monocytes are undergoing apoptosis by 48 h postisolation (data not shown). BH3 profiling predicted that HCMV induces Mcl-1 within infected monocytes to mediate survival, which is in accord with our previous studies showing that HCMV infection increases Mcl-1 expression to stimulate an antiapoptotic state (19, 48). In accord, HCMV-infected cells were sensitive to reported Mcl-1-specific inhibitors, including compound 21 and compound 10 (54) and MIM1 (55) (Fig. 3A). Compounds 21 and 10 were 2-fold-more effective at killing HCMV-infected cells than MIM1 (32% versus 16% at 30 μM), consistent with their stronger affinity for binding to Mcl-1 (Fig. 3B; Table 1). Similarly, the ability of pan-Bcl-2 inhibitors to stimulate the death of HCMV-infected monocytes (AT101>TW37) correlated with their dissociation constant (Ki) values for Mcl-1 (0.18 μM [AT101] versus 0.26 μM [TW37]) (56, 57). Further indicating a critical role for Mcl-1 in the block of mitochondrion-mediated apoptosis, the level of death induced by pan-Bcl-2 inhibitors was comparable to that for Mcl-1-specific inhibitors (compounds 10 and 21) with similar Mcl-1 Ki values and higher than that for MIM-1, which has a significantly weaker Mcl-1 binding affinity (Fig. 3B; Table 1). However, BH3 profiling also predicted a dependence on Bcl-2 for survival of HCMV-infected cells, although the comparable Bcl-2 expression levels for uninfected and infected cells suggest Bcl-2 to be a general survival factor for monocytes. In support, these cells underwent death upon treatment with high concentrations (>10 μM) of a Bcl-2/BclxL inhibitor (ABT737) (58) and a Bcl-2-selective inhibitor (ABT199) (59) (Fig. 3A), although the specificity of ABT199 is likely lost at high micromolar concentrations (Table 1). At 0.3 μM, Mcl-1 inhibitors were ∼4- to 8-fold-more effective than ABT199 at inducing death of HCMV-infected monocytes (Fig. 3C). Since Mcl-1, but not Bcl-2, was induced following infection (Fig. 2A), these data indicate that Mcl-1 functions as a viability “switch” responsible for determining the cell fate of HCMV-infected monocytes.
FIG 3.
BH3 profiling predicts sensitivity of latently infected cells to small-molecule inhibitors of antiapoptotic Bcl-2 family members. (A and B) HCMV-infected monocytes and EBV (Akata)- and KSHV (Bcbl-1)-infected B cells were treated with increasing concentrations of ABT199 (Bcl-2 inhibitor), ABT737 (Bcl-2/Bcl-xL inhibitor), AT101 (pan-Bcl-2 inhibitor), TW37 (pan-Bcl-2 inhibitor), MIM1 (Mcl-1 inhibitor), compound 10 (Mcl-1 inhibitor), and compound 21 (Mcl-1 inhibitor). (C) HCMV-infected monocytes were treated with 0.3 μM ABT199, MIM1, compound 10, and compound 21. (A to C) After 24 h of treatment with inhibitor, cell viability was measured by trypan blue exclusion. Shown are the means of the results of 3 to 6 independent experiments; error bars show standard deviation. Asterisks (*) represent P values of ≤0.05.
TABLE 1.
Dissociation constant values for small-molecule inhibitors of antiapoptotic Bcl-2 proteinsa
For EBV-infected cells, BH3 profiling showed a dependence on Bcl-xL to block cytochrome c release (Fig. 1E). Accordingly, the use of inhibitors with increasing binding potency toward Bcl-xL (TW37<AT101<ABT199<ABT737) (Table 1) induced increasing levels of cell death (8%, 13%, 20%, and 45%, respectively, at 10 μM) in Akata cells (Fig. 3A and B). Furthermore, although Mcl-1 was highly expressed, EBV-infected cells were resistant to the Mcl-1 inhibitors MIM1, compound 10, and compound 21, inducing only 8%, 3%, and 0.83% death, respectively, at 30 μM (Fig. 3A and B), which is in agreement with BH3 profiling analysis. Although the Bcl-2-selective inhibitor ABT199 also induced cell death at concentrations of >10 μM (Fig. 3A), its binding affinity to Bcl-xL is in fact greater than that of the paninhibitors AT101 and TW37 (Table 1); thus, ABT199-induced death is likely through Bcl-xL inhibition. Although these data are highly suggestive that EBV-infected cells are dependent on Bcl-xL for the inhibition of cell death, confirmation will require Bcl-xL-specific inhibitors, which are not currently available. Nonetheless, the development of Bcl-xL-selective small-molecule inhibitors may hold promise for the treatment of EBV-associated B-cell lymphomas.
The KSHV-infected cells demonstrate a distinct BH3 profile, with high Noxa and Hrk activities, and low Bad activity consistent with Mcl-1 and KsBcl-2 dependence for survival. Importantly, the three-dimensional structure of KsBcl-2 showed overall structural similarity to Bcl-xL, Bcl-2, and Mcl-1 (34, 60). In accord, only the treatment with the pan-Bcl-2 inhibitors TW37 and AT101 induced apoptosis of KSHV-infected cells (42% and 65%, respectively, at 30 μM) (Fig. 3A and B). Although ABT199 (a Bcl-2-selective inhibitor) also stimulated death at 30 μM (Fig. 3A), ABT199 likely functions as a pan-Bcl-2 inhibitor at high micromolar concentrations (Table 1). In contrast, KSHV-infected cells were resistant to the Bcl-xL/Bcl-2 inhibitor (ABT737) and Mcl-1 inhibitors (MIM1 and compound 21), clearly demonstrating that both Mcl-1 and KsBcl-2 are simultaneously required for the survival of KSHV-infected cells. However, compound 10 induced apoptosis at 30 μM, albeit to a lesser extent than the pan-Bcl-2 inhibitors (Fig. 3B). KsBcl-2 is most closely related to Mcl-1, based on function and sequence homology (34); thus, compound 10 may have the unique ability to bind both Mcl-1 and KsBcl-2 at high concentrations, although further studies are needed to assess this possibility. Regardless, compound 10 likely represents a leading drug from which specific derivatives could be developed to target B cells latently infected with KSHV.
CONCLUSION
To date, BH3 profiling has been used to predict the sensitivities of cancer cells to small-molecule antagonists of antiapoptotic Bcl-2 proteins; however, we have now performed BH3 profiling on cells latently infected with human herpesviruses. Our data demonstrate BH3 profiling analysis to be a viable functional approach to globally decipher the magnitude of dependence on individual antiapoptotic Bcl-2 proteins within latently infected cells. Moreover, our study provides a proof-of-principle for the use of BH3 profiling to predict the efficacy of Bcl-2 family antagonists at eliminating specific persistent virus and cell infection combinations, which could have immediate impact on the treatment of diseases associated with latent herpesvirus infections. Since KSHV and EBV are oncogenic viruses and BH3 profiling is currently being used to predict tumor sensitivity to Bcl-2 inhibitors, BH3 profiling could be used to design effective chemotherapeutic regimens against the cancers induced by these viruses (61, 62). In addition, BH3 profiling can be used to screen the efficacy of different Mcl-1 small-molecule inhibitors in inducing death of HCMV-infected monocytes and CD34+ stem cells. Effective Mcl-1 antagonists could then be administered to recipients at high risk for acute HCMV infection prior to solid organ transplantation or be utilized to remove and/or deplete HCMV-infected CD34+ stem cells in vitro prior to bone marrow transplantation. Overall, our study demonstrates BH3 profiling to be a feasible technique for determining the level of dependence on individual Bcl-2 proteins within latently infected cells and for predicting sensitivity to small-molecule Bcl-2 inhibitors.
ACKNOWLEDGMENTS
We thank Arturo Babachano-Guerrero and Christine King (Upstate Medical University, Syracuse, NY) for kindly providing Bcbl-1 cells and Carrie Coleman and Rosemary Rochford (Upstate Medical University, Syracuse, NY) for kindly providing Akata and LCL-10 cell lines.
This work was supported by the Sinsheimer Scholar Award and grants from the American Heart Association and the National Institute of Allergy and Infectious Diseases (R56AI110803-01) to G. C. Chan and by the National Cancer Institute (R01CA149442 to Z. Nikolovska-Coleska).
We have no conflicting financial interests.
REFERENCES
- 1.Bolovan-Fritts CA, Mocarski ES, Wiedeman JA. 1999. Peripheral blood CD14(+) cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood 93:394–398. [PubMed] [Google Scholar]
- 2.Mendelson M, Monard S, Sissons P, Sinclair J. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol 77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 3.Taylor-Wiedeman J, Sissons P, Sinclair J. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol 68:1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Söderberg-Nauclér C, Fish KN, Nelson JA. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119–126. doi: 10.1016/S0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 5.Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol 71:4882–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyashita EM, Yang B, Lam KM, Crawford DH, Thorley-Lawson DA. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 7.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404. doi: 10.1016/S1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 8.Du MQ, Liu H, Diss TC, Ye H, Hamoudi RA, Dupin N, Meignin V, Oksenhendler E, Boshoff C, Isaacson PG. 2001. Kaposi sarcoma-associated herpesvirus infects monotypic (IgM lambda) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood 97:2130–2136. doi: 10.1182/blood.V97.7.2130. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Lagunoff M. 2005. Establishment and maintenance of Kaposi's sarcoma-associated herpesvirus latency in B cells. J Virol 79:14383–14391. doi: 10.1128/JVI.79.22.14383-14391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romee R, DiPersio JF. 2014. CMV prophylaxis in hematopoietic-cell transplantation. N Engl J Med 371:576. doi: 10.1056/NEJMc1406756. [DOI] [PubMed] [Google Scholar]
- 11.van der Bij W, Speich R. 2001. Management of cytomegalovirus infection and disease after solid-organ transplantation. Clin Infect Dis 33(Suppl 1):S32–S37. doi: 10.1086/320902. [DOI] [PubMed] [Google Scholar]
- 12.Razonable RR, Paya CV, Smith TF. 2002. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid-organ transplant recipients. J Clin Microbiol 40:746–752. doi: 10.1128/JCM.40.3.746-752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Booss J, Dann PR, Griffith BP, Kim JH. 1989. Host defense response to cytomegalovirus in the central nervous system. Predominance of the monocyte. Am J Pathol 134:71–78. [PMC free article] [PubMed] [Google Scholar]
- 14.Rollag H, Asberg A, Ueland T, Hartmann A, Jardine AG, Humar A, Pescovitz MD, Bignamini AA, Aukrust P. 2012. Treatment of cytomegalovirus disease in solid organ transplant recipients: markers of inflammation as predictors of outcome. Transplantation 94:1060–1065. doi: 10.1097/TP.0b013e31826c39de. [DOI] [PubMed] [Google Scholar]
- 15.Sagedal S, Nordal KP, Hartmann A, Degre M, Holter E, Foss A, Osnes K, Leivestad T, Fauchald P, Rollag H. 2000. A prospective study of the natural course of cytomegalovirus infection and disease in renal allograft recipients. Transplantation 70:1166–1174. doi: 10.1097/00007890-200010270-00007. [DOI] [PubMed] [Google Scholar]
- 16.Thompson MP, Kurzrock R. 2004. Epstein-Barr virus and cancer. Clin Cancer Res 10:803–821. doi: 10.1158/1078-0432.CCR-0670-3. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan LD. 2013. Human herpesvirus-8: Kaposi sarcoma, multicentric Castleman disease, and primary effusion lymphoma. Hematology Am Soc Hematol Educ Program 2013:103–108. doi: 10.1182/asheducation-2013.1.103. [DOI] [PubMed] [Google Scholar]
- 18.Rudin CM, Thompson CB. 1997. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med 48:267–281. doi: 10.1146/annurev.med.48.1.267. [DOI] [PubMed] [Google Scholar]
- 19.Chan G, Nogalski MT, Bentz GL, Smith MS, Parmater A, Yurochko AD. 2010. PI3K-dependent upregulation of Mcl-1 by human cytomegalovirus is mediated by epidermal growth factor receptor and inhibits apoptosis in short-lived monocytes. J Immunol 184:3213–3222. doi: 10.4049/jimmunol.0903025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves MB, Breidenstein A, Compton T. 2012. Human cytomegalovirus activation of ERK and myeloid cell leukemia-1 protein correlates with survival of latently infected cells. Proc Natl Acad Sci U S A 109:588–593. doi: 10.1073/pnas.1114966108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole E, McGregor Dallas SR, Colston J, Joseph RS, Sinclair J. 2011. Virally induced changes in cellular microRNAs maintain latency of human cytomegalovirus in CD34+ progenitors. J Gen Virol 92:1539–1549. doi: 10.1099/vir.0.031377-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Rowe M, Lundgren E. 1996. Expression of the Epstein Barr virus transforming protein LMP1 causes a rapid and transient stimulation of the Bcl-2 homologue Mcl-1 levels in B-cell lines. Cancer Res 56:4610–4613. [PubMed] [Google Scholar]
- 23.Kim JH, Kim WS, Park C. 2012. Epstein-Barr virus latent membrane protein-1 protects B-cell lymphoma from rituximab-induced apoptosis through miR-155-mediated Akt activation and up-regulation of Mcl-1. Leuk Lymphoma 53:1586–1591. doi: 10.3109/10428194.2012.659736. [DOI] [PubMed] [Google Scholar]
- 24.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107–1115. doi: 10.1016/0092-8674(91)90007-L. [DOI] [PubMed] [Google Scholar]
- 25.Portis T, Longnecker R. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619–8628. doi: 10.1038/sj.onc.1207905. [DOI] [PubMed] [Google Scholar]
- 26.Gao J, Cai Q, Lu J, Jha HC, Robertson ES. 2011. Upregulation of cellular Bcl-2 by the KSHV encoded RTA promotes virion production. PLoS One 6:e23892. doi: 10.1371/journal.pone.0023892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham C, Matta H, Yang Y, Yi H, Suo Y, Tolani B, Chaudhary PM. 2013. Kaposi's sarcoma-associated herpesvirus oncoprotein K13 protects against B cell receptor-induced growth arrest and apoptosis through NF-κB activation. J Virol 87:2242–2252. doi: 10.1128/JVI.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellows DS, Howell M, Pearson C, Hazlewood SA, Hardwick JM. 2002. Epstein-Barr virus BALF1 is a BCL-2-like antagonist of the herpesvirus antiapoptotic BCL-2 proteins. J Virol 76:2469–2479. doi: 10.1128/jvi.76.5.2469-2479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med 3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 30.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. 2006. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Del Gaizo Moore V, Letai A. 2013. BH3 profiling–measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett 332:202–205. doi: 10.1016/j.canlet.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni Chonghaile T, Letai A. 2008. Mimicking the BH3 domain to kill cancer cells. Oncogene 27(Suppl 1):S149–S157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas LW, Lam C, Edwards SW. 2010. Mcl-1; the molecular regulation of protein function. FEBS Lett 584:2981–2989. doi: 10.1016/j.febslet.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 34.Flanagan AM, Letai A. 2008. BH3 domains define selective inhibitory interactions with BHRF-1 and KSHV BCL-2. Cell Death Differ 15:580–588. doi: 10.1038/sj.cdd.4402292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kvansakul M, Hinds MG. 2013. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis 4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan JA, Brunelle JK, Letai A. 2010. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A 107:12895–12900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitelaw DM. 1972. Observations on human monocyte kinetics after pulse labeling. Cell Tissue Kinet 5:311–317. [DOI] [PubMed] [Google Scholar]
- 38.Cheng EH, Nicholas J, Bellows DS, Hayward GS, Guo HG, Reitz MS, Hardwick JM. 1997. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci U S A 94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol 75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer NH, Tschachler E, Colombini S, Ensoli B, Sturzl M. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J Virol 71:7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz TF. 2000. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8): epidemiology and pathogenesis. J Antimicrob Chemother 45(Suppl T3):15–27. doi: 10.1093/jac/45.suppl_4.15. [DOI] [PubMed] [Google Scholar]
- 42.Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, Haase AT. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 71:715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andoniou CE, Andrews DM, Manzur M, Ricciardi-Castagnoli P, Degli-Esposti MA. 2004. A novel checkpoint in the Bcl-2-regulated apoptotic pathway revealed by murine cytomegalovirus infection of dendritic cells. J Cell Biol 166:827–837. doi: 10.1083/jcb.200403010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roulston A, Marcellus RC, Branton PE. 1999. Viruses and apoptosis. Annu Rev Microbiol 53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 45.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, Kieff ED, Mocarski ES, Chittenden T. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A 96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A 98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldmacher VS. 2002. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84:177–185. doi: 10.1016/S0300-9084(02)01367-6. [DOI] [PubMed] [Google Scholar]
- 48.Chan G, Nogalski MT, Yurochko AD. 2012. Human cytomegalovirus stimulates monocyte-to-macrophage differentiation via the temporal regulation of caspase 3. J Virol 86:10714–10723. doi: 10.1128/JVI.07129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardwick JM, Lieberman PM, Hayward SD. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol 62:2274–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson GR, Luka J, Petti L, Sample J, Birkenbach M, Braun D, Kieff E. 1987. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology 160:151–161. doi: 10.1016/0042-6822(87)90055-9. [DOI] [PubMed] [Google Scholar]
- 51.Austin PJ, Flemington E, Yandava CN, Strominger JL, Speck SH. 1988. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc Natl Acad Sci U S A 85:3678–3682. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodescot M, Perricaudet M. 1986. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res 14:7103–7114. doi: 10.1093/nar/14.17.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly GL, Long HM, Stylianou J, Thomas WA, Leese A, Bell AI, Bornkamm GW, Mautner J, Rickinson AB, Rowe M. 2009. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in Burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog 5:e1000341. doi: 10.1371/journal.ppat.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abulwerdi FA, Liao C, Mady AS, Gavin J, Shen C, Cierpicki T, Stuckey JA, Showalter HD, Nikolovska-Coleska Z. 2014. 3-Substituted-N-(4-hydroxynaphthalen-1-yl)arylsulfonamides as a novel class of selective Mcl-1 inhibitors: structure-based design, synthesis, SAR, and biological evaluation. J Med Chem 57:4111–4133. doi: 10.1021/jm500010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen NA, Stewart ML, Gavathiotis E, Tepper JL, Bruekner SR, Koss B, Opferman JT, Walensky LD. 2012. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem Biol 19:1175–1186. doi: 10.1016/j.chembiol.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammad RM, Goustin AS, Aboukameel A, Chen B, Banerjee S, Wang G, Nikolovska-Coleska Z, Wang S, Al-Katib A. 2007. Preclinical studies of TW-37, a new nonpeptidic small-molecule inhibitor of Bcl-2, in diffuse large cell lymphoma xenograft model reveal drug action on both Bcl-2 and Mcl-1. Clin Cancer Res 13:2226–2235. doi: 10.1158/1078-0432.CCR-06-1574. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, Nikolovska-Coleska Z, Yang CY, Wang R, Tang G, Guo J, Shangary S, Qiu S, Gao W, Yang D, Meagher J, Stuckey J, Krajewski K, Jiang S, Roller PP, Abaan HO, Tomita Y, Wang S. 2006. Structure-based design of potent small-molecule inhibitors of anti-apoptotic Bcl-2 proteins. J Med Chem 49:6139–6142. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- 58.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 59.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. 2013. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 60.Polster BM, Pevsner J, Hardwick JM. 2004. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim Biophys Acta 1644:211–227. doi: 10.1016/j.bbamcr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Pierceall WE, Kornblau SM, Carlson NE, Huang X, Blake N, Lena R, Elashoff M, Konopleva M, Cardone MH, Andreeff M. 2013. BH3 profiling discriminates response to cytarabine-based treatment of acute myelogenous leukemia. Mol Cancer Ther 12:2940–2949. doi: 10.1158/1535-7163.MCT-13-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pierceall WE, Warner SL, Lena RJ, Doykan C, Blake N, Elashoff M, Hoff DV, Bearss DJ, Cardone MH, Andritsos L, Byrd JC, Lanasa MC, Grever MR, Johnson AJ. 2014. Mitochondrial priming of chronic lymphocytic leukemia patients associates Bcl-xL dependence with alvocidib response. Leukemia 28:2251–2254. doi: 10.1038/leu.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. 2007. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell 12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]