Abstract
Replication of plus-stranded RNA [(+)RNA] viruses depends on the availability of coopted host proteins and lipids. But, how could viruses sense the accessibility of cellular resources? An emerging concept based on tombusviruses, small plant viruses, is that viruses might regulate viral replication at several steps depending on what cellular factors are available at a given time point. I discuss the role of phospholipids, sterols, and cellular WW domain proteins and eukaryotic elongation factor 1A (eEF1A) in control of activation of the viral RNA-dependent RNA polymerase (RdRp) and regulation of the assembly of viral replicase complexes (VRCs). These regulatory mechanisms might explain how tombusviruses could adjust the efficiency of RNA replication and new VRC assembly to the limiting resources of the host cells during infections.
INTRODUCTION
Plus-stranded RNA [(+)RNA] viruses, which are widespread pathogens of plants and animals, replicate in the cytosol of infected cells. These viruses assemble membrane-bound viral replicase complexes (VRCs), which consist of the viral RNA and viral proteins as well as coopted host proteins (1–3). After translation of the incoming viral (+)RNA in the cytosol, the VRC assembly takes place, followed by robust RNA synthesis driven by the viral RNA-dependent RNA polymerase (RdRp) located in the VRCs. After multiple cycles of translation/replication of the newly made (+)RNAs, the viral (+)RNA progeny become encapsidated and a few (+)RNAs move to neighboring cells to initiate new infections. The replication process is “expensive” for the cell because the virus steals away numerous proviral host proteins, subverts lipids and subcellular membranes, and uses up large amounts of amino acids, ATP, and other ribonucleotides for VRC assembly and RNA synthesis. The hijacked host proteins include translation factors, protein chaperones, RNA-modifying enzymes, ESCRT (endosomal sorting complexes required for transport) proteins, and cellular proteins involved in lipid biosynthesis (4, 5). The emerging picture is that VRC assembly is driven by many factors; thus, the assembly process is likely regulated by viral and host factors for optimal replication in infected cells.
The viral replication process could be so robust and rapid that in the case of several plant RNA viruses, the progeny viral RNAs could reach over a million copies in a single cell in ∼24 h. How does the virus “know” when to stop the assembly of new VRCs because the cell runs out of proviral factors and resources? Incomplete VRC assembly due to a shortage in one or several host factors during exponential replication (16 to 48 h) could have disastrous consequences, leading to lots of unfinished or truncated RNA products, high mutation and recombination rates, or high exposure of the viral double-stranded RNA (dsRNA) replication intermediate to the cellular RNA sensors that activate innate defense responses. To avoid this doom scenario, viruses might apply molecular sensors to monitor the status of the cell or the availability and abundance of proviral host factors to keep the replication process under control. This model will be discussed below based on new results from tomato bushy stunt virus (TBSV).
TBSV is among the best-characterized RNA viruses due to the use of a yeast (Saccharomyces cerevisiae) model (2). A dozen systematic genome-wide screens and global proteomics approaches in yeast or in vitro have led to the identification of ∼500 host proteins/genes involved in TBSV replication (4). TBSV codes for two replication proteins, the auxiliary p33 and the p92pol. p33 is an RNA chaperone and recruits the TBSV (+)RNA to the site of replication, which takes place at the cytosolic surface of peroxisomal membranes.
TBSV has been shown to coopt several host factors involved in the assembly of the membrane-bound VRCs, such as the heat shock protein 70 (Hsp70), the eukaryotic elongation factor 1A (eEF1A), Vps23p ESCRT (endosomal sorting complexes required for transport) protein, Bro1p ESCRT-associated protein, Vps4p AAA+ ATPase, and Cdc34p E2 ubiquitin-conjugating enzyme (4, 6–9). In addition, TBSV channels sterols and phospholipids (phosphatidylethanolamine [PE]) to enrich these lipids at the sites of replication, possibly via the use of cellular VAMP/synaptobrevin-associated protein, called VAP (Scs2p in yeast), and oxysterol-binding proteins (ORPs; Osh6 and Osh7 in yeast) and membrane contact sites between the endoplasmic reticulum (ER) and peroxisomes (Fig. 1) (10).
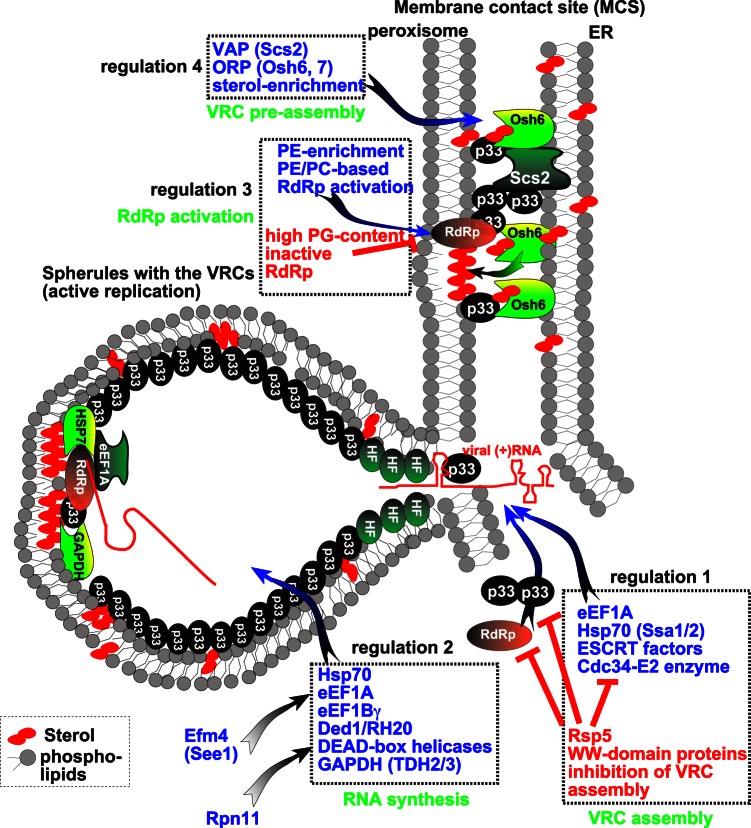
FIG 1.
Coopted host factors and regulation of tombusvirus replication. At the early stage of replication, the tombusvirus p33 and p92 replication proteins bind primarily to the abundant cellular susceptibility factors (proviral host factors, in blue), to other viral replication proteins, and the viral (+)RNA to recruit the viral (+)RNA to PE- and sterol-rich cellular membranes (located at or near membrane contact sites) to assemble functional VRCs. At the late stage of replication, the host factors have been depleted due to sequestration into previously assembled VRCs, and the new viral replication proteins bind to WW domain proteins, blocking new VRC assembly (regulation 1). A shortage in Efm4-methylated eEF1A leads to degradation of p33 and p92 RdRp (regulation 2). Alternatively, limitation in PE-rich (regulation 3) and sterol-rich (regulation 4) membranes or p92 RdRp binding to PG leads to inactive RdRp, inhibiting new viral RNA synthesis at the late stage of replication. PC, phosphatidylcholine.
Another group of subverted cellular proteins is involved in viral RNA synthesis within the VRCs. The list includes eEF1A; the eukaryotic elongation factor 1Bγ (eEF1Bγ); Hsp70; DDX3-, DDX5-, and eIF4AIII-like DEAD box helicases; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which facilitate either minus-strand or plus-strand synthesis during RNA replication (Fig. 1) (2, 11–13). Subversion of so many host factors for TBSV replication highlights the need for regulation of new rounds of VRC assembly or RdRp activity when cellular resources become limited or exhausted, which could be envisioned to occur at the late stage of replication due to their depletion in the previous rounds of VRC assembly.
Incorrect VRC assembly in the absence of selected host factors.
Observations are indeed accumulating that incorrect VRC assembly could occur with severe negative effects on TBSV replication. Multiple genome-wide screens with TBSV in yeast indicated that depletion of a proviral host factor does not necessarily prevent the assembly of the VRCs (4) but frequently leads to incorrect VRC assembly that changes some of the activities of the viral replicase. For example, depletion of multifunctional proteasomal Rpn11p metalloprotease in yeast results in inefficient recruitment of Ded1p DEAD box helicase into the tombusvirus VRCs (14). The low level of Ded1p leads to reduced replication and the production of high levels of truncated viral RNAs and recombinant viral RNAs (14). Also, depletion of ESCRT proteins (Vps24 or Vps4p) results in incorrect assembly of tombusvirus VRCs in yeast, which gives rise to spherules with wide openings, instead of the narrow “neck-like” opening in wild-type (wt) yeast (8). These open VRC structures are likely less protective of the viral dsRNA intermediates against the host antiviral machinery. Altogether, these observations suggest that it is unlikely that TBSV replication just stops when the cell runs out of host factors and other cellular resources, because that would lead to the production of lots of “junk” replication products. Accordingly, the late-stage replication products look similar to early-stage products of virus replication, suggesting that a regulation process exists that somehow prevents the formation of incomplete VRCs at the late stage in wild-type cells. Regulation of new rounds of VRC assembly could also facilitate the switch of the viral (+)RNA from replication to other viral steps, such as virion assembly.
Regulatory roles of host factors in TBSV replication.
How then does TBSV regulate new rounds of VRC assembly and RdRp activity at the latter stage of infection? The first indication that a regulation process exists in the case of TBSV has come from studies with the WW-domain-containing cellular proteins (15). The WW domain is a simple and highly conserved protein domain involved in protein-protein interactions and binds to ligands usually carrying proline-rich sequences.
The yeast Nedd4-type Rsp5p E3 ubiquitin ligase carrying the WW domain and several cellular WW domain proteins, including yeast Wwm1p and Prp40p and plant AtDrh1, AtFCA, and AtPrp40c, were shown elsewhere to bind to the tombusvirus replication proteins and inhibit their functions (15, 16). However, the cellular WW domain proteins bind to the replication proteins with lower affinity than the proviral host factors (17), making them poor competitors with coopted proviral host factors when these subverted cellular proteins are abundant. It was proposed that the WW domain proteins bind to newly translated viral replication proteins, blocking their functions, when the proviral factors have been exhausted (Fig. 1, regulation 1). Increasing WW domain protein amount by overexpression interfered with complex formation between p33 replication protein and several cellular host factors, such as eEF1A, Bro1p, Vps4p, and Cdc34p, involved in VRC assembly (17). Moreover, the WW domain inhibited the binding of p33 to the viral RNA and the oligomerization of p33, thus blocking tombusvirus replication. Another feature of the WW domain proteins is that they facilitate the degradation of excess amounts of viral replication proteins. The reduction of the p92pol level could prevent the functional interference of the replication protein with other, nonreplicative functions of the viral RNA (16).
Depletion of multiple WW domain proteins in yeast resulted in especially error-prone replicase at the late stage of replication (17). This is likely due to incorrect VRC assembly, possibly caused by (i) prior depletion of one or more proviral factors at the earlier rounds of VRC assembly and (ii) the fact that the depleted WW domain proteins could not block new VRC assembly. In summary, TBSV might be able to sense the availability of proviral host factors versus WW domain proteins to control if new rounds of VRC assembly could be continued or halted depending on replication protein-host protein interactions.
Additional RNA viruses might also take advantage of WW domain proteins for sensing the cellular environment, since the unrelated nodaviruses (insect RNA viruses) could also be inhibited by overexpression of yeast Rsp5p and Wwm1p WW domain proteins (16).
Another intriguing case for a putative cellular sensory function is based on the key role of the methylation status of eEF1A for TBSV replication (Fig. 1, regulation 2). When methylation-negative mutants of eEF1A were expressed in yeast, then the tombusvirus replication protein was degraded (18). Similarly, deletion of the METTL10-like Emf4 (See1) methyltransferase, required for methylation of eEF1A, reduced the stability of p33 replication protein (18). Based on these findings, I suggest that TBSV could measure the availability of methylated eEF1A in cells and would stop assembling new VRCs and promote the degradation of p33 replication proteins when free methylated eEF1A has been depleted. This might prevent the assembly of new VRCs missing the critical eEF1A required for proper VRC assembly and minus-strand RNA synthesis (6).
Regulatory roles of phospholipids and sterols in TBSV replication.
One of the major regulatory steps during TBSV replication is the activation of p92pol, which is initially inactive in the cytosol after translation (7). The RdRp activation takes place inside the assembled VRCs and requires a cis-acting element in the viral (+)RNA, the cellular Hsp70 chaperone, and neutral phospholipids (J. Pogany and P. D. Nagy, unpublished data). It is currently not known if Hsp70 could become limited during TBSV replication.
Interestingly, the negatively charged phosphatidylglycerol (PG) greatly inhibits the RNA binding capacity of p92pol and prevents its activation in vitro, while the neutral PE enhances RNA binding and activation of p92pol (Pogany and Nagy, unpublished). Therefore, the ratio of proviral PE to inhibitory PG at the site of replication likely serves as a signal for TBSV to activate p92pol (when PE is abundant) or inactivate it, when PE is less abundant and PG binds to p92pol. Overall, the TBSV-induced enrichment of PE at the sites of replication could become limiting at the late stage of replication, allowing PG to block the activation of the newly made p92pol. Thus, TBSV might utilize the cellular PE and PG levels to continue building active VRCs in a PE-rich microenvironment or stop the new VRC assembly or block RdRp activity in PG-rich sites, allowing (+)RNAs to perform nonreplicative functions (Fig. 1, regulation 3).
Another level of regulation of new VRC assembly could be the ability of TBSV to sense the capacity of the cell to synthesize phospholipids de novo that is required for membrane proliferation. For example, depletion of total phospholipids via overexpression of Opi1p transcription repressor of phospholipid synthesis genes led to reduced VRC assembly and instability of the viral replication proteins (19). In contrast, robust membrane proliferation in pah1Δ yeast (a lipin orthologue, whose deletion causes mainly proliferation of the ER membrane) or enhanced phospholipid synthesis in opi1Δ yeast resulted in efficient tombusvirus VRC assembly and a high level of TBSV replication (19, 20). Thus, TBSV might be able to sense the phospholipid status/synthesis of the host cell whether the microenvironment is suitable for building more new VRCs or not.
TBSV might also sense sterol levels in peroxisomal membranes, because deletion of oxysterol-binding ORPs (Osh3, -5, -6, and -7) or Scs2 VAP protein reduced the accumulation of both TBSV RNA and replication proteins in yeast (Fig. 1, regulation 4) (10). The ORPs and VAP proteins are needed for the formation of membrane contact sites between the ER and the peroxisomes to channel sterols to the sites of TBSV replication (10). In addition, pharmacological inhibition of sterol synthesis or depletion of sterol levels due to downregulation of ergosterol synthesis genes (ERG25 and ERG4) reduced the stability of the p92pol replication protein and inhibited the activity of the viral replicase obtained from yeast (21). Overall, “sensing” the redistributed sterol level in the peroxisomal membranes by p92pol might be another regulatory step during VRC assembly.
In addition to the above strategies, tombusviruses might also utilize replication protein ubiquitination or phosphorylation to sense intracellular processes. The detailed mechanisms on how these posttranslational modifications might regulate new TBSV VRC formation, however, have not yet been unraveled.
Model.
I propose that tombusviruses can sense the status of the host cells that control RdRp activation and assembly of new VRCs (Fig. 1). At the early stage, the tombusvirus replication proteins interact with the host susceptibility factors, which are abundant and accessible, to build active VRCs. As viral replication proceeds with multiple cycles of translation and new rounds of replication, the cell likely runs out of one or more susceptibility factors at the late stage of replication. Depletion of the susceptibility factors facilitates the interaction of the cellular WW domain proteins with the viral replication proteins, which then blocks the assembly of new VRCs and the formation of new p33-viral RNA complexes and leads to the degradation of p92pol RdRp. Degradation of RdRp is also promoted by the shortage of coopted methylated eEF1A. Lipid synthesis, membrane proliferation, and PE-versus-PG content at the site of replication also control RdRp activation and VRC assembly. Stopping the formation of new VRCs at the late stage of replication facilitates the switching of newly made viral (+)RNAs from the translation/replication cycle to encapsidation.
Summary.
Altogether, the ability of tombusviruses to sense the status of the infected cells determines if new rounds of VRCs are assembled or if the VRC assembly process is halted. This strategy could minimize the massive production of viral “junk” RNAs when host factors and resources have been exhausted.
ACKNOWLEDGMENTS
I apologize that I could not include other viruses in this short Gem article.
This research was supported by the NIH-NIAID (1R21AI109529).
REFERENCES
- 1.den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagy PD, Pogany J. 2011. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Castro IF, Volonte L, Risco C. 2013. Virus factories: biogenesis and structural design. Cell Microbiol 15:24–34. doi: 10.1111/cmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagy PD, Pogany J, Lin JY. 2014. How yeast can be used as a genetic platform to explore virus-host interactions: from ‘omics’ to functional studies. Trends Microbiol 22:309–316. doi: 10.1016/j.tim.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Panda D, Cherry S. 2012. Cell-based genomic screening: elucidating virus-host interactions. Curr Opin Virol 2:784–792. doi: 10.1016/j.coviro.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog 6:e1001175. doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc Natl Acad Sci U S A 105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barajas D, Martin IF, Pogany J, Risco C, Nagy PD. 2014. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog 10:e1004087. doi: 10.1371/journal.ppat.1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalev N, Nagy PD. 2014. The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog 10:e1004051. doi: 10.1371/journal.ppat.1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barajas D, Xu K, de Castro Martin IF, Sasvari Z, Brandizzi F, Risco C, Nagy PD. 2014. Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog 10:e1004388. doi: 10.1371/journal.ppat.1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasvari Z, Izotova L, Kinzy TG, Nagy PD. 2011. Synergistic roles of eukaryotic translation elongation factors 1Bgamma and 1A in stimulation of tombusvirus minus-strand synthesis. PLoS Pathog 7:e1002438. doi: 10.1371/journal.ppat.1002438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang RY, Nagy PD. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Kovalev N, Pogany J, Nagy PD. 2014. Template role of double-stranded RNA in tombusvirus replication. J Virol 88:5638–5651. doi: 10.1128/JVI.03842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasanth KR, Barajas D, Nagy PD. 2015. The proteasomal Rpn11 metalloprotease suppresses tombusvirus RNA recombination and promotes viral replication via facilitating assembly of the viral replicase complex. J Virol 89:2750–2763. doi: 10.1128/JVI.02620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barajas D, Li Z, Nagy PD. 2009. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J Virol 83:11751–11764. doi: 10.1128/JVI.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J, Barajas D, Nagy PD. 2012. An inhibitory function of WW domain-containing host proteins in RNA virus replication. Virology 426:106–119. doi: 10.1016/j.virol.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Barajas D, Kovalev N, Qin J, Nagy PD. 2015. Novel mechanism of regulation of tomato bushy stunt virus replication by cellular WW-domain proteins. J Virol 89:2064–2079. doi: 10.1128/JVI.02719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Gonzalez PA, Sasvari Z, Kinzy TG, Nagy PD. 2014. Methylation of translation elongation factor 1A by the METTL10-like See1 methyltransferase facilitates tombusvirus replication in yeast and plants. Virology 448:43–54. doi: 10.1016/j.virol.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Barajas D, Xu K, Sharma M, Wu CY, Nagy PD. 2014. Tombusviruses upregulate phospholipid biosynthesis via interaction between p33 replication protein and yeast lipid sensor proteins during virus replication in yeast. Virology 471–473:72–80. doi: 10.1016/j.virol.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang C, Barajas D, Qin J, Nagy PD. 2014. Inactivation of the host lipin gene accelerates RNA virus replication through viral exploitation of the expanded endoplasmic reticulum membrane. PLoS Pathog 10:e1003944. doi: 10.1371/journal.ppat.1003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma M, Sasvari Z, Nagy PD. 2010. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol 84:2270–2281. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]