ABSTRACT
Filoviruses, including both Ebola virus (EBOV) and Marburg virus (MARV), can infect humans and other animals, causing hemorrhagic fever with a high mortality rate. Entry of these viruses into the host is mediated by a single filoviral glycoprotein (GP). GP is composed of two subunits: GP1, which is responsible for attachment and binding to receptor(s) on susceptible cells, and GP2, which mediates viral and cell membrane fusion. Although numerous host factors have been implicated in the entry process, the initial attachment receptor(s) has not been well defined. In this report, we demonstrate that exostosin 1 (EXT1), which is involved in biosynthesis of heparan sulfate (HS), plays a role in filovirus entry. Expression knockdown of EXT1 by small interfering RNAs (siRNAs) impairs GP-mediated pseudoviral entry and that of infectious EBOV and MARV in tissue cultured cells. Furthermore, HS, heparin, and other related glycosaminoglycans (GAGs), to different extents, can bind to and block GP-mediated viral entry and that of infectious filoviruses. These results strongly suggest that HS and other related GAGs are attachment receptors that are utilized by filoviruses for entry and infection. These GAGs may have therapeutic potential in treating EBOV- and MARV-infected patients.
IMPORTANCE Infection by Ebola virus and Marburg virus can cause severe illness in humans, with a high mortality rate, and currently there is no FDA-approved vaccine or therapeutic treatment available. The ongoing 2014 outbreak in West Africa underscores a lack of our understanding in the infection and pathogenesis of these viruses and the urgency of drug discovery and development. In this study, we provide several pieces of evidence that demonstrate that heparan sulfate and other closely related glycosaminoglycans are the molecules that are used by filoviruses for initial attachment. Furthermore, we demonstrate that these glycosaminoglycans can block entry of and infection by filoviruses. Thus, this work provides mechanistic insights on the early step of filoviral infection and suggests a possible therapeutic option for diseases caused by filovirus infection.
INTRODUCTION
Filoviruses, including Ebola virus (EBOV) and Marburg virus (MARV), are long, filamentous enveloped viruses that cause hemorrhagic fevers in humans and nonhuman primates. Outbreaks of EBOV have occurred sporadically in Africa since the 1970s, with mortality rates of up to 90% (1). The ongoing and unprecedented 2014 Ebola epidemic in West Africa underscores the severity of the diseases associated with the infection and the challenge of dealing with it globally. Although several potential therapeutics were recently reported to be effective in treating nonhuman primates (2, 3), there are currently no approved antivirals or vaccines effective against filoviruses in humans, and treatments are solely symptom based (4, 5). However, development of antivirals against EBOV and MARV infection and diseases is hampered by a lack of understanding of the fundamental principles underlying the replication and pathogenesis of these viruses.
Infection by filoviruses is initiated by interactions of the viral glycoprotein GP with host factors on target cells. EBOV and MARV GPs are synthesized as GP0 precursors, with subsequent proteolytic cleavage into GP1 and GP2, which are linked together by disulfide bonds (1). A GP1-GP2 trimer on the virion surface mediates binding to viral receptors on the host surface via GP1 interactions (6–8), which is followed by macropinocytosis of the virion and virus-membrane fusion mediated by GP (9). Although several host factors have been implicated in filoviral entry (10–13), their cellular localization as well as inconsistencies in expression patterns suggests that other distinct attachment receptors have yet to be defined. Finding such factors would have a great impact on our understanding of filovirus entry and developing filovirus-specific antiviral treatments. To identify and characterize such host factors that are involved in filovirus entry, we have performed a genome-wide RNA interference (RNAi) screen against viral infection. In this report, we describe an important role of exostosin 1 (EXT1) and glycosaminoglycans (GAGs) in the initial attachment during MARV and EBOV infection. Furthermore, the potential therapeutic use of GAGs is discussed.
MATERIALS AND METHODS
Cells.
293T and A549 cells were obtained from the American Type Culture Collection (ATCC CCL-185). They were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 1× penicillin-streptomycin (Pen-Strep) and maintained at 37°C in a 5% CO2 atmosphere. Primary human pulmonary artery endothelial cells (HPAECs) were grown in EBM-2 medium (catalog number CC-3156; Lonza, Basel, Switzerland) supplemented with EGM-2MV growth factors (catalog number CC-4147; Lonza).
Infectious viruses.
EBOV and MARV expressing a green fluorescent protein (GFP) reporter were derived by reverse genetics as described by Towner et al. (14). All infectious virus assays were performed at the U.S. Army Medical Research Institute of Infectious Diseases at biosafety level 4. Infection by virus was determined by measuring GFP intensity in a Gemini EM spectrofluorometer (Molecular Devices, Sunnyvale, CA, USA).
Pseudovirus production.
293T cells were cotransfected with a replication-defective HIV vector (15) and the pcDNA 3.1+ encoding MARV GP, EBOV Zaire GP (16), or hemagglutinin (HA; H5) from influenza virus A/Viet Nam/1203/2004 and neuraminidase (NA; N1) from influenza virus A/Puerto Rico/8/1934 (17) by using a polyethylenimine (PEI)-based transfection protocol. Six hours posttransfection, the medium was changed to phenol red-free DMEM with 10% FBS and Pen-Strep. Forty-eight hours posttransfection, medium was collected, filtered through a 0.45-μm filter (Millipore, Billerica, MA, USA), and stored at 4°C.
RNAi screening.
The Silencer Select Human Druggable Genome siRNA Library V4, Human Druggable Genome siRNA Library V4 Extension Set, and Human Genome siRNA Library V4 Extension Set libraries were purchased from Applied Biosystems (Grand Island, NY, USA). A549 cells (1,000 cells/well) were reverse transfected with 10 nM siRNAs and 0.1 μl Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY, USA) in 384-well white Culturplates (PerkinElmer, Downers Grove, IL, USA) according to the RNAiMAX manufacturer's protocol with a Janus liquid handling system (PerkinElmer). Forty-eight hours posttransfection, the medium was removed and 30 μl of either Marburg or influenza pseudotyped virus was added. The medium was changed 24 h postinfection, and 48 h postinfection 15 μl of Neolite luciferase substrate (PerkinElmer) was mixed in; the mixture was incubated for 5 min, and luciferase activity was measured with a Envision plate reader (PerkinElmer).
siRNA transfection.
Small interfering RNAs (siRNAs) targeting EXT1 or nontargeting control or firefly luciferase control were obtained from Ambion (Grand Island, NY, USA). Reverse transfection of A549 cells with 10 nM siRNA and 0.3 μl Lipofectamine RNAiMAX was carried out in a 96-well plate according to the recommended protocol. Forty-eight hours posttransfection, medium was removed and 50 μl of infectious EBOV or MARV or 100 μl of pseudotyped virus was added. The medium was changed 24 h postinfection, and luciferase activity or GFP was read at 48 h postinfection.
Real-time PCR.
Expressions of target genes were determined 48 or 96 h after siRNA transfection by quantitative reverse transcription-PCR (qRT-PCR). Samples were prepared by using the SYBR green Cells-to-Ct kit (Ambion) according to the recommended protocol, and qRT-PCR was performed in the CFX96 real-time PCR system (Bio-Rad). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an internal control, and samples were normalized to samples transfected with nontargeting siRNA by using the ΔΔCT method (where CT is threshold cycle). Conditions were optimized to primer efficiencies according to the Bio-Rad CFX Manager software. Primer sequences were as follows: GAPDH, 5′-GAAGGTGAAGGTCGGAGTC and 3′-GAAGATGGTGATGGGATTTC; EXT1, 5′-GCTCTGCGCCCCTTCGTTC and 3′-TGCCTTTGTAGATGCTGGAGTTGG.
Cotransfection and Western assay.
293T cells were transfected with 10 nM siRNA with Lipofectamine RNAiMAX in Opti-MEM (Gibco, Grand Island, NY, USA). Twenty-four hours posttransfection, EXT1-c-Myc plasmid was transfected with PEI in Opti-MEM at various concentrations. The medium was changed to complete medium 16 h after plasmid transfection, and 48 h after plasmid transfection, cells were lysed and samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane. EXT1-c-Myc expression was probed by a monoclonal antibody (MAb) against the Myc tag and then probed with a peroxidase-conjugated secondary antibody. The bands were visualized by the chemiluminescence method according to the protocol of the supplier (Pierce, Rockford, IL, USA). In these experiments, mouse anti-β-actin (1:10,000 dilution) monoclonal antibodies were used as indicators for the cell lysate loading.
Compound blocking assay.
Heparan sulfate (H7640), heparin (H3393), chondroitin sulfate A (C9819), and chondroitin sulfate B (C3788) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Cells were seeded in a 96-well plate (A549 cells) or a 24-well plate (HPAECs) prior to experiment. Virus (50 μl of infectious virus or 100 μl/500 μl pseudovirus) was incubated with various concentrations of GAGs for 2 h at 37°C and added to cells in 96-well plates and 24-well plates, respectively. After another 2 h of incubation, virus was removed and fresh medium added. Luciferase activity or GFP was read 48 h postinfection.
ELISA and competition assays.
Enzyme-linked immune sorbent assays (ELISAs) were performed in streptavidin-coated 384-well plates (Pierce). Plates were incubated overnight with biotinylated heparin (Sigma) at 1.6 μg/well. Pseudovirus-containing supernatant was layered over a 30% sucrose–NTE (sodium Tris buffer with EDTA) cushion and spun at 55,000 rpm for 1 h in a Beckman SW55 rotor at 4°C. Virus pellets were resuspended in 200 μl Tris buffer. Concentrated virus was diluted in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA) and incubated in plates overnight at 4°C. Wells were washed three times with PBST (PBS, 0.1% Tween 20) and three times with PBS and then blocked with PBS with 1% BSA for 2 h at 4°C. Plates were incubated with primary antibody for 4 h at 4°C and washed again as described above. Plates were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody and washed again. The reaction was visualized by addition of 50 μl of Ultra TMB ELISA substrate (Pierce) for 20 min. The reaction was stopped with 50 μl of stop solution made of 2 M sulfuric acid, and absorbance at 450 nm was measured in an Envision plate reader (PerkinElmer). For competition ELISAs, compounds were incubated with resuspended virus for 1 h on ice before incubating in plates.
RESULTS
Knockdown of exostosin 1 expression impairs filoviral GP-mediated entry.
To identify the host proteins that play a role in filoviral entry, we developed a parallel high-throughput siRNA screening protocol (referred to as pHTS here) using an HIV-1-based lentiviral pseudotyping entry assay (7). This surrogate system consists of a replication-deficient HIV-1 core with a luciferase reporter and the glycoprotein(s) of a highly pathogenic enveloped virus such as Marburg virus, referred to as MARVpv here (8), or avian influenza virus H5N1 (AIVpv here) (17). This screening protocol (our unpublished data) was used to carry out a parallel screening with siRNA libraries (Ambion human siRNA libraries targeting 21,585 genes with three siRNAs for each gene). Briefly, A549 cells were reverse transfected with individual siRNAs in a 384-well format, and 48 h posttransfection, they were infected with AIVpv or MARVpv. Forty-eight hours postinfection, the luciferase activities of the infected cells were measured, and the data were analyzed to identify the putative hits. These putative hits (3,319) were further evaluated by confirmation screens to identify the virus-specific host proteins that play a potential role either for entry of avian influenza virus H5N1 or for entry of Marburg virus. Based on these analyses, exostosin 1 (EXT1) was identified as a host protein that plays a specific role in Marburg virus entry into the host cells.
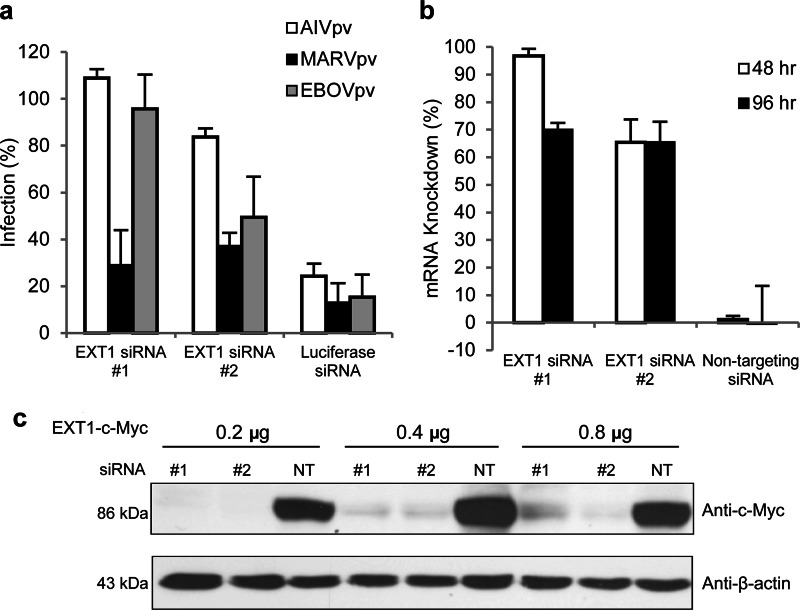
To further investigate the role of EXT1 in filoviral GP-mediated entry, two EXT1-specific siRNAs (labeled #1 and #2 in Fig. 1) were individually transfected to A549 cells, and the transfected cells were challenged with MARVpv, EBOVpv, or AIVpv to examine their effects on viral infection. While EXT1-specific siRNAs had little or no effect on infection of AIVpv, MARVpv infection was reduced by more than 60 to 70% of the nontargeting siRNA control (Fig. 1a). One of the EXT1-specific siRNAs (#2) also moderately reduced the infection by EBOVpv by ∼50% (Fig. 1a).
FIG 1.
Reductions in EXT1 mRNA and protein levels correlate with reduced infection of MARV and EBOV pseudotyped particles. (a) Introduction of siRNAs of EXT1 resulted in reduced infection by MARVpv and EBOVpv. The siRNAs of EXT1 (labeled as siRNA #1 and #2) were transfected to A549 cells, and their effects on pseudotyped Marburg virus (MARVpv), Ebola virus (EBOVpv), and influenza virus (AIVpv) were evaluated as described in Materials and Methods. A siRNA of firefly luciferase was used as a control in the experiment, and the data were normalized to nontargeting siRNA (NT). Error bars represent standard deviations. (b) Real-time PCR on cells transfected with EXT1 siRNAs showed mRNA knockdown of EXT1 at 48 and 96 h posttransfection, using NT siRNA as a control. Error bars represent standard deviations. (c) The siRNAs of EXT1 reduced the protein level of EXT1 in the target cells. An EXT1-C-Myc plasmid and EXT1 siRNAs were cotransfected to A549 cells, and the EXT1-C-Myc level in the cells was evaluated by Western analysis. In this experiment, different amounts of EXT1-C-Myc plasmid DNA (0.2 to 0.8 μg) were used. β-Actin was used as a control.
To confirm that EXT1 mRNA expression level was efficiently reduced by the EXT1-specific siRNAs, quantitative real-time PCR (qPCR) was performed on A549 cells transfected with either the EXT1-specific siRNAs or a nontargeting siRNA (as a control) at 48 and 96 h posttransfection. The level of EXT1 mRNA was reduced by approximately 70 to 100% by the EXT1-specific siRNAs, measured at 48 h and 96 h posttransfection (Fig. 1b).
To demonstrate that the EXT1 siRNAs could reduce the level of EXT1 protein expression in the cells, we first attempted to detect EXT1 protein in A594 and 293T cells with several commercially available antibodies but failed to detect EXT1 protein in these cells (data not shown), suggesting that the EXT1 protein level is quite low in these cells. Therefore, we devised an alternative protocol to demonstrate the specific siRNA knockdown of EXT1 protein. A plasmid containing the EXT1-C-myc-tagged version of EXT1 gene (18) was cotransfected with siRNAs to A549 cells, and the expression of the tagged EXT1 protein was examined by Western blotting. As shown in Fig. 1c, the EXT1-specific siRNAs 1 and 2 were able to effectively diminish the levels of EXT1-C-myc.
Together, these results demonstrate that EXT1 plays a specific role in Marburg virus GP-mediated (and to a lesser extent, in Ebola virus GP-mediated) viral entry.
HS biosynthesis-deficient cell lines are resistant to Marburg virus GP-mediated viral entry.
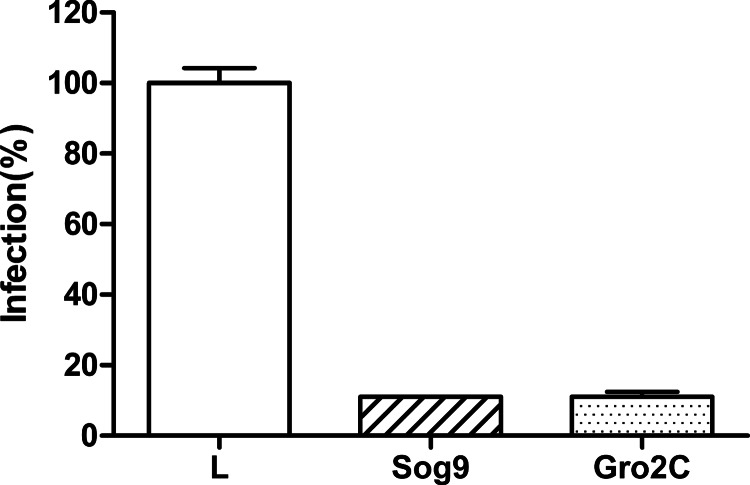
EXT1 plays a crucial role in the biosynthesis of heparan sulfate (HS), and it has been shown that the loss of this gene function leads to the reduction of HS on the cell surface (18, 19). A mouse cell line (L) and two L-cell-derived mutant cell lines that are defective in the biosynthesis of HS and other GAGs were challenged with MARVpv, and the infection was quantified by the luciferase level of the infected cells. Figure 2 shows that both HS-deficient cell lines Sog9 and Gro2C were resistant to the MARV GP-mediated viral entry in comparison to their nondeficient parent cell line, consistent with the notion that EXT1 (and GAGs) plays a role in MARV entry to the target cells.
FIG 2.
Heparan sulfate (HS) biosynthesis-deficient cell lines were resistant to MARVpv infection. The parental mouse L cells (L) and two lines of HS biosynthesis-deficient cells (Sog9 and Gro2C, which were derived from L cells) were challenged with MARVpv, and the infection was measured by the luciferase level of the infected cells. Error bars represent standard deviations.
GAGs block GP-mediated entry.
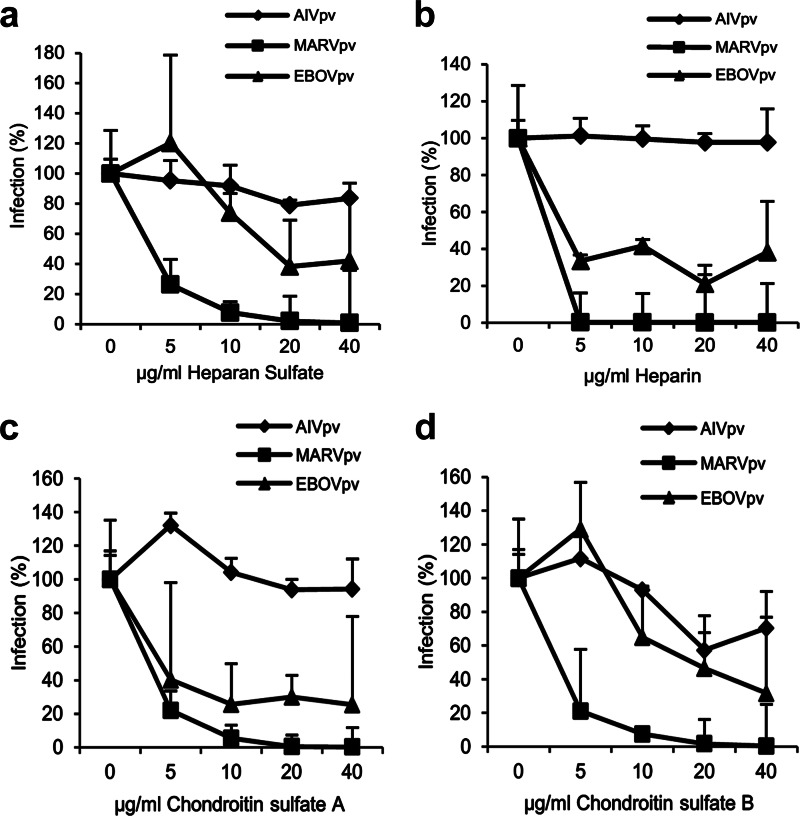
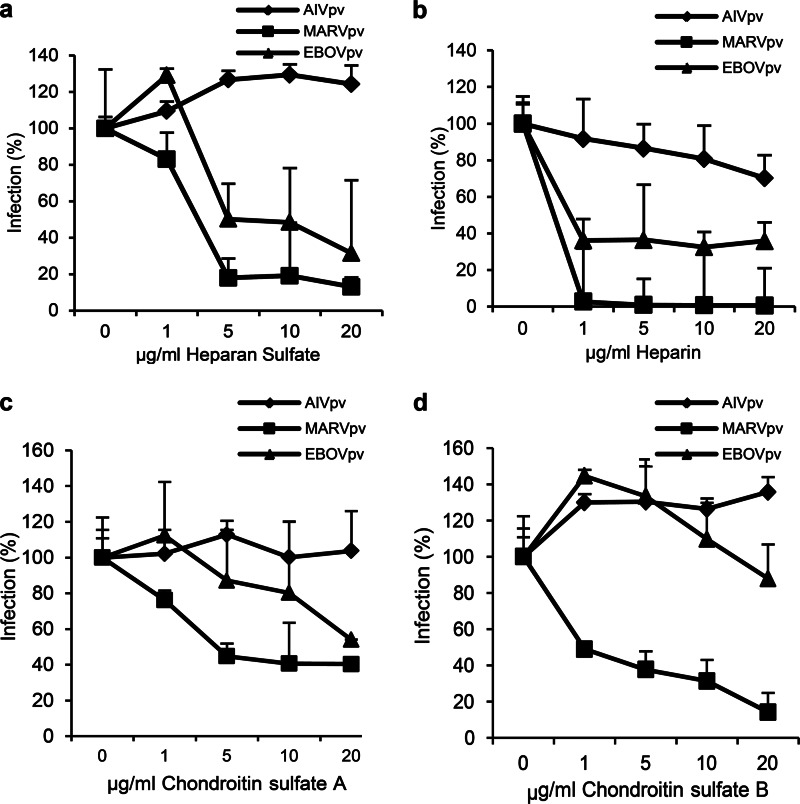
To further evaluate the role of GAGs in filoviral entry, four types of GAGs were tested in their ability to block GP-mediated viral entry: HS, heparin, chondroitin sulfate A (CSA), and chondroitin sulfate B (CSB). The experiments were performed as follows: the viral preparations of MARVpv, EBOVpv, or AIVpv were first mixed with GAGs at different concentrations for 2 h, and the mixtures were added to A549 target cells for an additional 2 h and then replaced with fresh medium. Cells were lysed, and the luciferase levels of the target cells were determined at 48 h postinfection. Infection of EBOVpv and MARVpv showed a dose-dependent reduction in the presence of HS, with approximately 60% of reduction for EBOVpv and 100% for MARVpv at 40 μg/ml of HS. In contrast, AIVpv was minimally affected by increasing concentrations of HS, as expected (Fig. 3a). Similar trends were observed for heparin, CSA, and CSB (Fig. 3b, c, and d, respectively).
FIG 3.
GAGs blocked infection by MARVpv and EBOVpv in A549 cells. The pseudotyped viruses MARVpv, EBOVpv, and AIVpv were mixed with different GAGs, HS (a), heparin (b), chondroitin sulfate A (c), and chondroitin sulfate B (d), at various concentrations; the mixtures were used to challenge A549 cells, and the effects of different GAGs were evaluated as described in Materials and Methods. Error bars represent standard deviations.
To ensure that the blocking effect of GAGs was not an artifact of a transformed cell line (A549 cells), each of the four GAGs was tested for its ability to block infection in primary human pulmonary artery endothelial cells (HPAECs). As with A549 cells, the greatest effect of the different GAGs was on blocking MARVpv infection, and their inhibitory effects on EBOVpv were less pronounced, while no or minimal effect was observed on AIVpv infection (Fig. 4).
FIG 4.
GAGs blocked infection of MARVpv and EBOVpv in primary human pulmonary artery endothelial cells (HPAECs). The pseudotyped viruses MARVpv, EBOVpv, and AIVpv were mixed with different GAGs, HS (a), heparin (b), chondroitin sulfate A (c), and chondroitin sulfate B (d) at various concentrations; the mixtures were used to challenge HPAECs, and the effects of different GAGs were evaluated as described in Materials and Methods. Error bars represent standard deviations.
Based on these results, we conclude that HS and related GAGs have specific anti-filoviral entry activity, implicating a direct interaction of GAGs with the filoviral glycoproteins.
Glycoprotein binds to heparin and heparan sulfate.
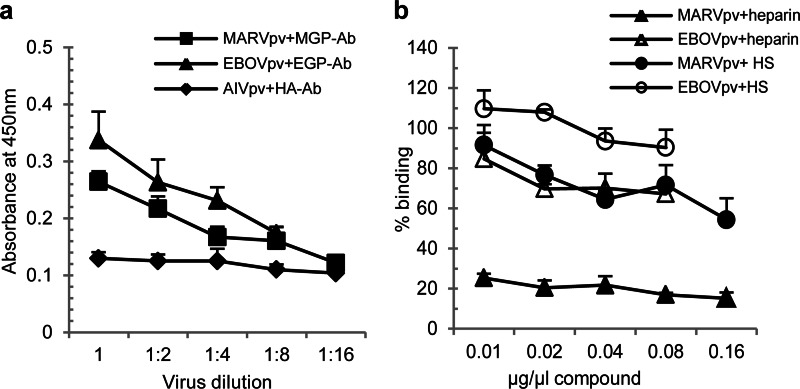
An ELISA that was modified from a published protocol for measuring binding of AIVpv particles to the receptor by us previously (20) was developed and performed to determine if the GAGs bind to GP. Briefly, plates were coated with biotinylated heparin, incubated with MARVpv, EBOVpv, or AIVpv, and washed, and then bound pseudovirions were detected with respective primary and HRP-conjugated secondary antibodies, and absorbance at 450 nM was measured. As expected, AIVpv did not show a dose-dependent binding. In contrast, both MARVpv and EBOVpv displayed a dose-dependent binding to heparin (Fig. 5a).
FIG 5.
MARVpv and EBOVpv were able to bind to heparin and HS. (a) An ELISA (described in Materials and Methods) was used to demonstrate specific binding of MARVpv and EBOVpv to heparin and HS. AIVpv was used here as a specificity control. Error bars represent standard deviations. (b) An ELISA-based competition experiment was used to demonstrate that “cold” heparin and HS can competitively block binding of MARVpv and EBOVpv to the heparin used to coat the plates. Error bars represent standard deviations.
To further evaluate different GAGs in their ability to bind filoviral GPs, a competition ELISA was used. Here, GAGs at different concentrations were first mixed with the pseudovirions and added to the heparin-coated plates, and the aforementioned ELISA protocol was then followed. As shown in Fig. 5b, MARVpv binding to the coated heparin was effectively blocked by unlabeled heparin, reducing the binding by approximately 80% even at the lowest concentration tested (0.01 μg/μl), while HS, at 0.16 μg/μl, was able to reduce binding by about 40%. In contrast, heparin had only a marginal effect on EBOVpv binding, reducing binding by roughly 30% at the highest concentration of heparin tested (0.16 μg/μl), while HS did not adversely affect binding. In addition, we tested CSA and CSB in their ability to compete against binding of MARVpv and EBOVpv to the heparin used to coat the plates, and we did not observe any significant effect (data not shown).
Based on these observations, we conclude that HS and other GAGs bind MARV GP and to a lesser extent EBOV GP. These results are in general consistent with the data presented above, implicating a role of HS and other GAGs in GP binding and filoviral infection.
Only HMWH is effective in blocking MARVpv infection.
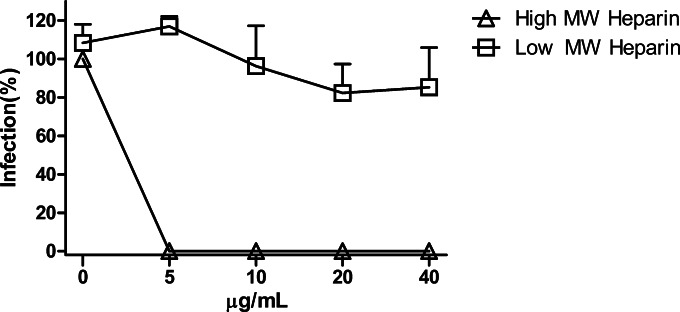
We decided to further explore the effect of heparin in MARVpv infection since it is the most effective GAG to bind GP. We tested two classes of heparin, high-molecular-weight heparin (HMWH) and low-molecular-weight heparin (LMWH). It is well known that LMWH is a class of anticoagulant medications. Porcine heparin (HMWH, ∼17 to 19 kDa) and heparin hexasaccharide (an LMWH, ∼1.5 kDa), at different concentrations, were first mixed with MARVpv and then added to the target cells (A549), and infection of MARVpv was determined 48 h postinfection. Heparin hexasaccharide, with all the concentrations tested (5 to 40 μg/ml), did not have any effect on blocking MARVpv infection, while porcine heparin, even at the lowest concentration (5 μg/ml), was able to completely block MARVpv infection (Fig. 6). These results demonstrate that HMWH, but not LMWH, is effective in blocking MARV entry.
FIG 6.
Only high-molecular-weight heparin (HMWH) could block MARVpv infection. Porcine heparin (HMWH, ∼17 to 19 kDa) and heparin hexasaccharide (LMWH, ∼1.5 kDa), at the concentrations indicated, were first mixed with MARVpv and then added to the target cells (A549), and infection by MARVpv was determined 48 h postinfection. Error bars represent standard deviations.
EXT1 and GAGs are involved in infection of infectious filoviruses.
The results described above, which demonstrated a role of EXT1 and GAGs in filoviral entry and infection, were from assays performed with pseudoviral particles. To validate these findings, infectious EBOV and MARV were used in the following experiments. First, to confirm the role of EXT1 in viral infection, A549 cells were transfected with EXT1 or NT siRNAs and infected with infectious MARV and EBOV carrying a GFP reporter. Introduction of siRNA 1 or 2 of EXT1 to A549 cells reduced MARV infection by 85% or 60%, respectively, compared to the NT siRNA control, while the same siRNAs reduced EBOV infection by 10% or 60%, respectively. These results are in agreement with the data above with the MARV and EBOV pseudovirions, demonstrating the role of EXT1 in filoviral entry (Fig. 7a).
FIG 7.
EXT1 and GAGs are involved in infection by infectious MARV and EBOV. (a) Knockdown expression of EXT1 by siRNAs reduced levels of infection by infectious MARV and EBOV. The EXT1 siRNAs were introduced to A549 cells, and their effects on infection of infectious MARV and EBOV were determined following a protocol as described in Materials and Methods. An NT siRNA was used as the control in this experiment. Error bars represent standard deviations. (b) Effects of different GAGs on blocking infectious MARV infection. Different GAGs, at different concentrations, were mixed with infectious MARV, and their effects on blocking MARV infection on A549 cells were evaluated. Error bars represent standard deviations. (c) Effects of different GAGs on blocking infectious EBOV infection. Error bars represent standard deviations.
Next, we examined the effects of different GAGs on blocking infectious MARV and EBOV infections. The most potent GAG to block infection by infectious MARV was HMWH, and a near-complete inhibition at all the concentrations tested (0.25 to 50 mg/ml) was observed, while other GAGs were less effective in blocking MARV infection, although they did display a dose-dependent inhibition on MARV (Fig. 7b). In contrast, these GAGs were less effective in inhibiting infection by infectious EBOV (Fig. 7c). Again, these results validated the data described above with the pseudovirions.
Together, these results demonstrate that EXT1 plays a role in filoviral infections, particularly infection by MARV, and that GAGs, particularly heparin and HS, can block filoviral entry and infection.
DISCUSSION
Here we report that expression of EXT1, a glycosyltransferase that is involved in the biosynthesis of heparan sulfate (HS) (18, 19), is required for efficient entry of the filoviruses into the host cells. We show that HS and heparin, a related glycosaminoglycan (GAG), are able to bind GP. These and other related GAGs effectively block MARV, and to a lesser extent EBOV, infection in primary cells and transformed cell lines. These results strongly suggest that HS and related GAGs are involved in initial attachment of filoviruses to the target cells and thus play an important role in mediating filoviral entry.
Numerous host factors, including the asialoglycoprotein receptor, the folate receptor-α, the dendritic cell-specific or liver lymph node-specific ICAM3-grabbing nonintegrins, human macrophage galactose-specific and N-acetylgalactosamine-specific C-type lectins, cathepsin B and L, and the Tyro3 receptor kinase family, have all been implicated in Ebola/Marburg virus entry (21–26). Recently, T-cell immunoglobulin and mucin domain 1 (TIM1) and Niemann-Pick disease type C1 (NPC1) receptors have been shown to be critical in EBOV/MARV entry (10–12). Nevertheless, the entry mechanism of filovirus is still poorly understood. To identify the other host factors that are involved in filoviral entry and infection, we performed a genome-wide siRNA screen, and several host genes were identified as being critical for filoviral entry, including NPC1 (data not shown) and EXT1 genes, the latter of which is the focus of the current study. The results presented here reveal a role of EXT1 and GAGs in mediating filoviral entry and infection. We have demonstrated that knockdown of EXT1 expression by siRNAs reduced the GP-mediated filoviral entry of MARV (and to a lesser extent, of EBOV) and that the mouse cell lines that are defective in the biosynthesis of HS and other related GAGs were impaired in MARV GP-mediated entry (Fig. 1 and 2). Furthermore, we have demonstrated that HS, heparin, and other related GAGs can block EBOV and MARV GP-mediated viral entry in tissue cultured cells and primary human cells (Fig. 3 and 4). In addition, we have demonstrated that EBOV GP can bind HS and heparin directly (Fig. 5). Importantly, the role of EXT1 and GAGs in filovirus infection has been validated with infectious EBOV and MARV in tissue culture (Fig. 7). Since HS and the related GAGs have been shown to be used by numerous viruses, including both DNA viruses such as herpes simplex viruses and human papillomavirus and RNA viruses such as respiratory syncytial virus (RSV) and HIV (27–38), in the initial attachment to the host cells, we hypothesize that HS and related GAGs act as an early attachment factor to initiate internalization and entry for filoviruses. This conclusion is generally consistent with a recent report that demonstrated a role of HS and heparin in binding and entry of filoviruses (39). Furthermore, we believe that EBOV and MARV may preferentially utilize different but related GAGs as the attachment receptors. This conclusion is based on our observations that knockdown expression of EXT1 had a more pronounced effect on MARV infection than on EBOV infection and that heparin, HS, and the other related GAGs were more effective in blocking infection by MARV than that by EBOV. These facts may reflect the complex nature of filoviruses using GAGs as the attachment receptors during the early stage of infection. Further studies are needed to decipher the subtle distinction in GAG usage as an attachment receptor between MARV and EBOV.
The 2014 West Africa Ebola outbreak underlines the global challenge of treating Ebola (and Marburg) virus infections since there is no clinically approved treatment or vaccine for EBOV and MARV. Thus, it is worthwhile to evaluate the therapeutic potential of heparin, HS, and the other related GAGs on anti-filovirus infection based on the results presented here and reported by another group.
Although it is known that EXT1 is involved in HS synthesis (18) and that expression knockdown of EXT1 can impair the entry of filoviruses, particularly of MARV (described in this study), we observed that compared to HS, heparin shows substantially stronger inhibition for both MARV and EBOV entry. Structurally, HS and heparin share a very similar scaffold, with the main difference being in the quantity of attached sulfate groups (40). Regarding their expression, heparin is expressed sparingly in the body, being found exclusively in mast cells, whereas HS is expressed ubiquitously throughout the body and in the extracellular matrix. This suggests that HS is likely utilized as the attachment receptors for filoviruses due to its availability.
Another interesting aspect of the findings reported here is the potential role of heparin as a therapeutic option against filoviral infections and diseases. It is known that disseminated intravascular coagulation (DIC) is a common occurrence in the human and nonhuman primate filovirus hemorrhagic fever (FHF) cases (27), which may be caused by the tissue factors released due to filoviral infection (41). Low-molecular-weight heparins (LMWHs) are known to prevent blood coagulation and are used routinely in clinical settings as an anticoagulant. Interestingly, there is an anecdotal case study reported in 1975 in which three individuals presented with FHF due to a Marburg virus outbreak in Africa. The primary patient died, revealing extensive intravascular coagulation. Heparin was administered prophylactically to the remaining two patients, who both survived (42). The use of heparin to treat intravascular coagulation is debated, however, and this case study alone is insufficient to make confident conclusions on the extent of the effect of heparin. Nevertheless, it is an intriguing prospect. However, since the results reported here demonstrated that only high-molecular-weight heparin (HMWH), but not LMWH, is effective in blocking MARV GP-mediated viral entry, we speculate that LMWHs and HMWHs may be effective in preventing MARV infection and diseases via different mechanisms. These findings along with the evidence presented above suggest that heparin and related GAGs have the potential to be effective and readily available antivirals for MARV and EBOV infections. Their use as prophylactics for patients in outbreak scenarios or as treatments for those in early stages of disease is an exciting concept and should be pursued in further research.
ACKNOWLEDGMENTS
We thank Gary Cohen for mouse L, Sog9, and Gro2C cell lines and Steven Dudek and Tingting Zhou for HPAECs.
This research was partially supported by National Institutes of Health grants AI059570 and AI77767 to L.R.
REFERENCES
- 1.Feldmann F, Sanchez A, Geisbert JB. 2013. Filoviridae: Marburg and Ebola viruses, p 923–956. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, Honnold S, Bantia S, Kotian P, Chen X, Taubenheim BR, Welch LS, Minning DM, Babu YS, Sheridan WP, Bavari S. 2014. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kortepeter MG, Bausch DG, Bray M. 2011. Basic clinical and laboratory features of filoviral hemorrhagic fever. J Infect Dis 204(Suppl 3):S810–S816. doi: 10.1093/infdis/jir299. [DOI] [PubMed] [Google Scholar]
- 5.Roddy P, Colebunders R, Jeffs B, Palma PP, Van Herp M, Borchert M. 2011. Filovirus hemorrhagic fever outbreak case management: a review of current and future treatment options. J Infect Dis 204(Suppl 3):S791–S795. doi: 10.1093/infdis/jir297. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem 281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 7.Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J Virol 79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manicassamy B, Wang J, Rumschlag E, Tymen S, Volchkova V, Volchkov V, Rong L. 2007. Characterization of Marburg virus glycoprotein in viral entry. Virology 358:79–88. doi: 10.1016/j.virol.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 9.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. 2010. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 6(9):e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Cin PD, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 77:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB Jr, Chiorini J, Maury W. 2011. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci U S A 108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. 2005. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 15.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manicassamy B, Rong L. 2009. Expression of Ebolavirus glycoprotein on the target cells enhances viral entry. Virol J 6:75. doi: 10.1186/1743-422X-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, Rumschlag-Booms E, Wang J, Xiao H, Yu J, Wang J, Guo L, Gao GF, Cao Y, Caffrey M, Rong L. 2009. Analysis of hemagglutinin-mediated entry tropism of H5N1 avian influenza. Virol J 6:39. doi: 10.1186/1743-422X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick C, Leduc Y, Martindale D, Mattison K, Esford LE, Dyer AP, Tufaro F. 1998. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat Genet 19:158–161. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 19.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. 1998. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem 273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Tscherne DM, McCullough C, Caffrey M, Garcia-Sastre A, Rong L. 2012. Residue Y161 of influenza virus hemagglutinin is involved in viral recognition of sialylated complexes from different hosts. J Virol 86:4455–4462. doi: 10.1128/JVI.07187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol 76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker S, Spiess M, Klenk HD. 1995. The asialoglycoprotein receptor is a potential liver-specific receptor for Marburg virus. J Gen Virol 76(Part 2):393–399. [DOI] [PubMed] [Google Scholar]
- 23.Chan SY, Empig CJ, Welte FJ, Speck RF, Schmaljohn A, Kreisberg JF, Goldsmith MA. 2001. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 106:117–126. doi: 10.1016/S0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- 24.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y. 2006. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J Virol 80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takada A, Fujioka K, Tsuiji M, Morikawa A, Higashi N, Ebihara H, Kobasa D, Feldmann H, Irimura T, Kawaoka Y. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol 78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 28.de Boer SM, Kortekaas J, de Haan CA, Rottier PJ, Moormann RJ, Bosch BJ. 2012. Heparan sulfate facilitates Rift Valley fever virus entry into the cell. J Virol 86:13767–13771. doi: 10.1128/JVI.01364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris J, Werling D. 2003. Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response. Cell Microbiol 5:671–680. doi: 10.1046/j.1462-5822.2003.00313.x. [DOI] [PubMed] [Google Scholar]
- 31.Heil ML, Albee A, Strauss JH, Kuhn RJ. 2001. An amino acid substitution in the coding region of the E2 glycoprotein adapts Ross River virus to utilize heparan sulfate as an attachment moiety. J Virol 75:6303–6309. doi: 10.1128/JVI.75.14.6303-6309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson T, Ellard FM, Ghazaleh RA, Brookes SM, Blakemore WE, Corteyn AH, Stuart DI, Newman JW, King AM. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol 70:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letian T, Tianyu Z. 2010. Cellular receptor binding and entry of human papillomavirus. Virol J 7:2. doi: 10.1186/1743-422X-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roderiquez G, Oravecz T, Yanagishita M, Bou-Habib DC, Mostowski H, Norcross MA. 1995. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol 69:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shieh MT, WuDunn D, Montgomery RI, Esko JD, Spear PG. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol 116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smit JM, Waarts BL, Kimata K, Klimstra WB, Bittman R, Wilschut J. 2002. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki forest viruses with liposomes containing lipid-conjugated heparin. J Virol 76:10128–10137. doi: 10.1128/JVI.76.20.10128-10137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spear PG. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol 6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 38.WuDunn D, Spear PG. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol 63:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvador B, Sexton NR, Carrion R Jr, Nunneley J, Patterson JL, Steffen I, Lu K, Muench MO, Lembo D, Simmons G. 2013. Filoviruses utilize glycosaminoglycans for their attachment to target cells. J Virol 87:3295–3304. doi: 10.1128/JVI.01621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gandhi NS, Mancera RL. 2008. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des 72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 41.Geisbert TW, Young HA, Jahrling PB, Davis KJ, Kagan E, Hensley LE. 2003. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J Infect Dis 188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- 42.Gear JS, Cassel GA, Gear AJ, Trappler B, Clausen L, Meyers AM, Kew MC, Bothwell TH, Sher R, Miller GB, Schneider J, Koornhof HJ, Gomperts ED, Isaacson M, Gear JH. 1975. Outbreake of Marburg virus disease in Johannesburg. Br Med J 4:489–493. doi: 10.1136/bmj.4.5995.489. [DOI] [PMC free article] [PubMed] [Google Scholar]