ABSTRACT
Herpesvirus nascent capsids, after assembly in the nucleus, must acquire a variety of tegument proteins during maturation. However, little is known about the identity of the tegument proteins that are associated with capsids in the nucleus or the molecular mechanisms involved in the nuclear egress of capsids into the cytoplasm, especially for the two human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), due to a lack of efficient lytic replication systems. Murine gammaherpesvirus 68 (MHV-68) is genetically related to human gammaherpesviruses and serves as an excellent model to study the de novo lytic replication of gammaherpesviruses. We have previously shown that open reading frame 33 (ORF33) of MHV-68 is a tegument protein of mature virions and is essential for virion assembly and egress. However, it remains unclear how ORF33 is incorporated into virions. In this study, we first show that the endogenous ORF33 protein colocalizes with capsid proteins at discrete areas in the nucleus during viral infection. Cosedimentation analysis as well as an immunoprecipitation assay demonstrated that ORF33 is associated with both nuclear and cytoplasmic capsids. An immunogold labeling experiment using an anti-ORF33 monoclonal antibody revealed that ORF33-rich areas in the nucleus are surrounded by immature capsids. Moreover, ORF33 is associated with nucleocapsids prior to primary envelopment as well as with mature virions in the cytoplasm. Finally, we show that ORF33 interacts with two capsid proteins, suggesting that nucleocapsids may interact with ORF33 in a direct manner. In summary, we identified ORF33 to be a tegument protein that is associated with intranuclear capsids prior to primary envelopment, likely through interacting with capsid proteins in a direct manner.
IMPORTANCE Morphogenesis is an essential step in virus propagation that leads to the generation of progeny virions. For herpesviruses, this is a complicated process that starts in the nucleus. Although the process of capsid assembly and genome packaging is relatively well understood, how capsids acquire tegument (the layer between the capsid and the envelope in a herpesvirus virion) and whether the initial tegumentation process takes place in the nucleus remain unclear. We previously showed that ORF33 of MHV-68 is a tegument protein and functions in both the nuclear egress of capsids and final virion maturation in the cytoplasm. In the present study, we show that ORF33 is associated with intranuclear capsids prior to primary envelopment and identify novel interactions between ORF33 and two capsid proteins. Our work provides new insights into the association between tegument proteins and nucleocapsids at an early stage of the virion maturation process for herpesviruses.
INTRODUCTION
The family Herpesviridae consists of three subfamilies, i.e., the Alphaherpesvirinae, the Betaherpesvirinae, and the Gammaherpesvirinae. All herpesvirus virions share a structure: an icosahedral capsid shell containing a double-stranded DNA genome is surrounded by a tegument layer, which is encased by a lipid bilayer spiked with viral glycoproteins (1–3). Herpesvirus virion morphogenesis is an ordered process. According to the widely accepted envelopment-deenvelopment-reenvelopment model, nucleocapsids, after assembly in the nucleus, undergo a process of primary envelopment at the inner nuclear membrane, resulting in the formation of primary virions in the perinuclear space. The primary envelope then fuses with the outer nuclear membrane, releasing capsids into the cytoplasm. Final envelopment and the formation of mature virions are believed to occur at trans-Golgi network-derived vesicles (4, 5).
The tegument is a unique structure of herpesvirus particles. An increasing amount of knowledge gained from viral genetics studies, biochemical analysis, as well as electron microscopy (EM) has indicated that tegument proteins play critical roles in virion morphogenesis in all herpesvirus subfamilies (6–12). At present, knowledge about how capsids acquire specific tegument proteins and the molecular mechanisms of tegument protein involvement in the virion maturation process has largely come from studies on alphaherpesviruses, including herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV) (9). Tegument addition is generally thought to begin with primary envelopment at the inner nuclear membrane, where capsids acquire a primary tegument (13). A final set of tegument proteins is added in the cytoplasm following nuclear egress and finally at the trans-Golgi network-derived vesicles (5). Compared to the large number of tegument proteins that are recruited onto capsids in the cytoplasm following nuclear egress or at the stage of secondary envelopment, little tegument is found in primary virions in the perinuclear space (5, 13, 14). Furthermore, specific tegument proteins have been shown to concentrate in areas called “assemblons,” formed during capsid assembly in the nucleus. However, whether binding of tegument protein to capsids occurs at these areas is still a mystery (15–17).
The process of virion morphogenesis, especially tegumentation, of gammaherpesviruses is poorly understood, largely because the productive replication of the two human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) is limited in cultured cells. Murine gammaherpesvirus 68 (MHV-68) is genetically related to these two human gammaherpesviruses, on the basis of their genomic sequences and other biological properties (18, 19). The ability to initiate and complete efficient productive infections in fibroblast and epithelial cell lines makes MHV-68 an excellent model to study the lytic replication of gammaherpesviruses. By taking advantage of its genome cloned as a bacterial artificial chromosome (BAC) (20, 21), the roles of several MHV-68 tegument proteins in virus lytic replication have been revealed using a genetic approach (8, 12, 22–24).
MHV-68 open reading frame 33 (ORF33) encodes a tegument protein and is expressed with true late kinetics (8). By construction and analyses of an ORF33-null mutant, we show that ORF33 is essential for viral lytic replication in cultured cells. However, viral genome replication, viral gene expression, and capsid assembly are not affected in the absence of ORF33. Based on an analysis of thin sections obtained from cells infected by an ORF33-null mutant under a transmission electron microscope (TEM), we subsequently found that the nuclear egress of nucleocapsids into the cytoplasm is markedly inhibited. Maturation of virions is finally arrested at a cytoplasmic stage of partially tegumented nucleocapsids (8). The prominent role of ORF33 in the translocation of capsids from the nucleus to the cytoplasm prompted us to examine whether this protein is associated with capsids in the nucleus since, until now, the limited number of tegument proteins or glycoproteins of alphaherpesviruses associated with nuclear capsids have all been found to function in this process (14, 25).
In this study, we took multiple approaches and demonstrate that ORF33 is accumulated in the nucleus at discrete areas and is associated with capsids in the nucleus prior to primary envelopment. Furthermore, we identify novel interactions between ORF33 and two capsid proteins.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells, Vero cells, and 293T cells were cultured in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Wide-type (WT) MHV-68 was originally obtained from Ren Sun (University of California, Los Angeles). The working stock was generated by infecting BHK-21 cells at a multiplicity of infection (MOI) of 0.03. To infect BHK-21 cells or Vero cells, the viral inoculum in DMEM was incubated with cells for 1 h with occasional swirling. The inoculum was then removed and replaced with fresh DMEM plus 10% fetal bovine serum. The titer of virus was measured by a plaque assay in BHK-21 cells as previously described (8). The 33Stop BAC was described previously (8).
Plasmid construction.
Plasmid pHA-33 was described previously (8). All plasmids with a 3× FLAG-CBP tag (Tag-25N, Tag-25C, Tag-26, Tag-43, and Tag-33) used in this study were gifts from Ren Sun (University of California, Los Angeles). Plasmids pHA-62 and pHA-65 were constructed by inserting the coding sequence of ORF62 and ORF65 into the EcoRI and XhoI sites of the pCMV-HA vector (Clontech), respectively. The sequences of the primers used for plasmid construction are available upon request.
Separation and analyses of capsids.
The nucleus and cytoplasm of MHV-68-infected BHK-21 cells were isolated using a CelLytic NuCLEAR extraction kit (product code NXTRACT; Sigma-Aldrich) according to the instructions in the manual accompanying the kit, with some modifications. Briefly, eight 15-cm plates of confluent BHK-21 cells were infected at an MOI of 5. At 12 h postinfection, the cells were digested with trypsin, collected by centrifugation at 1,000 × g for 5 min, resuspended in 6 ml of lysis buffer (containing dithiothreitol [DTT] and protease inhibitors; product code L9036; Sigma-Aldrich), and incubated on ice for 5 min. Then, 10% Igepal CA-630 solution was added to the swollen cells to a final concentration of 0.1%, and the mixture was vortexed vigorously for 10 s. The cytoplasmic fraction was separated from the nuclei by centrifugation at 1,000 × g for 10 min. The nuclei were washed three times and resuspended in 0.5 ml of extraction buffer (product code E2525; Sigma-Aldrich) containing DTT and protease inhibitors. Capsids were released from the purified nuclei by freezing (80°C) and then thawing (37°C) three times. Insoluble materials from the nuclear and cytoplasmic fractions were cleared by centrifugation at 8,000 × g for 30 min. The capsids remaining in the soluble supernatant of the nuclear and cytoplasmic fractions were pelleted through a 1.7-ml 30% (wt/vol) sucrose cushion in TNE buffer (20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA) by centrifugation at 83,500 × g for 1 h in a P40T rotor (Hitachi). The pellets were then resuspended in 500 μl of TNE buffer, sonicated for 2 min at moderate power, and layered onto a discontinuous sucrose density gradient consisting of 20 to 45% (wt/vol) sucrose in TNE buffer. The gradients were then centrifuged at 74,000 × g for 1 h in a P40T rotor (Hitachi). All centrifugation steps were carried out at 4°C. Fractions of 850 μl each were collected from the top of the gradient. A total of 14 fractions, named fractions 1 to 14, were obtained from the top to the bottom. Trichloroacetic acid (TCA) was added to a final concentration of 13%, and the samples were incubated overnight at 4°C. The precipitated proteins were collected by centrifugation at 18,000 × g for 30 min, washed with 100% ethanol, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for Western blotting.
Immunoprecipitation of capsids and trypsin treatment assay.
The nucleus and cytoplasm of MHV-68-infected BHK-21 cells were isolated as described above. ORF33 and its associated capsids were immunoprecipitated with a mouse monoclonal antibody against ORF33 or mouse IgG as a control. For trypsin treatment, a portion of the immunoprecipitates in each fraction was diluted into 200 μl of trypsin buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM CaCl2) with 1% Triton X-100 supplementation, trypsin was added to a final concentration of 4 μg/ml, and the mixture was incubated at 37°C for 1 h. The reaction was stopped by adding 0.5 mM phenylmethylsulfonyl fluoride and protease inhibitors. Both immunoprecipitates and trypsin-treated samples were separated by electrophoresis in SDS–10% polyacrylamide gels. The gels were then electrotransferred onto nitrocellulose membranes and subjected to immunoblotting analysis.
Antibodies, immunoprecipitation, and immunoblotting.
Bacterially expressed and purified His-ORF33 protein was provided to the Experimental Animal Center, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, to generate mouse monoclonal antibodies against MHV-68 ORF33. Rabbit polyclonal anti-ORF26 and anti-ORF65 antibodies were kind gifts from Ren Sun (University of California, Los Angeles). Mouse monoclonal antibody M2 or rabbit polyclonal antibody against a FLAG epitope and mouse monoclonal antibody or polyclonal antibody against a hemagglutinin (HA) epitope were purchased from Sigma. 293T cells seeded onto a 6-cm plate were transfected with 7.5 μg of total DNA. At 48 h after transfection, cells were washed once with ice-cold phosphate-buffered saline (PBS) and then solubilized in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1 mM EDTA) containing a protease inhibitor cocktail. Lysates were cleared by centrifugation for 15 min at 16,000 × g. Ten percent of the supernatant was used as an input control. Soluble proteins were mixed with 20 μl anti-FLAG M2 agarose (Sigma), and the mixture was rotated at 4°C for 4 h. The beads were washed three times with lysis buffer before use. Immune complexes were washed five times with lysis buffer, and supernatant was depleted. Bound proteins were then subjected to immunoblotting analysis. For immunoblotting, samples were heated to 100°C and subjected to SDS-PAGE. Proteins on gels were transferred onto a polyvinylidene difluoride membrane (Millipore) and incubated sequentially with appropriate primary antibody and secondary antibody (anti-rabbit or anti-mouse immunoglobulin G conjugated with horseradish peroxidase), and the proteins were detected by use of an enhanced chemiluminescence system (Millipore).
Indirect immunofluorescence assay (IFA).
A total of 1 × 105 Vero cells were seeded on glass coverslips in a 24-well plate. Cells were infected with MHV-68 at an MOI of 3 and were fixed at 12 h postinfection (hpi) or 24 hpi. Formaldehyde fixation was carried out at 4°C for 15 min with freshly made 4% formaldehyde in PBS. After washing with PBS, formaldehyde-fixed samples were permeabilized with 0.2% Triton X-100 dissolved in PBS at 4°C for 5 min. The cells were washed again with PBS and incubated in 5% normal goat serum dissolved in PBS for 30 min at room temperature (RT). Mouse anti-ORF33 antibody and rabbit anti-ORF65 and anti-ORF26 antibodies were applied as indicated below and incubated for 1 h at RT. Primary antibody was removed by washing the cells three times with PBS for 10 min each time with rocking. The cells were then incubated with appropriate fluorescently labeled secondary antibodies (antimouse antibody labeled with Alexa Fluor 568 and antirabbit antibody labeled with Alexa Fluor 488; Invitrogen) for 1 h at RT. After three washes with 1‰ Tween 20 dissolved in PBS (PBST) and two washes with H2O, the cells were then incubated with DAPI (4′,6-diamidino-2-phenylindole) for 10 min. After two washes with H2O, the prepared coverslips were mounted on microscope slides with Fluoromount reagent (Sigma). All images were obtained by using a confocal microscope (Olympus).
Immunogold TEM.
BHK-21 cells were seeded a day before infection with MHV-68 at 50% confluence in 10-cm plates. The cells were infected at an MOI of 5. Sample preparation and immunostaining were performed as described previously, with some modifications (12). Briefly, at 11 h after infection, the cells were fixed with 2% paraformaldehyde plus 0.05% glutaraldehyde in 0.1 M phosphate buffer (PB; pH 7.2) for 15 min at 4°C on the plate and embedded in 10% gelatin in 0.1 M PB. Small blocks were infiltrated in 2.3 M sucrose plus 20% polyvinylpolypyrrolidone (PVP) in PB overnight at 4°C and quickly plunged into liquid nitrogen. Sections approximately 60 nm thick were cut with a Leica UC6/FC6 ultramicrotome and picked up with a wire loop filled with 2.3 M sucrose. Immunostaining was performed as follows: the sections were washed in bovine serum albumin (BSA) buffer (1% BSA and 0.15% glycine in PBS), followed by blocking in normal goat serum (1:20 dilution in BSA buffer) for 30 min. Subsequently, the sections were reacted for 2 h at RT with mouse monoclonal anti-ORF33 antibody (or BSA buffer as a negative control) and then for 1 h at RT with goat anti-mouse IgG conjugated with 10-nm colloidal gold particles (Sigma). After a brief wash in BSA buffer and PB, the sections were treated with 2.5% glutaraldehyde for 5 min. Finally, the sections were stained with 2% neutral uranyl acetate sealed with methyl cellulose. The sections were examined with a 120- or 200-kV electron microscope (Tecnai; FEI).
RESULTS
ORF33 accumulates in the nucleus and colocalizes with capsid proteins at discrete areas during MHV-68 infection.
Our previous data demonstrated that ORF33 localizes evenly in both the cytoplasm and the nucleus in cells transiently expressing exogenously supplied ORF33. After infection with MHV-68, it seemed that ORF33 was prone to translocate from the cytoplasm to the nucleus (8). To determine the localization of endogenous ORF33 during MHV-68 infection, we developed a mouse monoclonal antibody against ORF33. We first verified the specificity of the antibody using MHV-68-infected cells. A band corresponding to 36-kDa proteins, consistent with the predicted molecular mass of ORF33, was detected in lysate from virus-infected cells but not in that from mock-infected cells (data not shown). To more rigorously verify the specificity of the antibody, we transfected pHA-33, the MHV-68 WT BAC, or the 33Stop BAC into either Vero cells or BHK-21 cells and performed Western blotting using the monoclonal antibody for ORF33 or an antibody for capsid protein ORF26. Both the MHV-68 WT BAC and the 33Stop BAC were successfully transfected into the cells, and viral lytic proteins, such as ORF26, were expressed (Fig. 1A, middle, lanes 3, 4, 7, and 8). A 37-kDa band or a 36-kDa band protein was detected in lysates from pHA-33- or WT BAC-transfected cells, respectively, consistent with the predicted molecular mass of HA-tagged ORF33 (Fig. 1A, top, lanes 1 and 5) or native ORF33 (Fig. 1A, top, lanes 3 and 7). In contrast, no band was detected in lysates from 33Stop BAC-transfected cells (8) (Fig. 1A, top, lanes 4 and 8) or mock-transfected cells (Fig. 1A, top, lanes 2 and 6), demonstrating the specificity of the anti-ORF33 monoclonal antibody.
FIG 1.
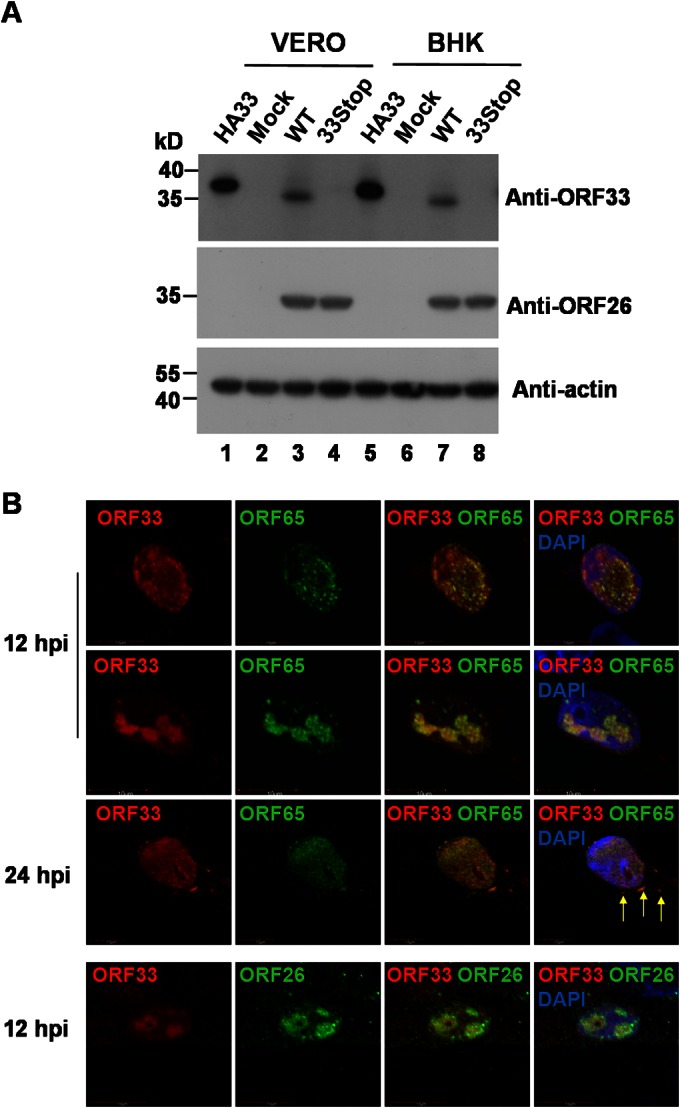
MHV-68 ORF33 accumulates in the nucleus and colocalizes with capsid proteins during virus infection. (A) Verification of the specificity of the anti-ORF33 monoclonal antibody. Vero and BHK-21 cells were transfected with pHA-33, the WT BAC, or the 33Stop BAC. At 36 h posttransfection, cells were lysed and subjected to Western blotting with the anti-ORF33 monoclonal antibody, a polyclonal antibody against ORF26, or a monoclonal antibody specific for β-actin. Lysates from mock-transfected cells were used as a negative control. (B) ORF33 colocalizes with capsid proteins during viral infection. Vero cells were infected with MHV-68 at an MOI of 3. At 12 and 24 h postinfection, cells were fixed and subjected to an indirect immunofluorescence assay using the anti-ORF33 monoclonal antibody and a rabbit polyclonal antibody against ORF65 or ORF26. The anti-ORF33 antibody was detected by use of an Alexa Fluor 568-conjugated secondary antibody (red channel). The anti-ORF65 and amti-ORF26 antibodies were detected by use of an Alexa Fluor 488-conjugated secondary antibody (green channel). Arrows, cytoplasmically localized ORF33.
We then examined the localization of endogenous ORF33 in comparison to that of two capsid proteins, ORF65 (a small capsid protein) and ORF26 (the triplex-2 protein). Vero cells were infected with MHV-68 and then subjected to an indirect immunofluorescence assay performed with the anti-ORF33 monoclonal antibody and an anti-ORF65 or anti-ORF26 rabbit polyclonal antibody. As shown in Fig. 1B, endogenous ORF33 localized predominantly in distinct compartments within the nucleus, where capsid protein ORF65 (rows 1, 2, and 3) or ORF26 (row 4) accumulated at the indicated time points. At 24 h postinfection, endogenous ORF33 also colocalized with ORF65 at discrete areas in the cytoplasm (Fig. 1B, row 3, arrows). We have previously demonstrated that ORF33 plays an important role during nuclear egress of capsids (8). Since ORF33 is predominantly colocalized with capsid proteins in the nucleus during viral infection, we reasoned that ORF33 might be associated with capsids in the nucleus.
ORF33 is associated with both nuclear and cytoplasmic capsids.
To address whether ORF33 is associated with nuclear capsids, a cosedimentation assay was performed. The nuclear fraction was separated from the cytoplasmic extract of MHV-68-infected cells. Clean separation of the two fractions was demonstrated by Western blotting probing for a cytoplasmic marker, caspase 3, or a nuclear marker, lamin A/C, respectively (Fig. 2A). Nuclear and cytoplasmic capsids were first pelleted through a sucrose cushion prior to sucrose gradient sedimentation. Fractions were collected, concentrated, and separated on an SDS-polyacrylamide gel prior to immunoblotting analyses with antibodies against ORF33 and capsid protein ORF26. ORF26 was used as an indicator of the location of the capsids in the sucrose gradient. We found that ORF33 was present in fractions containing capsids collected from both the nuclear and cytoplasmic extracts (Fig. 2B). These data suggest that ORF33 is associated with both nuclear and cytoplasmic capsids.
FIG 2.
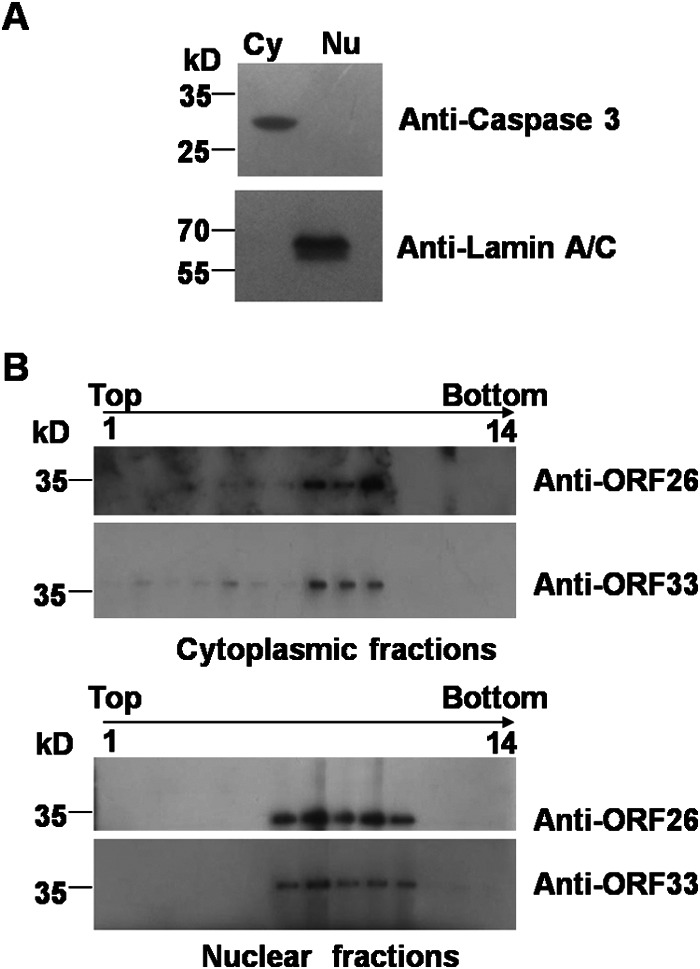
Cosedimentation of ORF33 with both nuclear and cytoplasmic capsids. (A) BHK-21 cells were infected with MHV-68 at an MOI of 5. At 12 h postinfection, the cells were harvested and separated into nuclear (Nu) and cytoplasmic (Cy) fractions, as described in Materials and Methods. A small portion of the two fractions was analyzed by immunoblotting with antibodies against lamin A/C or caspase 3. (B) Nuclear and cytoplasmic capsids were first pelleted through a 30% sucrose cushion prior to 20 to 50% (wt/vol) sucrose gradient sedimentation. Fractions were collected, and proteins were concentrated and separated on SDS–12% polyacrylamide gels prior to immunoblotting analyses with antibodies against ORF33 or capsid protein ORF26.
To further confirm the association of ORF33 with both nuclear and cytoplasmic capsids, a virion coimmunoprecipitation assay was also employed. We reasoned that if ORF33 is indeed associated with nuclear and cytoplasmic capsids, then capsids in both the nuclear and cytoplasmic extracts could be immunoprecipitated with the anti-ORF33 antibody. Again, nuclear and cytoplasmic extracts were cleanly separated, as demonstrated by the absence of cross contaminations (Fig. 3A). The nuclear and cytoplasmic extracts were individually incubated with the anti-ORF33 antibody or mouse IgG. Then, the immunoprecipitates were equally divided into two parts. One part was used to determine whether capsid proteins were precipitated. Both the ORF65 and ORF26 capsid proteins were successfully pulled down with the anti-ORF33 antibody in either the nuclear or the cytoplasmic immunoprecipitates (Fig. 3B, lanes 2 and 4). In contrast, ORF33, ORF65, or ORF26 was not immunoprecipitated with mouse IgG (Fig. 3B, lanes 1 and 3). To verify that it was assembled capsids and not free ORF65 and ORF26 proteins that were precipitated with the anti-ORF33 antibody, we took advantage of the property that capsid proteins, once assembled into virions, are much less sensitive than tegument proteins to trypsin treatment in the presence of detergent (8, 26). The remaining halves of the immunoprecipitates pulled down with the anti-ORF33 antibody were treated with trypsin in the presence of Triton X-100. As shown in Fig. 3B, both ORF65 and ORF26 in the two immunoprecipitates were resistant to trypsin digestion, demonstrating that both cytoplasmic and nuclear capsids are indeed pulled down with the anti-ORF33 antibody (Fig. 3B, middle and bottom, lanes 5 and 6). As a tegument protein, ORF33 is more sensitive to trypsin digestion and therefore was digested to an undetectable level (Fig. 3B, top, lanes 5 and 6). Taken together, these data further confirmed that ORF33 is associated with both nuclear and cytoplasmic capsids.
FIG 3.
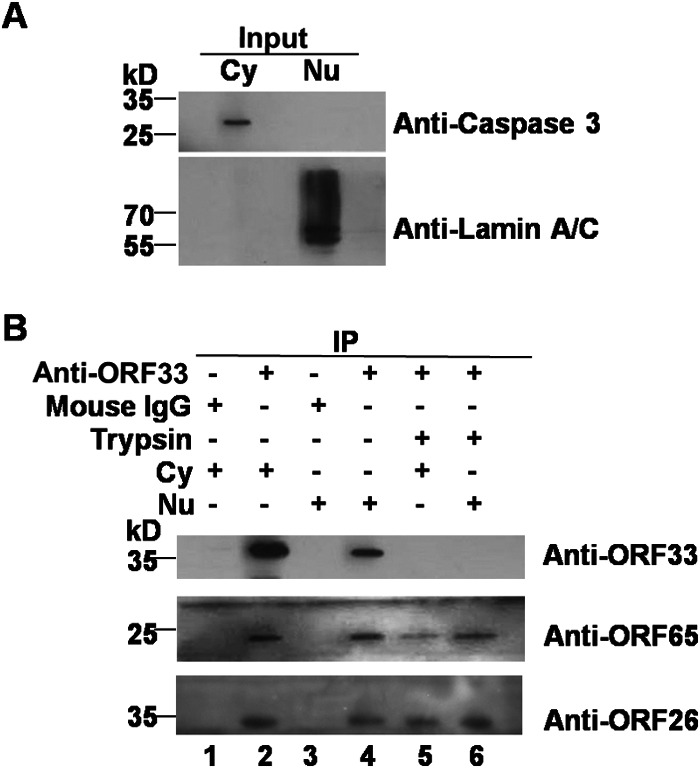
Immunoprecipitation of both nuclear and cytoplasmic capsids by the anti-ORF33 antibody. (A) BHK-21 cells were infected with MHV-68 at an MOI of 5 for 12 h. Nuclear and cytoplasmic fractions were separated as described in Materials and Methods and analyzed as described in the legend to Fig. 2A. (B) Nuclear and cytoplasmic fractions were incubated with the mouse anti-ORF33 monoclonal antibody or mouse IgG. Each precipitated sample pulled down with the anti-ORF33 antibody was divided into two parts. One part was analyzed by Western blotting to detect viral proteins using antibodies against ORF33, ORF65, or ORF26 (lanes 2 and 4). The other part was treated with trypsin for 1 h in the presence of 1% Triton X-100 prior to Western blotting analyses (lanes 5 and 6). Precipitates pulled down by mouse IgG were also analyzed by Western blotting (lanes 1 and 3). Cy, cytoplasmic extract; Nu, nuclear extract; IP, immunoprecipitation.
ORF33 is associated with capsids prior to primary envelopment.
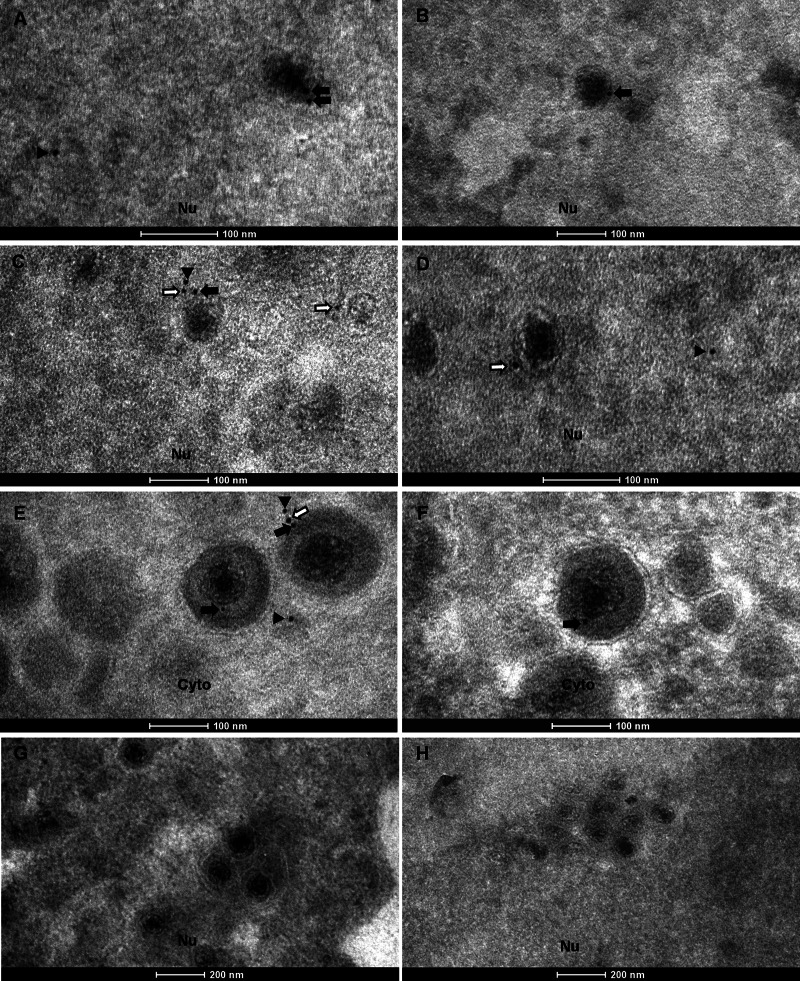
Although the data presented above demonstrated that ORF33 is associated with both nuclear and cytoplasmic capsids, it was unclear whether ORF33 interacts with capsids prior to or during nuclear egress of capsids. We therefore used electron microscopy to further examine the association of ORF33 with capsids at different stages of virion assembly. BHK-21 cells infected with MHV-68 were analyzed by an immunogold labeling assay using the anti-ORF33 monoclonal antibody, and viral particles in the nucleus and cytoplasm were examined (Fig. 4). In the nucleus, capsids were labeled with gold particles prior to arriving at the inner nuclear membrane, indicating that ORF33 is associated with intranuclear capsids (Fig. 4A to D, black and white arrows). We noticed that some gold particles were located at a distance (usually less than 20 nm) from the capsids (Fig. 4C and D, white arrows). This may reflect the diameter of the primary and secondary antibodies used in the labeling assay, as previous work showed that, in an indirect labeling system, colloidal gold is located 15 to 30 nm from the antigenic site to which the primary antibody binds (27). ORF33 was also associated with viral particles in the cytoplasm, as shown by labeling of cytoplasmic virions (Fig. 4E and F). As a negative control, sections of cells transfected with the 33Stop BAC were also examined. Consistent with our previous observation (8), capsid formation was not affected in the absence of ORF33, but the nuclear egress of capsids was partially inhibited, as shown by the much more frequent clustering of capsids in the nucleus than in the cytoplasm. However, these capsids were not labeled with gold particles, demonstrating the specificity of the antibody in the immunogold labeling assay (Fig. 4G and H).
FIG 4.
ORF33 is associated with nuclear capsids prior to primary envelopment as well as with mature virions in the cytoplasm. (A to F) BHK-21 cells were infected with WT MHV-68 at an MOI of 5 and analyzed 11 h later by immunoelectron microscopy with the anti-ORF33 monoclonal antibody, followed by a secondary antibody conjugated with 10-nm gold particles. Gold particles were detected both on or adjacent to capsids in the nucleus (Nu) (A to D) and on or adjacent to virions in the cytoplasm (Cyto) (E and F). (G and H) BHK-21 cells transfected with the 33Stop BAC were used as a negative-control sample. No gold particles were detected on the capsids. Black arrows, gold particles closely associated with capsids or virions; white arrows, gold particles adjacent to capsids or virions; arrowheads, free gold particles.
When we examined capsids in the nucleus, electron-dense areas with abundant ORF33 staining were frequently observed, consistent with the dense fluorescent compartments observed in Fig. 1B. Examples of these areas are shown in Fig. 5A and B (arrowheads). These ORF33-rich nuclear regions were often surrounded by immature capsids (Fig. 5A and B, arrows). These nuclear areas that accumulated with immature capsids and tegument protein are reminiscent of the assemblons that formed during capsid assembly in alphaherpesviruses, as previously reported (17). ORF33-rich cytoplasmic regions, consistent with the dense fluorescent compartments revealed in the confocal images (Fig. 1B, row 3, arrows), were also detected. An example of these areas is shown in Fig. 5C (white arrowhead).
FIG 5.
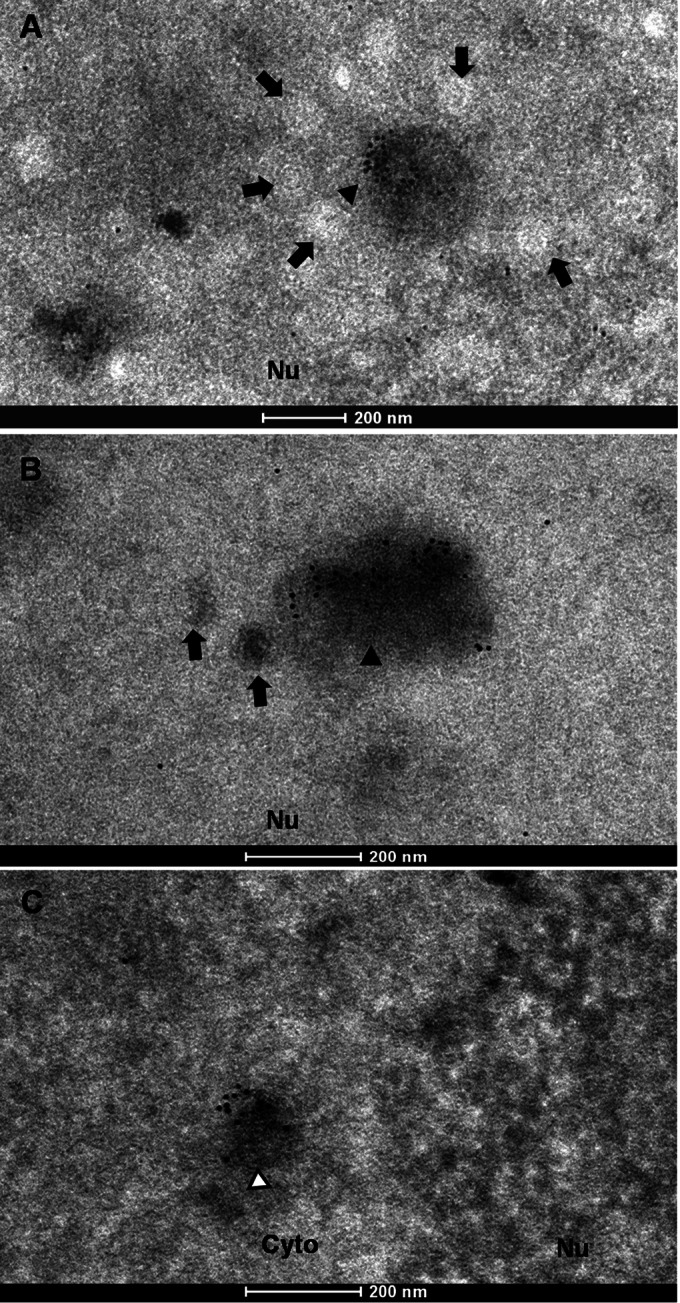
Immunogold labeling of ORF33-rich regions in the nucleus and cytoplasm. Samples were treated and viewed as described in the legend to Fig. 4. Arrows, nuclear capsids; black arrowheads, ORF33-rich nuclear areas; white arrowheads, ORF33-rich cytoplasmic area.
During analysis of these images, we noticed that the labeling efficiencies of both the intranuclear capsids and the cytoplasmic virions were low. Quantitative analyses of images from multiple regions showed that although only 10.4% of intranuclear capsids and 15.6% of cytoplasmic virions were labeled, the labeling was highly specific, as no gold particle was found to be associated with intranuclear capsids or cytoplasmic virions in 33Stop BAC-transfected samples (Table 1). Obviously, those intranuclear capsids that have just completed assembly and DNA packaging but that have not yet acquired ORF33 would not be labeled. Other reasons for the low labeling efficiency may include a weak affinity of the anti-ORF33 antibody to ORF33 under the experimental condition for EM analysis and/or the low occupancy of ORF33 on or near capsids and virions, since most labeled capsids and virions had only 1 or 2 gold particles. In addition, depending on the relative location of the ORF33 protein within the embedded sample block, it may be difficult for the anti-ORF33 antibody to gain access to ORF33 and/or for gold particles to penetrate the medium used to embed samples for electron microscopy analysis. For these reasons, we do not think that it is suitable to quantify the average number of gold particles per particle. Instead, we analyzed the same EM images for the distribution of the gold particles and found that, in the presence of WT virus, 40% of the gold particles were localized in ORF33-rich nuclear regions, 22% were associated with nuclear capsids, 5% were localized in ORF33-rich cytoplasmic regions, and 26% were associated with cytoplasmic virions. The remaining gold particles were randomly distributed (Table 2). In contrast, only 8 free gold particles were enumerated from multiple regions of equal sizes from 33Stop BAC-transfected samples, which may be caused by rare nonspecific binding of gold-conjugated secondary antibody. Importantly, none of these gold particles was associated with capsids or virions (Table 2). Taken together, these data demonstrate that ORF33 is associated with nuclear capsids prior to primary envelopment as well as with mature virions in the cytoplasm.
TABLE 1.
Percentage of virions labeled with gold particles in EM images
| Virus | Type of virion | Total no. of virions counteda | % of virions labeled with gold particles |
|---|---|---|---|
| WT MHV-68 | Nuclear capsids | 241 | 10.4 |
| Cytoplasmic virions | 232 | 15.6 | |
| 33Stop BAC | Nuclear capsids | 539 | 0 |
| Cytoplasmic virions | 98 | 0 |
The total number of virion particles enumerated from images of 15 nuclear regions and 15 cytoplasmic regions.
TABLE 2.
Distribution of gold particles in EM images
| Virus | No. of gold particlesa | Fraction (%) of total gold particles in the indicated regions |
||||
|---|---|---|---|---|---|---|
| ORF33- rich nuclear regions | Nuclear capsids | ORF33-rich cytoplasmic regions | Cytoplasmic virions | Free gold particles | ||
| WT MHV-68 | 244 | 40 | 22 | 5 | 26 | 7 |
| 33Stop BAC | 8 | 0 | 0 | 0 | 0 | 100 |
The total number of gold particles was enumerated from the same images used for the analysis whose results are presented in Table 1.
ORF33 interacts with capsid proteins ORF25 and ORF26.
Herpesvirus virion assembly is driven by multiple protein-protein interactions (28–31). Since ORF33, as an inner tegument protein (8), is associated with intranuclear capsids (Fig. 2 to and 4), we hypothesized that ORF33 may do so through interacting with individual capsid proteins. We thus tested the interactions between ORF33 and the five capsid proteins conserved in all herpesviruses, ORF25, ORF26, ORF43, ORF62, and ORF65. For better expression, the major capsid protein ORF25 was divided into two parts (designated ORF25N for amino acids [aa] 1 to 808 and ORF25C for aa 777 to 1365, respectively). Expression plasmids for HA-tagged ORF33 (HA-33) or 3× FLAG-CBP-tagged ORF33 (Tag-33) and individual capsid proteins with a 3× FLAG-CBP tag or HA tag were cotransfected into 293T cells, and the cells were harvested for coimmunoprecipitation assays. We found that ORF33 interacted with ORF25C and ORF26 (Fig. 6A) but not with the others (Fig. 6A and B). Therefore, ORF33 may associate with intranuclear capsids through directly interacting with the major capsid protein ORF25 and/or the triplex-2 protein ORF26.
FIG 6.
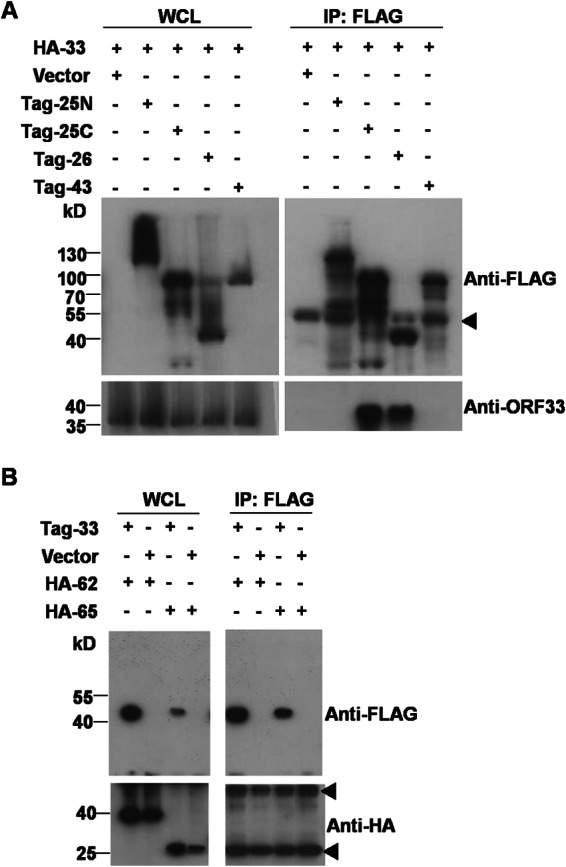
ORF33 interacts with capsid proteins ORF25 and ORF26. 293T cells were cotransfected with expression plasmids, as indicated. At 48 h posttransfection, whole-cell lysates (WCL) were prepared, and the expression of each protein was examined by Western blotting. The whole-cell lysates were further subjected to immunoprecipitation with anti-Flag M2 agarose and analyzed by Western blotting using anti-HA, anti-FLAG, or anti-ORF33 antibodies. Arrowheads, antibody heavy and light chains.
DISCUSSION
As a structural linker between the capsid and the envelope, the tegument plays multiple roles in virus lytic replication. To date, most of our knowledge regarding how the tegument layer is formed within a virion has come from studies on alphaherpesviruses. The dual roles of ORF33 of a gammaherpesvirus in virion morphogenesis at separate stages (8) prompted us to investigate how this tegument is associated with capsids. Here we found that ORF33 not only colocalized with capsid proteins during MHV-68 infection in an immunofluorescence assay (Fig. 1B) but also was associated with both nuclear and cytoplasmic capsids in cosedimentation and immunoprecipitation assays (Fig. 2 and 3). Immunogold labeling assays showed that ORF33 accumulated in discrete areas of the nucleus surrounded by immature capsids (Fig. 5). Consistently, ORF33 was detected on capsids prior to primary envelopment (Fig. 4A to D). These data indicate that ORF33 may associate with capsids at these ORF33-rich areas, although we cannot rule out the possibility that the association of ORF33 with capsids and the protein's accumulation in the nucleus are two independent events. Nonetheless, we identified ORF33 as a tegument protein that is associated with intranuclear capsids prior to primary envelopment.
Herpesvirus assembly involves multiple protein-protein interactions (29–32). A previous study on the genome-wide protein interaction network of MHV-68 showed that ORF33 interacts with tegument proteins ORF38 and ORF45 (33). We have confirmed these interactions (34) (data not shown). However, our immunogold labeling results showed that, unlike ORF33, both ORF38 and ORF45 are packaged onto capsids in the cytoplasm (34) (data not shown). Thus, their interactions with ORF33 may participate in virion morphogenesis in the cytoplasm, but these interactions are unlikely to contribute to the association of ORF33 with nucleocapsids. Here we identified two novel interacting partners for ORF33: the major capsid protein ORF25 and the triplex-2 protein ORF26. ORF25, which makes up the capsomere, has three domains: the floor, middle, and upper domains (35). Binding of the inner tegument protein(s) to the capsid is mediated by the upper domain of the major capsid proteins and the triplex protein(s) (1, 35, 36). We previously showed that capsid formation is not affected in the absence of ORF33, and ORF33 is an inner tegument protein of mature MHV-68 virions (8). Since ORF33 is associated with capsids in the nucleus, the interactions identified strongly indicated that ORF33, as an inner tegument protein, may interact with nascent capsids in a direct manner.
Our previous work demonstrated that ORF33 plays an important role in the nuclear egress of capsids (8), but the underlying mechanism remained unclear. An essential step in the nuclear egress process is the primary envelopment of nucleocapsids. For this to take place, the nuclear lamina is dissolved by virus genome- and host genome-encoded kinases to make the inner nuclear membrane accessible to the capsids. Two viral proteins of HSV-1, UL31 and UL34, make up the nuclear egress complex and play a key role in this process (37, 38). These two proteins are conserved (UL53 and UL50 in human cytomegalovirus [HCMV], BFLF2 and BFRF1 in EBV, and ORF69 and ORF67 in KSHV or MHV-68), but how capsids are directed or linked to the nuclear egress complex is still largely unknown (37–40). The interaction between HSV-1 UL31 and members of the capsid vertex-specific complex (CVSC) has been shown to be important to bridge capsids and the nuclear egress complex (41–43). As an inner tegument protein that is associated with capsids in the nucleus and plays a functional role in the nuclear egress of capsids, MHV-68 ORF33 may regulate the nuclear egress of capsids through interacting with key molecules in this process. Alternatively, ORF33 may be involved in the intranuclear movement of capsids, which, after assembly in the nucleus, must move toward the inner nuclear membrane for envelopment. This is a rapid and active transport process dependent on energy and temperature. Components of the cellular transportation machine, such as nuclear actin and the motor protein myosin, were suggested to participate in the movement of capsids (44). However, whether there exists a virion protein(s) that directly interacts with the motor proteins is still a mystery. As a tegument protein that is associated with intranuclear capsids and partially impairs the nuclear egress of capsids, ORF33 may participate in the transportation of capsids within the nucleus.
ORF33 is a tegument protein conserved among all herpesvirus subfamilies (9). Its homologue in alphaherpesviruses, UL16, has been extensively studied. HSV-1 UL16 is also a virion-associated tegument protein and functions in virion maturation (26, 45). Both ORF33 and UL16 accumulate in the nucleus during virus infection and colocalize with intranuclear capsid proteins (16) (Fig. 1), and both proteins are critical for virion maturation in the cytoplasm through interacting with its binding partners (34, 45, 46). However, UL16 and ORF33 differ in several aspects: (i) UL16 was reported to participate in viral DNA packaging (47), but this is not the case for ORF33 (8). (ii) Unlike ORF33, UL16 is dispensable for the replication of HSV-1 in cell culture (8, 48). (iii) Through biochemical analysis of capsids isolated from either the nucleus or the cytoplasm, researchers from one group showed that UL16 bound to cytoplasmic capsids with dynamic kinetics but was not present on nuclear capsids. However, the authors also acknowledged that the association between UL16 and nuclear capsids might be too weak to be detected under the lysis conditions used in their studies (26). (iv) UL16 has been reported to interact with a panel of viral proteins (32, 45, 46, 49). Among these, only the interaction between UL16 and UL11 is conserved in gammaherpesviruses (in the form of ORF33 and ORF38). ORF33 also interacts with ORF45, but there is no homologue of ORF45 in alphaherpesviruses (41).
The primary sequence similarity of these two proteins is only 18.2% when their sequences are aligned by use of the Clustal (version 2.1) program. To better compare these two proteins, we generated the three-dimensional (3D) structure models of ORF33 (for which the structure with PDB accession number 1NOV was used as the template) and UL16 (for which the structure with PDB accession number 1F8V was used as the template) using the FR-t5-M program (50). We found that the two proteins share more similarity in their 3D structures (root mean square deviation = 3.64) than in their primary sequences, but there are obvious differences in local regions (data not shown). These local differences may help explain the functional differences between ORF33 and UL16. Unfortunately, all efforts to map the specific domain(s) mediating the interaction of UL16 or ORF33 with its binding partners failed (49, 51) (data not shown), impeding further analysis.
The homologue of ORF33 in betaherpesviruses is UL94. Like gammaherpesvirus ORF33, HCMV UL94 also plays an essential role in virus lytic infection (10). A recent study demonstrated that UL94 participates in secondary envelopment through interacting with another tegument protein (UL99), the homologue of HSV-1 UL11 and MHV-68 ORF38 (30). Interestingly, UL94 also accumulates in the nucleus during virus infection (52). However, whether UL94 is associated with intranuclear capsids and plays a functional role in the nuclear egress of HCMV capsids remains to be determined.
ACKNOWLEDGMENTS
We thank Shufeng Sun at the Center for Biological Imaging (CBI), Institute of Biophysics, for help with EM sample preparation; Yan Teng and Chunli Jiang at CBI for help with confocal image analysis; Ren Sun at the University of California, Los Angeles, for providing the pTag plasmids used in the study; Wentao Dai at the Shanghai Academy of Science and Technology for bioinformatics data analysis; and members of the H. Deng laboratory for helpful discussions.
This work was supported by grants from the National Natural Science Foundation of China (no. 81171582 and no. 81325012).
REFERENCES
- 1.Dai W, Jia Q, Bortz E, Shah S, Liu J, Atanasov I, Li X, Taylor KA, Sun R, Zhou ZH. 2008. Unique structures in a tumor herpesvirus revealed by cryo-electron tomography and microscopy. J Struct Biol 161:428–438. doi: 10.1016/j.jsb.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 3.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 4.Mettenleiter TC, Klupp BG, Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol 9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MS, Loftus MS, Kedes DH. 2014. Maturation and vesicle-mediated egress of primate gammaherpesvirus rhesus monkey rhadinovirus require inner tegument protein ORF52. J Virol 88:9111–9128. doi: 10.1128/JVI.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998. doi: 10.1007/s13238-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J Virol 83:10582–10595. doi: 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res 145:173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Phillips SL, Bresnahan WA. 2012. The human cytomegalovirus (HCMV) tegument protein UL94 is essential for secondary envelopment of HCMV virions. J Virol 86:2523–2532. doi: 10.1128/JVI.06548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathish N, Zhu FX, Yuan Y. 2009. Kaposi's sarcoma-associated herpesvirus ORF45 interacts with kinesin-2 transporting viral capsid-tegument complexes along microtubules. PLoS Pathog 5:e1000332. doi: 10.1371/journal.ppat.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Guo H, Reyes N, Lee S, Bortz E, Guo F, Sun R, Tong L, Deng H. 2012. Distinct domains in ORF52 tegument protein mediate essential functions in murine gammaherpesvirus 68 virion tegumentation and secondary envelopment. J Virol 86:1348–1357. doi: 10.1128/JVI.05497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow H, Klupp BG, Mettenleiter TC. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J Virol 78:1314–1323. doi: 10.1128/JVI.78.3.1314-1323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol 76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebrun M, Thelen N, Thiry M, Riva L, Ote I, Conde C, Vandevenne P, Di Valentin E, Bontems S, Sadzot-Delvaux C. 2014. Varicella-zoster virus induces the formation of dynamic nuclear capsid aggregates. Virology 454-455:311–327. doi: 10.1016/j.virol.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Nalwanga D, Rempel S, Roizman B, Baines JD. 1996. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- 17.Ward PL, Ogle WO, Roizman B. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol 70:4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. 1990. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol 71(Pt 6):1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 19.Simas JP, Efstathiou S. 1998. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol 6:276–282. doi: 10.1016/S0966-842X(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 20.Wu TT, Liao HI, Tong L, Leang RS, Smith G, Sun R. 2011. Construction and characterization of an infectious murine gammaherpesivrus-68 bacterial artificial chromosome. J Biomed Biotechnol 2011:926258. doi: 10.1155/2011/926258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. doi: 10.1128/JVI.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortz E, Wang L, Jia Q, Wu TT, Whitelegge JP, Deng H, Zhou ZH, Sun R. 2007. Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm. J Virol 81:10137–10150. doi: 10.1128/JVI.01233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia Q, Chernishof V, Bortz E, McHardy I, Wu TT, Liao HI, Sun R. 2005. Murine gammaherpesvirus 68 open reading frame 45 plays an essential role during the immediate-early phase of viral replication. J Virol 79:5129–5141. doi: 10.1128/JVI.79.8.5129-5141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song MJ, Hwang S, Wong WH, Wu TT, Lee S, Liao HI, Sun R. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A 102:3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A 104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meckes DG Jr, Wills JW. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol 81:13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann R, Walther P, Muller M. 1996. Immunogold labeling in scanning electron microscopy. Histochem Cell Biol 106:31–39. doi: 10.1007/BF02473200. [DOI] [PubMed] [Google Scholar]
- 28.Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. 2014. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology 454-455:67–77. doi: 10.1016/j.virol.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J Virol 84:1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips SL, Cygnar D, Thomas A, Bresnahan WA. 2012. Interaction between the human cytomegalovirus tegument proteins UL94 and UL99 is essential for virus replication. J Virol 86:9995–10005. doi: 10.1128/JVI.01078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozen R, Sathish N, Li Y, Yuan Y. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J Virol 82:4742–4750. doi: 10.1128/JVI.02745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Salwinski L, Zhang C, Chu D, Sampankanpanich C, Reyes NA, Vangeloff A, Xing F, Li X, Wu TT, Sahasrabudhe S, Deng H, Lacount DJ, Sun R. 2011. An integrated approach to elucidate the intra-viral and viral-cellular protein interaction networks of a gamma-herpesvirus. PLoS Pathog 7:e1002297. doi: 10.1371/journal.ppat.1002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen S, Guo H, Deng H. 2014. Murine gammaherpesvirus-68 ORF38 encodes a tegument protein and is packaged into virions during secondary envelopment. Protein Cell 5:141–150. doi: 10.1007/s13238-013-0005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt WJ, Zhu H, Liu F, Zhou ZH. 2013. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog 9:e1003525. doi: 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai X, Gong D, Wu T, Sun R, Zhou ZH. 2014. Organization of capsid-associated tegument components in Kaposi's sarcoma-associated herpesvirus. J Virol 88:12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol 74:10063–10073. doi: 10.1128/JVI.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A 104:7241–7246. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farina A, Feederle R, Raffa S, Gonnella R, Santarelli R, Frati L, Angeloni A, Torrisi MR, Faggioni A, Delecluse HJ. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J Virol 79:3703–3712. doi: 10.1128/JVI.79.6.3703-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sam MD, Evans BT, Coen DM, Hogle JM. 2009. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J Virol 83:2996–3006. doi: 10.1128/JVI.02441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leelawong M, Guo D, Smith GA. 2011. A physical link between the pseudorabies virus capsid and the nuclear egress complex. J Virol 85:11675–11684. doi: 10.1128/JVI.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang K, Wills E, Lim HY, Zhou ZH, Baines JD. 2014. Association of herpes simplex virus pUL31 with capsid vertices and components of the capsid vertex-specific complex. J Virol 88:3815–3825. doi: 10.1128/JVI.03175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lanerolle P, Johnson T, Hofmann WA. 2005. Actin and myosin I in the nucleus: what next? Nat Struct Mol Biol 12:742–746. doi: 10.1038/nsmb983. [DOI] [PubMed] [Google Scholar]
- 45.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A 109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh PC, Han J, Chadha P, Meckes DG Jr, Ward MD, Semmes OJ, Wills JW. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J Virol 85:9425–9436. doi: 10.1128/JVI.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol 143:863–880. doi: 10.1007/s007050050338. [DOI] [PubMed] [Google Scholar]
- 48.Baines JD, Roizman B. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol 65:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harper AL, Meckes DG Jr, Marsh JA, Ward MD, Yeh PC, Baird NL, Wilson CB, Semmes OJ, Wills JW. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol 84:2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai W, Song T, Wang X, Jin X, Deng L, Wu A, Jiang T. 2014. Improvement in low-homology template-based modeling by employing a model evaluation method with focus on topology. PLoS One 9:e89935. doi: 10.1371/journal.pone.0089935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh PC, Meckes DG Jr, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol 82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Cui Z, Zhang Z, Wei H, Zhou Y, Wang M, Zhang XE. 2009. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388:68–77. doi: 10.1016/j.virol.2009.03.007. [DOI] [PubMed] [Google Scholar]