ABSTRACT
Hepatitis C virus (HCV) NS3 is a multifunctional protein composed of a protease domain and a helicase domain linked by a flexible linker. Protease activity is required to generate viral nonstructural (NS) proteins involved in RNA replication. Helicase activity is required for RNA replication, and genetic evidence implicates the helicase domain in virus assembly. Binding of protease inhibitors (PIs) to the protease active site blocks NS3-dependent polyprotein processing but might impact other steps of the virus life cycle. Kinetic analyses of antiviral suppression of cell culture-infectious genotype 1a strain H77S.3 were performed using assays that measure different readouts of the viral life cycle. In addition to the active-site PI telaprevir, we examined an allosteric protease-helicase inhibitor (APHI) that binds a site in the interdomain interface. By measuring nucleotide incorporation into HCV genomes, we found that telaprevir inhibits RNA synthesis as early as 12 h at high but clinically relevant concentrations. Immunoblot analyses showed that NS5B abundance was not reduced until after 12 h, suggesting that telaprevir exerts a direct effect on RNA synthesis. In contrast, the APHI could partially inhibit RNA synthesis, suggesting that the allosteric site is not always available during RNA synthesis. The APHI and active-site PI were both able to block virus assembly soon (<12 h) after drug treatment, suggesting that they rapidly engage with and block a pool of NS3 involved in assembly. In conclusion, PIs and APHIs can block NS3 functions in RNA synthesis and virus assembly, in addition to inhibiting polyprotein processing.
IMPORTANCE The NS3/4A protease of hepatitis C virus (HCV) is an important antiviral target. Currently, three PIs have been approved for therapy of chronic hepatitis C, and several others are in development. NS3-dependent cleavage of the HCV polyprotein is required to generate the mature nonstructural proteins that form the viral replicase. Inhibition of protease activity can block RNA replication by preventing expression of mature replicase components. Like many viral proteins, NS3 is multifunctional, but how PIs affect stages of the HCV life cycle beyond polyprotein processing has not been well studied. Using cell-based assays, we show here that PIs can directly inhibit viral RNA synthesis and also block a late stage in virus assembly/maturation at clinically relevant concentrations.
INTRODUCTION
Chronic infection with the hepatitis C virus (HCV) is a leading cause of end-stage liver disease and hepatocellular carcinoma. HCV is an RNA virus with a cytoplasmic life cycle, and therapies that prevent virus replication can ultimately eradicate the virus from the host, reducing both the risk of development of liver disease and the risk of cancer. The former standard of care for chronic hepatitis C was dual therapy with pegylated alpha interferon and ribavirin, but this was lengthy, poorly tolerated, and effective in only <50% of persons infected with the most common HCV genotypes. Over the past decade, intensive research efforts directed at understanding the HCV life cycle have resulted in the development of small-molecule inhibitors targeting specific viral proteins, including the nonstructural 3 (NS3) protease and the NS5B RNA-dependent RNA polymerase (1). Some of these direct-acting antiviral (DAA) drugs have already been approved for use in therapy, and several other DAAs are in clinical development.
The NS3 protein has emerged as a key target for antiviral drug development. The genome of HCV encodes a single polyprotein that is co- and posttranslationally cleaved into 10 individual proteins by cellular and viral proteases. The HCV NS3 protein, together with its cofactor, NS4A, is a serine protease that is required to cleave the polyprotein at four sites in order to generate viral proteins essential for replication of the RNA genome. In addition, NS3 cleaves the adaptor proteins MAVS (2) and TRIF (3) to block activation of interferon gene expression through the retinoic acid-inducible gene I (RIG-I) and Toll-like receptor 3 (TLR3) pathways. Thus, the NS3 protease is a particularly attractive target for antiviral intervention since its inhibition not only interferes with polyprotein processing but also restores antiviral signaling (4, 5). The first direct-acting antiviral drugs to be approved for the therapy of chronic hepatitis C, boceprevir (6) and telaprevir (7), are both peptidomimetic linear ketoamides that target the active site of the protease domain of NS3. Further development of protease inhibitors (PIs) with macrocycles at either P1-P3 or P2-P4 resulted in improved antiviral potency. Recently, simeprevir (8) became the first macrocyclic PI to be approved for the treatment of chronic hepatitis C in the United States (9). Several other PIs are in clinical development, including more potent, pan-genotypic PIs, such as grazoprevir (10).
Although the protease activity of NS3 has been the focus of drug development efforts, NS3 is a bifunctional enzyme with separate protease and helicase domains connected by a flexible linker. The helicase domain has NTPase and 3′-5′ RNA unwinding activity (11). The ATP-dependent RNA unwinding activity of the NS3 helicase is essential for HCV RNA synthesis (12), and genetic and biochemical studies have implicated the NS3 helicase domain in viral assembly, independently of its role in RNA synthesis (13).
The two domains can be separated and their enzymatic activities can be studied in vitro, but their attachment in the full-length NS3 has been shown to strongly influence their individual properties. For example, the isolated helicase domain preferentially unwinds DNA substrates, but the presence of the protease domain in full-length NS3 can alter substrate selectivity and enhance RNA binding and unwinding (14). Conversely, the helicase domain has been shown to stimulate protease activity in the context of full-length protein (15), and polynucleotides, especially polyuracil, have been shown to stimulate the protease activity of the full-length NS3 but not the activity of the isolated protease (16). A recent mutational analysis has shown that the linker region connecting the two domains is not required for protease or helicase activity but is critical for replication and infectivity (17). These data support a role for the linker either in modulating the conformation of full-length NS3 or in mediating interactions between NS3 and other viral or host proteins during the HCV life cycle.
Recently, a novel class of allosteric protease/helicase inhibitors was identified using fragment-based screening and structure-guided design (18). These inhibitors bind at the interface between the protease and helicase domains of NS3, a region proposed to modulate the activities of the two enzyme domains in vivo (19).
To characterize the mechanism of action of allosteric NS3 inhibitors, a representative compound from this class, AT23708, was compared to the active-site PI telaprevir in multiple assays of antiviral activity that specifically examined different steps in the viral life cycle, including polyprotein synthesis, RNA synthesis, and the intracellular assembly and release of infectious genotype 1a virus H77S.3. These assays not only allowed characterization of the allosteric NS3 inhibitor but also revealed unexpected antiviral activities of the PI telaprevir on NS3-dependent stages of the viral life cycle, in addition to polyprotein processing.
Both classes of inhibitor were able to inhibit a late stage in virus assembly or maturation within 12 h following addition of drug to infected cells. Additionally, RNA synthesis was almost completely blocked by telaprevir and partially blocked by AT23708 within 12 h, a time point when there is little detectable reduction in intracellular NS5B levels. These data show that protease inhibitors targeting the multifunctional NS3 protein have an early effect on RNA synthesis, in addition to polyprotein processing.
MATERIALS AND METHODS
Cell culture and inhibitors.
The Huh7 subclone 2-3c (20) is a cured replicon cell line that is highly permissive for HCV replication and shows very weak RIG-I signaling following challenge with Sendai virus (21). 2-3c cells were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum, 100 U penicillin, 100 U streptomycin, 1 mM l-glutamine, 1× nonessential amino acids, 1 μM vitamin E, 1 mM sodium pyruvate, and 10 mM HEPES buffer, pH 7.4. For virus production assays, the concentration of HEPES buffer, pH 7.4, was increased to 50 mM. AT23708, an allosteric NS3 inhibitor (18), and the active-site PI telaprevir (7) were synthesized at Astex Pharmaceuticals (Cambridge, United Kingdom). The nonnucleoside polymerase inhibitor HCV796 (22) was a gift from Anita Howe (Merck Research Laboratories, Kenilworth, NJ). Stock solutions of telaprevir, AT23708, and HCV796 were prepared in dimethyl sulfoxide (DMSO).
Virus infections and antiviral assays.
H77S.3 is a cell culture-adapted infectious molecular clone of a genotype 1a HCV isolate (23, 24). H77S.3/GLuc2A is a modified H77S.3 isolate that expresses Gaussia luciferase (GLuc) as a fusion with its polyprotein. In this genome, the GLuc sequence, followed by that of a foot-and-mouth disease virus 2A autoprotease, is inserted in frame between the p7 and NS2 sequences of H77S.3. The cleavage events required for production of mature secreted GLuc are independent of HCV protease activity. Plasmids encoding the H77S.3 or H77S.3/GLuc2A genome were linearized by XbaI digestion and transcribed in vitro using a T7 MEGAscript kit (Ambion). RNA products were DNase treated and purified using an RNeasy minikit (Qiagen) and electroporated into Huh7 cells as described previously (25). Electroporated cells were cultured for 7 days to allow HCV replication to reach a steady state before seeding to different plate formats for specific assays of the virus life cycle. GLuc assays of HCV genome replication and measurement of infectious HCV production by a focus-forming-unit (FFU) assay have been described previously (26). To measure the effects of protease inhibitors on cell proliferation and viability, a WST-1 assay was used according to the manufacturer's protocol (Roche).
Western blot analyses of viral protein abundance.
Cells were washed twice in 1× phosphate-buffered saline (PBS), directly lysed in 1× Laemmli sample buffer containing 5% β-mercaptoethanol, and passed through a QIAshredder apparatus (Qiagen) to reduce sample viscosity. Samples corresponding to 1/10 of a 12-well plate were resolved by SDS-PAGE on a 4 to 15% Tris-glycine gradient gel (Bio-Rad), and total protein was transferred to a low-fluorescence polyvinylidene difluoride membrane using a Transblot Turbo transfer system (Bio-Rad) according to the manufacturer's instructions. Blots were probed with primary antibodies against HCV NS5B (rabbit polyclonal antibody; catalog number ab65410; Abcam) and β-actin (mouse monoclonal antibody; catalog number AC-15; Sigma-Aldrich). Secondary antibodies were IRDye 800CW goat anti-mouse IgG and IRDye 680 goat anti-rabbit IgG. Blots were visualized by two-color detection using an Odyssey infrared imaging system, and bands were quantified using Odyssey (v3.0) software (LI-COR, Inc.). For each dose of inhibitor, the abundance of NS5B was normalized to the abundance of β-actin, used as a loading control in the same lane.
Statistical analyses.
To determine the 50% effective concentrations (EC50s) and 90% effective concentrations (EC90s) of telaprevir and AT23708 in the different assays (GLuc, immunoblot quantitation of residual NS5B abundance, FFU, and RNA synthesis assays), data were fit to a four-parameter dose-response curve with variable slope using Prism (v6.0) software for Windows (GraphPad Software, Inc.). Values reported are the estimated concentration ± 95% confidence interval. Maximum achievable inhibitory activities (maximum-effect [Emax] values) at different time points were compared by the Mann-Whitney test using Prism (v6.0) software.
Measurement of RNA synthesis inhibition.
To determine the impact of NS3 inhibitors on viral RNA synthesis, nascent RNA was labeled by incubating HCV-infected cells with 5-ethynyl uridine (EU) from 2 to 12 h after addition of inhibitor. Total RNA was purified from cells using the RNeasy minikit (Qiagen). EU-labeled RNA was conjugated to biotin and isolated from the total RNA using a Click-iT nascent RNA capture kit (Invitrogen). Newly synthesized, EU-labeled HCV RNA and total cell-associated HCV RNA were quantified using HCV-specific primers and a 2-step quantitative reverse transcription-PCR (qRT-PCR) assay described previously (23).
Rate-zonal gradient analyses.
Cells were transfected with H77S.3 as described above and treated for 12 h with either 0.1% DMSO or NS3 inhibitors at concentrations that represent 5× EC90s of those drugs, as measured by the GLuc assay. After drug treatment, cells were harvested by trypsinization, washed twice in PBS, and resuspended in 400 μl PBS. Lysates were prepared by multiple freeze-thaw cycles as described previously (26) and subjected to rate-zonal centrifugation. Clarified lysate (350 μl) was loaded onto a 10 to 50% sucrose gradient prepared in TNE (10 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA) and centrifuged for 1 h at 40,000 rpm in an SW55 Ti rotor at 4°C. For each gradient, 20 fractions were collected from the top of the gradients and analyzed for HCV RNA by qRT-PCR (23) and for infectivity by FFU assay.
RESULTS
Kinetics of antiviral suppression by different classes of inhibitors that target NS3.
As a first step to characterize how inhibitors that bind at different sites on the NS3 protein impact different aspects of the HCV life cycle, the kinetics of antiviral activity were monitored by measuring the Gaussia luciferase (GLuc) activity secreted from cells infected with genotype 1a cell culture-infectious H77S.3/GLuc2A at different time points following treatment with either telaprevir (a linear ketoamide that binds the active site of the NS3 protease domain) or AT23708 (an allosteric protease-helicase inhibitor [APHI] that binds NS3 at the interface of the protease and helicase domains). H77S.3/GLuc2A is a modified version of H77S.3 in which the GLuc-coding sequence has been inserted between p7 and NS2, followed by the foot-and-mouth disease virus 2A protein-coding sequence. When H77S.3/GLuc2A RNA is transfected into Huh7 cells, the RNA can replicate and the GLuc translated from the replicating genomes is secreted into the medium. Secreted GLuc is a measure of polyprotein synthesis, but this correlates well with intracellular RNA levels. Thus, HCV RNA abundance and polyprotein synthesis can be monitored by measuring GLuc activity in medium collected at 12- or 24-h intervals following transfection.
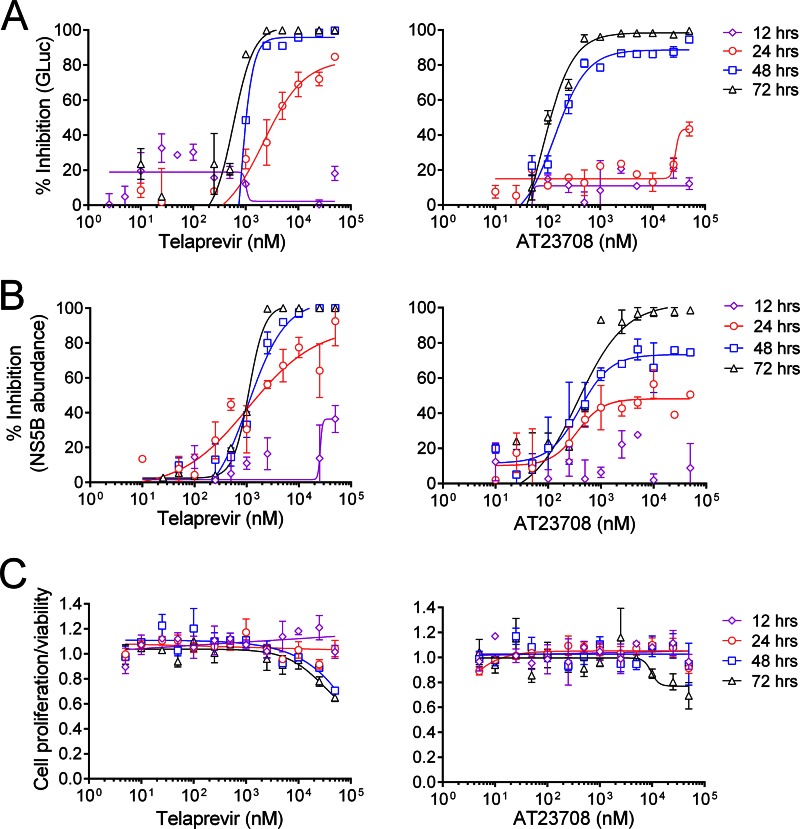
In this assay, telaprevir and AT23708 displayed different kinetics of antiviral suppression (Fig. 1A and Table 1). This difference in kinetics can be seen most easily by comparing the maximum achievable inhibition (Emax) values for both drugs in the GLuc assay at different time points (Table 2). Emax values were significantly higher for telaprevir than AT23708 at both 24 h (81% versus 38%; P = 0.0022) and 48 h (99% versus 90%; P = 0.0022).
FIG 1.
(A) Kinetic analysis of the suppression of H77S.3/GLuc2A virus replication by the active-site protease inhibitor telaprevir or the allosteric NS3 inhibitor AT23708. Huh7 cells were electroporated with H77S.3/GLuc2A RNA genomes and passaged for 1 week before they were challenged with an inhibitor over a range of concentrations. At 12, 24, 48, and 72 h following addition of inhibitor, the cell culture media were harvested and replaced with fresh medium containing inhibitor. The percent inhibition of GLuc activity in cell culture medium is plotted on the y axis, where 0% is the GLuc activity in cells treated with medium containing DMSO vehicle only (no inhibitor) and 100% inhibition is a reduction of GLuc activity to background levels (determined by measurement of GLuc activity in medium from cells transfected with a replication-incompetent RNA genome, H77S-AAG/GLuc2A). The data shown represent the mean ± SEM from 3 replicates of an experiment representative of multiple experiments. Data were fit to dose-response curves as described in Materials and Methods. EC50 and EC90 values for telaprevir and AT23708 in the GLuc assay are shown in Table 1. (B) Reduction of intracellular NS5B abundance following addition of telaprevir or AT23708 to Huh7 cells infected with H77S.3. NS5B and β-actin were detected in cell lysates by Western blotting. NS5B levels were normalized using β-actin as a loading control. Normalized NS5B abundance was expressed as percent inhibition, where 0% is the NS5B abundance in cells treated with the DMSO vehicle only and 100% inhibition is a reduction of NS5B levels to background levels (determined using lysates from uninfected cells). The data shown represent the mean ± SEM percent inhibition from 3 independent experiments. EC50 and EC90 values for telaprevir and AT23708 in the Western blot analyses are shown in Table 3. (C) Effect of telaprevir and AT23708 on cell proliferation and viability measured by a WST-1 cell proliferation assay. The data shown are normalized to the values obtained from cells grown in medium containing 0.1% DMSO vehicle and represent the mean ± SEM from 3 independent experiments.
TABLE 1.
EC50s and EC90s of NS3 inhibitors against H77S.3/GLuc2A virus determined by GLuc assay
| Inhibitor | EC50 (nM) |
EC90 (nM) |
||||||
|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 72 h | 12 h | 24 h | 48 h | 72 h | |
| Telaprevir | >50,000a | 2,180 (1,100–4,317)b | 749 (614–914) | 584 (406–840) | >50,000 | >50,000 | 1,366 (940–1,985) | 1,564 (677–3,612) |
| AT23708 | >50,000 | 50,000 | 889 (525–1,503) | 274 (131–573) | >50,000 | >50,000 | 6,519 (907–46,860) | 1,023 (189.5–5,521) |
The highest concentration tested.
The values in parentheses are 95% confidence intervals.
TABLE 2.
Maximum inhibitory activity (Emax) observed at different time points in different assays for telaprevir and AT23708
| Drug and assay |
Emaxa (%) |
|||
|---|---|---|---|---|
| 12 h | 24 h | 48 h | 72 h | |
| Telaprevir | ||||
| GLuc assay | 16.67 ± 3.08 | 80.83 ± 2.09 | 99.01 ± 0.28 | 99.94 ± 0.02 |
| NS5B reduction by Western blotting | 34.29 ± 6.18 | 84.99 ± 7.61 | 100 ± 0.01 | 100 ± 0.27 |
| Virus production by FFU assay | 100 ± 0.01 | 100 ± 0.01 | 100 ± 0.01 | 100 ± 0.01 |
| AT23708 | ||||
| GLuc assay | 19.16 ± 3.10 | 38.06 ± 3.39 | 89.99 ± 2.87 | 99.54 ± 0.39 |
| NS5B reduction by Western blotting | 27.92 ± 0.97 | 48.39 ± 2.24 | 77.64 ± 1.45 | 100 ± 0.44 |
| Virus production by FFU assay | 99.89 ± 0.07 | 100 ± 0.01 | 99.71 ± 0.26 | 100 ± 0.01 |
Data represent the mean ± standard error of the mean Emax values from ≥3 experiments.
WST-1 assays were performed in parallel to determine the impact of telaprevir and AT23708 on cell viability and proliferation (Fig. 1C). No change in cellular proliferation was observed at 12 or 24 h for either inhibitor. At 48 and 72 h, a modest 20 to 30% reduction in cell viability/proliferation was observed only at the highest concentrations tested (25 or 50 μM) for both inhibitors. The 50% cytotoxic concentrations (CC50s) for both inhibitors were greater than 50 μM, the highest concentration tested.
Reduction of intracellular nonstructural protein levels.
Active-site protease inhibitors such as telaprevir inhibit the NS3-dependent cleavage events that generate the mature NS proteins required for RNA genome replication. AT23708 also inhibits NS3-dependent proteolytic processing in vitro (18). Western blot analyses were used to compare the rates at which each inhibitor reduces intracellular nonstructural protein levels.
Huh7 cells were electroporated with H77S.3 RNA genomes and cultured for 1 week to allow virus replication to stabilize. Infected cells were mock treated (0.1% DMSO) or treated with telaprevir or AT23708 at concentrations ranging from 25 nM to 50 μM for 72, 48, 24, or 12 h prior to harvest, at which time cell lysates were prepared and subjected to SDS-PAGE and Western blotting. Membranes were probed for HCV NS5B or core protein with actin or β-tubulin as a loading control.
The rates of reduction of NS5B protein abundance by both telaprevir and AT23708 were low (Fig. 1B and Table 3). In agreement with the findings of the GLuc assays (Fig. 1A), the rate of reduction of NS5B abundance was slightly higher following addition of telaprevir than following addition of AT23708. As was the case for the GLuc assay, the difference in kinetics can be seen most easily by comparing the Emax values for both drugs in the NS5B Western blot assay at different time points (Table 2). Emax values were significantly higher for telaprevir than AT23708 at both 24 h (85% versus 48%; P = 0.0022) and 48 h (100% versus 78%; P = 0.0159). Importantly, at 12 h after addition of either drug there was little detectable reduction in intracellular NS5B abundance. This observation allowed us to perform further analyses focusing on RNA synthesis and infectious virus production at early time points after addition of drug when protease inhibition is not yet resulting in significant decreases in NS5B abundance.
TABLE 3.
EC50s and EC90s of NS3 inhibitors against H77S.3 virus assessed by determination of NS5B reduction by Western blotting
| Inhibitor | EC50 (nM) |
EC90 (nM) |
||||||
|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 72 h | 12 h | 24 h | 48 h | 72 h | |
| Telaprevir | 25,760 | 1,076 (241–4,803)a | 608 (97–3,823) | 1,099 (908–1,329) | >50,000b | 28,729 (327–50,000) | 1,878 (112–50,000) | 2,366 (1,455–3,846) |
| AT23708 | >50,000 | 351 (120–1,027) | 399 (192–830) | 364 (161–822) | >50,000 | >50,000 | >50,000 | 1,750 (357–8,588) |
The values in parentheses are 95% confidence intervals.
The highest concentration tested.
Inhibition of RNA synthesis by DAAs targeting NS3.
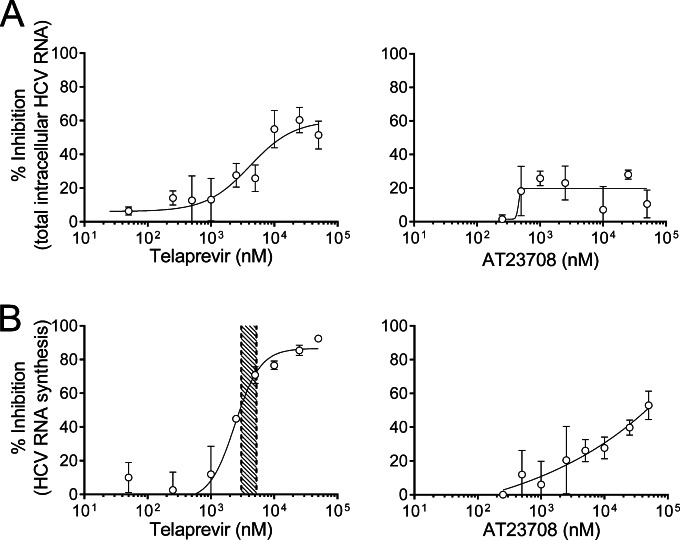
To directly investigate inhibition of RNA synthesis, nascent RNA was labeled by incubating HCV-infected cells with 5-ethynyl uridine (EU) from 2 to 12 h after addition of inhibitor. Total RNA was extracted from the cells, and following isolation of EU-labeled RNA, newly synthesized HCV RNA genomes were quantified by qRT-PCR. Total residual HCV RNA abundance was also quantified by qRT-PCR at 12 h after inhibitor addition (Fig. 2A). The rate of reduction of newly synthesized HCV RNA (Fig. 2B) was higher than that of total residual HCV RNA for both inhibitors, suggesting that they primarily act to reduce new RNA synthesis and do not accelerate degradation of existing viral RNA genomes. Telaprevir was able to almost completely inhibit HCV RNA synthesis (Emax, 92%) within 12 h of drug addition at high but clinically relevant concentrations (Fig. 2B, left). These data suggest that telaprevir can directly interfere with NS3 activity during RNA synthesis independently of its inhibitory activity on proteolytic processing, since at 12 h after addition of drug there was little reduction of intracellular NS5B (Fig. 1B).
FIG 2.
(A) Reduction of total residual HCV RNA in H77S.3-infected Huh7 cells by telaprevir or AT23708 at 12 h. (B) Inhibition of new HCV RNA synthesis in H77S.3-infected Huh7 cells by telaprevir or AT23708 between 2 and 12 h. The shaded area in the left-hand panel represents a previously reported range of telaprevir concentrations (minimum concentration, 3.0 μM; maximum concentration, 5.2 μM) in the plasma of persons treated with 750 mg telaprevir every 8 h (4). The data shown represent the mean ± SEM percent inhibition from 3 experiments and were fit to a dose-response curve, as described in the legend to Fig. 1.
In contrast to telaprevir, AT23708 showed only partial inhibition of HCV RNA synthesis at 12 h after addition (Emax, 53%; Fig. 2B, right). The difference between telaprevir and AT23708 in the early kinetics of RNA synthesis inhibition correlates with the difference in kinetics observed in the GLuc assay and is consistent with a model in which binding of AT23708 to NS3 is conformation dependent (see Discussion).
Inhibition of virus production by DAAs targeting NS3.
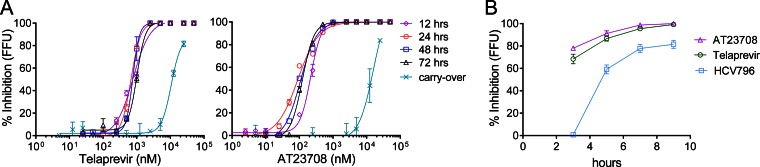
To compare the inhibition of virus production by the active-site PI and allosteric NS3 inhibitor, an FFU assay was used to measure the amount of infectious virus produced from H77S.3-infected cells at different 12- to 24-h intervals following addition of the inhibitors. Inhibition of virus production was rapid and occurred sooner than inhibition of RNA genome replication for both telaprevir and AT23708 (Fig. 3A and Table 4). The difference between the timing of inhibition of RNA synthesis and the timing of virus production was particularly striking for AT23708. Complete inhibition of virus production was observed with AT23708 by 12 h after addition, a time point when there was incomplete inhibition of RNA synthesis (Fig. 2) and no reduction in intracellular nonstructural protein levels (Fig. 1B). Telaprevir was also able to block virus production at 12 h after addition (Fig. 3), but at this time point, telaprevir could also inhibit RNA synthesis (Fig. 2B). Several further lines of evidence suggest that telaprevir has an additional effect on virus production. First, the EC50 at 12 h in the virus production (FFU) assay was 777 nM (Table 4), whereas it was 2,400 nM in the RNA synthesis assay (Fig. 2). Furthermore, at very early time points following addition of inhibitor at 5 times the EC90, both telaprevir and AT23708 were able to block virus production faster than the RNA-dependent RNA polymerase inhibitor HCV796 (Fig. 3B). These data suggest that telaprevir is also able to inhibit virus production, in addition to blocking RNA synthesis and polyprotein processing. When the results of the different assays were compared, Emax values (Table 2) were significantly greater when they were measured in the virus production assay than when they were measured in the GLuc or NS5B Western blot assays for telaprevir at 12 and 24 h and for AT23708 at 12, 24, and 48 h (P < 0.01 in every case).
FIG 3.
Rapid inhibition of virus production by telaprevir and AT23708. (A) Reductions in infectious virus released into supernatants between 0 and 12, 12 and 24, 24 and 48, and 48 and 72 h after infected cell cultures were challenged with a range of concentrations of telaprevir or AT23708. Infectious virus release was quantified by the FFU assay. The reduction in virus infectivity caused by residual inhibitor present in the inoculum was measured and is plotted as the carryover. Results are shown as percent inhibition, where 0% is the number of FFU per milliliter released from infected cells grown in the absence of inhibitor (DMSO vehicle only) and 100% inhibition is a reduction of the level of virus production to below the limit of detection (<10 FFU/ml). The data shown represent the mean ± SEM from 2 to 3 independent experiments and were fit to a dose-response curve, as described in the legend to Fig. 1. EC50 and EC90 values for telaprevir and AT23708 in the FFU assay are shown in Table 4. (B) Very early inhibition of virus production by telaprevir and AT23708 compared to that by the nonnucleoside RNA-dependent RNA polymerase inhibitor HCV796. Inhibitors were added at 2-h intervals to H77S.3-infected cells at concentrations equivalent to 5× EC90 in the 72 h GLuc assay. At 6 h, medium was removed from all cultures and replaced with fresh medium containing the inhibitors. Supernatants were harvested 3 h later, and infectious virus was quantified by the FFU assay.
TABLE 4.
EC50s and EC90s of NS3 inhibitors against H77S.3 virus determined by FFU assay
| Inhibitor | EC50 (nM) |
EC90 (nM) |
||||||
|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 72 h | 12 h | 24 h | 48 h | 72 h | |
| Telaprevir | 777 (686–880)a | 677 (626–733) | 765 (695–843) | 738 (566–962) | 2,778 (2,085–3,701) | 1,389 (1,180–1,635) | 1,255 (1,080–1,458) | 2,493 (1,333–4,664) |
| AT23708 | 218 (200–237) | 91 (81–101) | 108 (97–121) | 123 (98–105) | 491 (413–583) | 428 (328–558) | 364 (285–466) | 294 (253–342) |
The values in parentheses are 95% confidence intervals.
A potential caveat in this experiment is that infectious virus production is measured in medium containing inhibitor. In the virus production assay, H77S.3-infected cells were treated with different concentrations of protease inhibitor and medium was harvested at various times after treatment. Virus released into the medium was used to inoculate naive cells for 4 h before the inoculum was washed off and the medium was replaced with fresh medium. After a further 3 days of culture, inoculated cells were stained for core protein to quantify infectious foci. Although the inoculum was washed out in this experimental design, it is possible that residual amounts of inhibitor were being carried over during the 4-h inoculation period and blocking cleavage of polyprotein translated from newly uncoated viral RNA following entry. To determine the contribution of drug carryover in this assay, the virus inoculum produced from cells grown in the absence of drug was spiked with an equal volume of conditioned cell culture medium from mock-infected cells treated for 12 h with a range of inhibitor concentrations, this mixture was used to inoculate naive cells for 4 h, and virus infectivity was assessed by FFU assay (Fig. 3A, carryover). Inhibition of virus infectivity by drug carryover was observed only at concentrations of 12.5 μM or higher for both telaprevir and AT23708, concentrations approximately 20- or 100-fold higher, respectively, than the concentrations required to block virus production. These data confirm that the rapid decrease in the number of FFU following protease inhibitor treatment is a genuine effect on virus production and not simply due to the inhibition of virus infectivity caused by drug carryover.
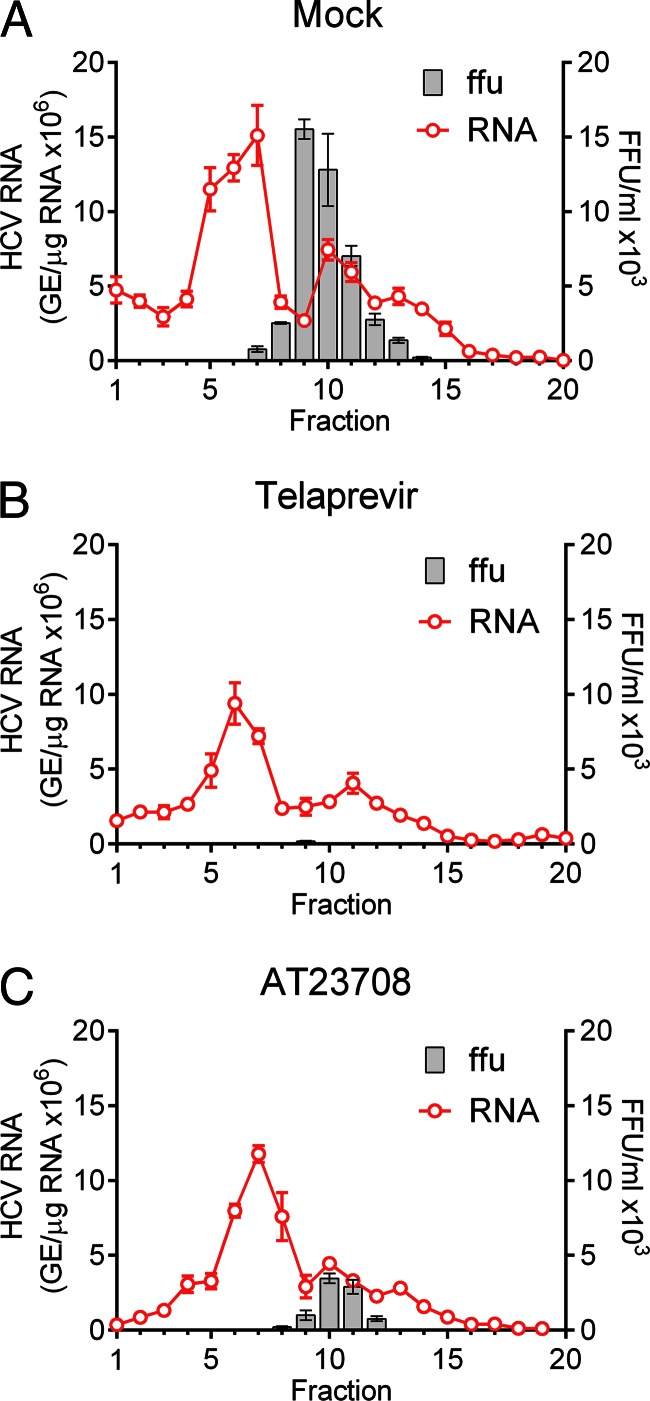
During the assembly of HCV, NS3 is recruited to lipid droplets along with the NS5A protein (27). Our previous studies suggested that production of HCV particles is a dynamic process that can be rapidly inhibited by DAAs that target viral proteins involved in assembly, such as NS5A (26). Whether inhibitors that target NS3 impact viral assembly or maturation is not known, but genetic and biochemical studies have suggested a role for the helicase domain of NS3 in virus assembly (13). Thus, it is possible that inhibitors targeting NS3 could have a direct effect on intracellular assembly. To assess this, infected cells were treated with a high concentration (5 μM; >5× EC90 in the GLuc assay at 72 h) of telaprevir or AT23708 for 12 h. The cells were then lysed by repeated freeze-thaw cycles in the absence of detergent, and intracellular viral particles were separated by rate-zonal ultracentrifugation through a sucrose gradient. Fractions collected from the top of the gradient were analyzed by qRT-PCR to determine the HCV genomic RNA content and the FFU assay to determine the infectious virus content (Fig. 4). Although the total amount of viral RNA in lysates from inhibitor-treated cells was reduced by 40 to 45% compared with that in lysates from cells treated only with DMSO, the presence of discrete peaks of HCV RNA in fractions 7 and 10 of the gradients suggested the continued assembly of RNA-containing particles in the presence of inhibitor. Infectious virus was clearly present and sedimenting with the second RNA peak in fractions 9 and 10 of the gradient loaded with lysate from mock-treated cells. Importantly, however, there was a sharp reduction in the amount of infectious virus in the fractions from cells treated with AT23708 or telaprevir.
FIG 4.
Rate-zonal centrifugation of cell lysates derived from H77S.3-transfected cells that were either mock treated (A) or treated with 5 μM telaprevir (B) or 5 μM AT23708 (C) for 12 h. Fractions collected from the top of the gradients were tested for HCV RNA by qRT-PCR and for infectivity by the FFU assay.
Collectively, the data suggest that NS3 inhibitors block a late step in the intracellular assembly and/or maturation of virus, resulting in the early and nearly complete inhibition of production of infectious virus.
DISCUSSION
In this study, the active-site PI telaprevir was compared with a novel allosteric NS3 inhibitor in cell-based assays that focus on different aspects of the HCV life cycle. Many viral proteins are multifunctional, and DAAs may hit one or multiple functions of their target protein. The use of multiple assays to probe the HCV life cycle in detail can help provide an understanding of how DAAs interfere with the life cycle and illuminate the role of specific proteins in virus replication.
Protease inhibitors can directly interfere with RNA synthesis.
In addition to blocking the NS3-dependent cleavage of the HCV polyprotein, the active-site PI telaprevir was found to inhibit new RNA synthesis and virus production prior to egress at a late stage during assembly or particle maturation. Since the HCV RNA-dependent RNA polymerase NS5B is generated by NS3-dependent cleavage of the polyprotein, inhibition of RNA synthesis by a PI such as telaprevir could be a consequence of depletion of intracellular NS5B. However, inhibition of RNA synthesis by telaprevir occurred at 2 to 12 h after addition of the drug, a time point when there is little reduction in NS5B abundance, suggesting that telaprevir can directly inhibit RNA synthesis. This activity of an active-site PI is unexpected but is consistent with a role for the protease domain during RNA synthesis. Previous biochemical studies have shown that the protease domain of NS3 is required for efficient unwinding of double-stranded RNA (dsRNA) by the helicase (28), and recently, Aydin et al. (19) proposed a model in which the protease domain of NS3 binds to dsRNA as a clamp while the helicase unwinds the double-stranded template to allow access by NS5B to the negative strand for RNA synthesis. It is tempting to speculate that binding of telaprevir to the protease domain may inhibit RNA synthesis by reducing helicase activity, but a previous study has reported that telaprevir does not inhibit the helicase activity of full-length NS3 in vitro (29). In vivo, however, NS3 functions as part of a multiprotein replicase complex where binding of telaprevir may result in disruption of protein-protein and protein-RNA interactions to block a function of NS3 in RNA synthesis. An alternative explanation is that the majority of new RNA synthesis is performed by newly synthesized NS5B. Further experiments will be required to test these hypotheses.
The APHI AT23708, already known to act as an inhibitor of polyprotein processing, was also able to inhibit RNA synthesis at early time points following addition of drug, but unlike the inhibition caused by telaprevir, the inhibition was only partial. This difference may be explained by the conformation-dependent binding of the APHI to NS3, in which the APHI targets a site in the interdomain interface that is present in the compact crystallographic conformation of NS3 but not in the extended conformation, which biochemical studies suggest represents the active conformation during RNA synthesis (30). Partial inhibition of RNA synthesis by AT23708 is consistent with NS3 adopting the extended conformation at least some of the time during RNA synthesis.
Protease inhibitors can interfere with virus assembly/maturation.
Both telaprevir and AT23708 were able to rapidly inhibit virus production from infected cells. Inhibition occurred at early time points (12 h after drug addition), when there was little reduction of residual HCV RNA in the cells and no reduction in intracellular levels of nonstructural proteins. Analyses at very early time points showed the onset of inhibition by 3 h following addition of drug and 90% inhibition by 9 h. The different kinetics of inhibition of virus assembly compared to the kinetics of inhibition of RNA synthesis and reduction of NS5B abundance suggest that (i) assembly is a highly dynamic process and particles are exported rapidly from the cell and (ii) the pool of NS3 involved in virus assembly is distinct from the pool of NS3 involved in RNA synthesis and polyprotein processing.
Our data show that inhibitors targeting NS3 at either the active site or an allosteric site in the interdomain interface can block virus production at a late step during assembly/maturation of the virus particles. While this specific effect of NS3/4A inhibitors has not been demonstrated previously, it is not surprising, as NS3 is recruited to lipid droplets together with NS5A and appears to function directly in viral assembly (13, 27). Some variants associated with resistance to NS3/4A inhibitors also impair viral fitness by negatively impacting steps involved in the release of infectious virus (23). Our data are in agreement with those from a previous study (31) that focused on daclatasvir but also included data obtained using mathematical modeling analyses of rates of viral RNA decline in patient serum following telaprevir treatment suggesting that telaprevir can block virus production. Those modeling analyses predicted that telaprevir can inhibit virus assembly/secretion more effectively than polymerase inhibitors but that this effect is weaker than that for NS5A inhibitors, such as daclatasvir.
How might inhibitors targeting NS3 block assembly? It is possible that binding of telaprevir to the active site or AT23708 to the allosteric site inhibits virus release by interfering with an activity of the protease in viral assembly/release. NS3 is a multifunctional protein, and the activity of its helicase is regulated through its protease-helicase domain interface (19). The binding of a small molecule to the protease domain or at the interdomain interface might thus alter helicase activity during these final steps in the viral life cycle. The interaction of inhibitors with NS3 may prevent an interaction of the helicase domain with core protein, which genetic (32) and biochemical (33) studies suggest is required for particle assembly. The effect of telaprevir and AT23708 on viral release could also occur indirectly via NS5A, as NS5A hyperphosphorylation is dependent upon NS3 protease activity (34), and its absence might affect NS5A functions involved in viral assembly and release. Further experiments will be required to distinguish between these possibilities.
The protease activity of NS3/4A is responsible for blunting RIG-I-dependent host innate immune responses through cleavage of the signaling adaptor molecule MAVS (2). Two previous studies have demonstrated that small-molecule inhibition of NS3 can restore host cell innate immune signaling by preventing the cleavage of MAVS (4, 5). Kinetic analyses demonstrated that restoration of signaling was faster with a protease inhibitor (2 to 4 days, depending upon the concentration) than with a polymerase inhibitor (7 days) (4). Importantly, the rapid effects (<12 h) on virus production and RNA synthesis observed in the present study precede (and are thus likely to be independent of) the restoration of innate immune signaling observed in previous studies. PI-mediated restoration of MAVS signaling may be slower than direct PI effects on the viral life cycle because it requires several events, including synthesis of a new MAVS protein, trafficking of MAVS and accumulation at the mitochondrion-associated membranes (35), interaction with RIG-I molecules engaged with the HCV pathogen-associated molecular pattern, and subsequent downstream signaling to promote the expression of antiviral genes (36).
The HCV genome encodes only 10 mature proteins, and many of these proteins have to be multifunctional to perform all of the activities required to complete the viral life cycle. For example, NS5A is involved in the assembly of new replicase complexes but is also required for virion assembly, and inhibitors targeting NS5A can block both of these processes (26, 31, 37). NS3 has been termed the Swiss Army knife of HCV because of its multiple roles in the viral life cycle (38). The current study has provided novel insight into how NS3 inhibitors impact specific steps in the viral life cycle and demonstrates that their mechanism of action is more complex than simply blocking NS3-dependent polyprotein cleavage to prevent replicase complex formation.
ACKNOWLEDGMENTS
We thank Zongdi Feng and Asuka Hirai-Yuki for expert advice on rate-zonal centrifugation analyses and Stanley Lemon for helpful discussions.
D.R.M. was funded by a research grant from Astex Pharmaceuticals and the University Cancer Research Fund of the University of North Carolina at Chapel Hill.
REFERENCES
- 1.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalkeri G, Lin C, Gopilan J, Sloan K, Rijnbrand R, Kwong AD. 2013. Restoration of the activated Rig-I pathway in hepatitis C virus (HCV) replicon cells by HCV protease, polymerase, and NS5A inhibitors in vitro at clinically relevant concentrations. Antimicrob Agents Chemother 57:4417–4426. doi: 10.1128/AAC.00399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang Y, Ishida H, Lenz O, Lin TI, Nyanguile O, Simmen K, Pyles RB, Bourne N, Yi M, Li K, Lemon SM. 2008. Antiviral suppression vs restoration of RIG-I signaling by hepatitis C protease and polymerase inhibitors. Gastroenterology 135:1710–1718.e1712. doi: 10.1053/j.gastro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Malcolm BA, Liu R, Lahser F, Agrawal S, Belanger B, Butkiewicz N, Chase R, Gheyas F, Hart A, Hesk D, Ingravallo P, Jiang C, Kong R, Lu J, Pichardo J, Prongay A, Skelton A, Tong X, Venkatraman S, Xia E, Girijavallabhan V, Njoroge FG. 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother 50:1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin K, Perni RB, Kwong AD, Lin C. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob Agents Chemother 50:1813–1822. doi: 10.1128/AAC.50.5.1813-1822.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin TI, Lenz O, Fanning G, Verbinnen T, Delouvroy F, Scholliers A, Vermeiren K, Rosenquist A, Edlund M, Samuelsson B, Vrang L, de Kock H, Wigerinck P, Raboisson P, Simmen K. 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother 53:1377–1385. doi: 10.1128/AAC.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenquist A, Samuelsson B, Johansson PO, Cummings MD, Lenz O, Raboisson P, Simmen K, Vendeville S, de Kock H, Nilsson M, Horvath A, Kalmeijer R, de la Rosa G, Beumont-Mauviel M. 2014. Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem 57:1673–1693. doi: 10.1021/jm401507s. [DOI] [PubMed] [Google Scholar]
- 10.Summa V, Ludmerer SW, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, Dimuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2012. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167. doi: 10.1128/AAC.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang PS, Jankowsky E, Planet PJ, Pyle AM. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J 21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam AM, Frick DN. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J Virol 80:404–411. doi: 10.1128/JVI.80.1.404-411.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol 82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beran RK, Serebrov V, Pyle AM. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem 282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 15.Beran RK, Pyle AM. 2008. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J Biol Chem 283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgenstern KA, Landro JA, Hsiao K, Lin C, Gu Y, Su MS, Thomson JA. 1997. Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of a hepatitis C virus NS3-NS4A complex isolated from transfected COS cells. J Virol 71:3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlway A, Pirakitikulr N, Ding SC, Yang F, Luo D, Lindenbach BD, Pyle AM. 2014. The linker region of NS3 plays a critical role in the replication and infectivity of hepatitis C virus. J Virol 88:10970–10974. doi: 10.1128/JVI.00745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saalau-Bethell SM, Woodhead AJ, Chessari G, Carr MG, Coyle J, Graham B, Hiscock SD, Murray CW, Pathuri P, Rich SJ, Richardson CJ, Williams PA, Jhoti H. 2012. Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function. Nat Chem Biol 8:920–925. doi: 10.1038/nchembio.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aydin C, Mukherjee S, Hanson AM, Frick DN, Schiffer CA. 2013. The interdomain interface in bifunctional enzyme protein 3/4A (NS3/4A) regulates protease and helicase activities. Protein Sci 22:1786–1798. doi: 10.1002/pro.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J Virol 78:1513–1524. doi: 10.1128/JVI.78.3.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K, Chen Z, Kato N, Gale M Jr, Lemon SM. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem 280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 22.Kneteman NM, Howe AY, Gao T, Lewis J, Pevear D, Lund G, Douglas D, Mercer DF, Tyrrell DL, Immermann F, Chaudhary I, Speth J, Villano SA, O'Connell J, Collett M. 2009. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752. doi: 10.1002/hep.22717. [DOI] [PubMed] [Google Scholar]
- 23.Shimakami T, Welsch C, Yamane D, McGivern DR, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi M, Hu F, Joyce M, Saxena V, Welsch C, Chavez D, Guerra B, Yamane D, Veselenak R, Pyles R, Walker CM, Tyrrell L, Bourne N, Lanford RE, Lemon SM. 2014. Evolution of a cell culture-derived genotype 1a hepatitis C virus (H77S.2) during persistent infection with chronic hepatitis in a chimpanzee. J Virol 88:3678–3694. doi: 10.1128/JVI.03540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. 2011. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol 85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, Lemon SM. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147:453–462.e457. doi: 10.1053/j.gastro.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 28.Frick DN, Rypma RS, Lam AM, Gu B. 2004. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem 279:1269–1280. doi: 10.1074/jbc.M310630200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndjomou J, Kolli R, Mukherjee S, Shadrick WR, Hanson AM, Sweeney NL, Bartczak D, Li K, Frankowski KJ, Schoenen FJ, Frick DN. 2012. Fluorescent primuline derivatives inhibit hepatitis C virus NS3-catalyzed RNA unwinding, peptide hydrolysis and viral replicase formation. Antiviral Res 96:245–255. doi: 10.1016/j.antiviral.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding SC, Kohlway AS, Pyle AM. 2011. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J Virol 85:4343–4353. doi: 10.1128/JVI.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A 110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones DM, Atoom AM, Zhang X, Kottilil S, Russell RS. 2011. A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J Virol 85:12351–12361. doi: 10.1128/JVI.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mousseau G, Kota S, Takahashi V, Frick DN, Strosberg AD. 2011. Dimerization-driven interaction of hepatitis C virus core protein with NS3 helicase. J Gen Virol 92:101–111. doi: 10.1099/vir.0.023325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neddermann P, Clementi A, De Francesco R. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol 73:9984–9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horner SM, Liu HM, Park HS, Briley J, Gale M Jr. 2011. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A 108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horner SM, Gale M Jr. 2013. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med 19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger C, Romero-Brey I, Radujkovic D, Terreux R, Zayas M, Paul D, Harak C, Hoppe S, Gao M, Penin F, Lohmann V, Bartenschlager R. 2014. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology 147:1094–1105.e25. doi: 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. 2011. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J Viral Hepat 18:305–315. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]