ABSTRACT
Highly pathogenic avian influenza viruses (HPAIVs) of hemagglutinin H5 and H7 subtypes emerge after introduction of low-pathogenic avian influenza viruses (LPAIVs) from wild birds into poultry flocks, followed by subsequent circulation and evolution. The acquisition of multiple basic amino acids at the endoproteolytical cleavage site of the hemagglutinin (HA) is a molecular indicator for high pathogenicity, at least for infections of gallinaceous poultry. Apart from the well-studied significance of the multibasic HA cleavage site, there is only limited knowledge on other alterations in the HA and neuraminidase (NA) molecules associated with changes in tropism during the emergence of HPAIVs from LPAIVs. We hypothesized that changes in tropism may require alterations of the sialyloligosaccharide specificities of HA and NA. To test this hypothesis, we compared a number of LPAIVs and HPAIVs for their HA-mediated binding and NA-mediated desialylation of a set of synthetic receptor analogs, namely, α2-3-sialylated oligosaccharides. NA substrate specificity correlated with structural groups of NAs and did not correlate with pathogenic potential of the virus. In contrast, all HPAIVs differed from LPAIVs by a higher HA receptor-binding affinity toward the trisaccharides Neu5Acα2-3Galβ1-4GlcNAcβ (3′SLN) and Neu5Acα2-3Galβ1-3GlcNAcβ (SiaLec) and by the ability to discriminate between the nonfucosylated and fucosylated sialyloligosaccharides 3′SLN and Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SiaLex), respectively. These results suggest that alteration of the receptor-binding specificity accompanies emergence of the HPAIVs from their low-pathogenic precursors.
IMPORTANCE Here, we have found for the first time correlations of receptor-binding properties of the HA with a highly pathogenic phenotype of poultry viruses. Our study suggests that enhanced receptor-binding affinity of HPAIVs for a typical “poultry-like” receptor, 3′SLN, is provided by substitutions in the receptor-binding site of HA which appeared in HA of LPAIVs in the course of transmission of LPAIVs from wild waterfowl into poultry flocks, with subsequent adaptation in poultry. The identification of LPAIVs with receptor characteristics of HPAIVs argues that the sialic acid-binding specificity of the HA may be used as a novel phenotypic marker of HPAIVs.
INTRODUCTION
Avian influenza is a highly contagious infection with influenza A viruses, with a worldwide occurrence in aquatic wild bird populations which represent the major natural hosts of these viruses. Influenza A viruses are classified into subtypes according to their surface glycoproteins: 18 hemagglutinin (HA) and 11 neuraminidase (NA) antigenic subtypes have been identified (1, 2, 3). Avian influenza viruses (AIVs) are subdivided into groups of high and low pathogenicity. The presence of multiple basic amino acids at the endoproteolytical cleavage site of the HA is a molecular indicator for high pathogenicity, at least for infections of gallinaceous poultry (4, 5, 6, 7, 8). Influenza viruses of HA subtypes H5, H7, and H9 are commonly identified in terrestrial gallinaceous poultry (9, 10). Only low-pathogenic AIVs (LPAIVs) of subtypes H5 and H7 naturally evolve into highly pathogenic AIVs (HPAIVs), which cause severe infections with high rates of mortality in poultry (11, 12, 13, 14) and can also be transmitted from birds to humans (15, 16, 17, 18).
The two viral glycoproteins, HA and NA, expressed on the surface of influenza virions play an important role in determining the pathogenic properties of the virus. Influenza virus infection is initiated by interactions between the viral HA and sialic acid-containing oligosaccharides on target cells. The NA cleaves off the terminal sialic acid residues from the host cell, promoting the release of virus progeny and preventing the formation of virus aggregates at the budding site. Furthermore, NA desialylates natural inhibitors of virus binding (such as mucins) and, thus, facilitates virus entry into target cells (19, 20). It was reported that NA expression directly enhances HA-mediated membrane fusion and infectivity (21). A functional match of HA and NA is a prerequisite for successful influenza virus infection and replication (20, 22, 23).
Influenza A viruses are transmitted occasionally from aquatic birds to other species. Rarely, they adapt to new hosts and circulate, forming stable host-specific virus lineages. An adaptation to sialic acid-containing receptors in a new host species may be required for successful interspecies transmission owing to host-specific differences in cell surface sialylation in target tissues of different species. Thus, it is well documented that a switch in the receptor specificity of the avian precursor is essential for the emergence of new stable virus lineages in pigs and humans (24, 25, 26, 27).
AIVs of domestic birds also originate from the natural reservoir in aquatic birds (28, 29, 30). Earlier, it was generally assumed for a long time that all avian viruses have similar receptor-binding specificities and, therefore, that there is no receptor-mediated restriction on interspecies transmission among different bird species (31, 32). This hypothesis was first challenged by the finding that HA and NA of H5 and H7 poultry viruses differ from those of aquatic birds by additional N-linked glycans at the top of HA and large deletions in the stalk of NA (33). These changes in HA and NA were detected in many independent lineages of poultry viruses. It was concluded that these changes may be a consequence of the adaptation of viruses of aquatic birds to cellular receptors in domestic gallinaceous birds (12, 30, 33, 34).
It was also shown that influenza viruses of different avian species may differ in their fine HA receptor-binding properties such as recognition of the inner parts (cores) of the carbohydrate chains, including differences in recognition of sulfated and/or fucosylated carbohydrate structures (35, 36, 37, 38, 39, 40). These findings suggested that sialic acid receptors in different birds are not identical and that distinctions in receptors determine differences in the viral fine HA receptor specificity.
Sporadically, LPAIVs of poultry-adapted H5 or H7 AIVs evolve into HPAIVs, usually through acquisition of multiple basic amino acids at the cleavage site of the HA (reviewed in reference 41). Typically, LPAIVs cause asymptomatic infections in wild aquatic birds, but when they are introduced into domestic poultry, infection may be asymptomatic or produce clinical signs and lesions in the respiratory, digestive, and reproductive systems. In contrast, the HPAIVs cause high morbidity and mortality in gallinaceous poultry, producing systemic disease with necrosis and inflammation in multiple visceral organs, nervous and cardiovascular systems, and the integument (42). Thus, HPAIVs and LPAIVs possess different tissue tropisms. Apart from the well-studied significance of the multibasic HA cleavage site, there is only limited knowledge of other alterations in the HA and NA molecules associated with changes in tropism during the emergence of HPAIVs from LPAIVs. We hypothesized that changes in tropism may require alterations in the sialyloligosaccharide specificities of HA and NA. To test this hypothesis, we analyzed HA receptor-binding and NA receptor-destroying activities of a panel of HPAIVs and LPAIVs using a set of synthetic receptor analogs, α2-3-sialylated glycopolymers, and determined that alteration of the receptor-binding specificity accompanies emergence of the HPAIVs from their low-pathogenic precursors.
MATERIALS AND METHODS
Materials.
Ninety-six-well polyvinyl chloride microtiter plates were obtained from Corning (USA), and horseradish peroxidase-streptavidin conjugate (Strept-POD) and 2,2′-azino-di-(3-ethylbenzthiazolinesulfonic acid) (ABTS) were obtained from Roche Diagnostics GmbH (Germany). High-molecular-mass (∼1,000 kDa) biotinylated sialylglycopolymers (SGPs), which are analogs of natural influenza virus receptors (43, 44), and SGPs with ω-aminoglycoside spacers and boron-dipyrromethene (BODIPY)-labeled SGPs were synthesized as described previously (45, 46, 47, 48). The structures and designations of their oligosaccharide moieties are presented in Fig. 1.
FIG 1.
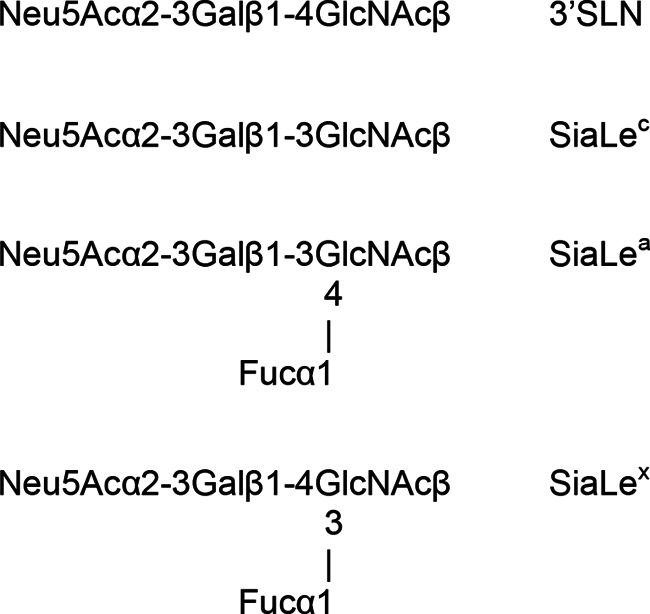
Structure of sialyloligosaccharide moieties of biotinylated sialylglycopolymers and BODIPY-labeled neuraminidase substrates.
DEAE-Toyopearl 650M was obtained from Tosoh Bioscience GmbH (Germany). Black polystyrene 96-well microtiter plates were purchased from Nunc (Denmark), and thin-well 0.2-ml Thermo microtubes were from ABgene (United Kingdom).
Viruses.
The viruses listed in Fig. 2 were from the repositories of the following: Friedrich Loeffler Institute, Riems, Germany; Federal Institute for Health Protection of Consumers and Veterinary Medicine, Berlin, Germany; Institute of Virology, Philipps University, Marburg, Germany; and Robert Koch Institute, Berlin, Germany. Earl Brown (University of Ottawa, Ottawa, Canada) kindly provided the A/turkey/Ontario/6213/66 virus strain. The viruses were grown in 9-day-old embryonated chicken eggs. All HPAIVs were inactivated by treatment with beta-propiolactone (0.2% for 16 h at 4°C) or formalin (0.02% for 72 h at 37°C). Work with highly pathogenic isolates was performed under biosafety level 3 (BSL-3) conditions. To assess the effect of inactivation on the properties of the viruses, we prepared two H5 LPAIVs and divided them into two portions; one was treated with beta-propiolactone, and the other was used as a native control.
FIG 2.
Avian influenza viruses used and evolutionary relationships of H5 and H7 subtype avian influenza viruses. A phylogenetic tree for amino acid sequences of the H5 and H7 HA1 protein was generated by the minimum-evolution method using MEGA software, version 5.2. The scale bar represents 0.01 or 0.005 units of amino acid per site for H5 or H7, respectively; black circles indicate LPAIVs. The tree is based on HA sequences presented in this study GenBank accession numbers are given (ns, not sequenced).
For virus purification, allantoic fluid was first clarified by low-speed centrifugation. The supernatant was layered on a top of 30% sucrose prepared in TN buffer (0.1 M NaCl, 0.02 M Tris, pH 7.2). The virus then was pelleted by high-speed centrifugation, resuspended in TN buffer containing 50% glycerol, and stored at −20°C.
Solid-phase receptor-binding assay.
Receptor-binding specificity of HA was investigated using a direct binding assay as described previously (49, 50). In brief, 96-well polyvinyl chloride microplates were coated with purified virus (40 μl/well) with a titer of 16 hemagglutinating units (HAU) for 16 h at 4°C. The plates were then washed with ice-cold washing buffer ([WB] 0.01% Tween 80 in phosphate-buffered saline [PBS]). Serial 2-fold dilutions of biotinylated sialylglycopolymers in reaction buffer ([RB] 0.02% Tween 80–0.02% bovine serum albumin in PBS) containing a 10 μM concentration of the neuraminidase inhibitor 2,2-didehydro-2,4-didesoxy-4-amino-N-acetyl-d-neuraminic acid were added into the wells (20 μl/well), and the plates were incubated at 4°C for 2 h. After the plates were washed, Strept-POD solution in RB (100 mU/ml) was added at 25 μl/well, and the plates were incubated at 4°C for 1 h. After the plates were washed, the peroxidase activity in the wells was assayed with ABTS (Roche Diagnostics GmbH) substrate solution. The optical density at 405 nm was measured using a Tecan reader (Tecan Group, Ltd., Switzerland). The affinity constants (Kaffs) were determined from slopes of Scatchard plots using micromolar concentrations of sialic acid for the calculations (49, 51). The data are presented as Kaff μM−1 ± standard deviation (SD). Mean values were calculated based on three independent experiments performed on different days.
Substrate specificity of influenza virus NA.
The fluorescent assay for studying the substrate specificity of NA was described previously (48, 52, 53). Purified influenza virus (3 μl; 1 to 20 HAU) was added to a microtube containing 0.7 nmol of a BODIPY-labeled sialyloligosaccharide in 4 μl of 0.1 M Na-acetate buffer (pH 5.0). The microtube was shaken at 37°C for the appropriate time period (10 to 20 min). To stop the reaction, the tube was heated at 70°C for 10 min, and the mixture was diluted with 63 μl of distilled water. The obtained solution was analyzed immediately or stored at −20°C. The reaction mixture consisted of the neutral BODIPY-labeled oligosaccharide and negatively charged BODIPY-labeled uncleaved substrate. Neutral BODIPY-labeled product was separated using DEAE-Toyopearl anion exchanger microcartridges which were prepared as described previously (48). A 20-μl aliquot of the diluted reaction mixture was loaded onto a moist surface of Toyopearl in the microcartridge and gently forced through it into a well of a 96-well black plate. After that the microcartridge was washed with 450 μl of water (in three portions of 150 μl each), and the eluate was collected into three wells of the plate. Then the microcartridge was washed with 450 μl of 0.5 M sodium acetate buffer, and the eluate was collected into the next three wells. A negative control, the mixture containing 3 μl of TN buffer instead of the virus suspension, was processed in the same way. Fluorescence was measured at 485/535 nm using a Tecan reader (Tecan Group, Ltd., Switzerland). The values of the aqueous eluates give the yield of the formed product (IP), whereas the same procedure for buffer eluates gives the amount of a nonreacted substrate (IS). The yield of the enzymatic reaction (Y) and the concentration of the obtained product (C) can be calculated from the equations Y = (IP × 100)/(IP + IS) and C = (IP × S0)/(IP + IS), respectively, where S0 is the initial molar concentration of the substrate. The assay conditions (virus concentrations in reaction mixtures and incubation time) were selected in such a way that it was possible to evaluate an initial rate of an enzyme reaction from a linear range of the product accumulation-versus-time plot. Virus NA activity for each sialoside was calculated as the slope of the linear region of the V0-versus-S0 kinetic curve [where V0 is the initial rate of the desialylation, calculated as V0 = (IP × S0)/(IP + IS)T, where T is incubation time]. Substrate specificity of each virus NA was determined by comparison of V0/S0 values obtained for all substrates under the same conditions. Data are presented as (V0/S0 ± SD)103 per min per HAU of virus. The mean values of V0/S0 were calculated from two or three independent experiments.
Sequencing.
PCR products of the amplified HA gene of AIVs were sequenced by automated nucleotide cycle sequencing (sequences of primers are available on request). PCR products were purified using an MSB Spin PCRapace kit (Invitek) and sequenced using a BigDye Terminator, version 3.1, cycle sequencing kit and a 3130xl capillary sequencer (Applied Biosystems, Darmstadt, Germany). The sequences were analyzed and edited using the BioEdit sequence alignment editor, version 7.0.9.0 (Tom Hall, Ibis Biosciences, Carlsbad, CA). Phylogenetic trees were generated based on amino acid sequences of H5 and H7 HAs determined in this study using MEGA software, version 5.2, with the minimum-evolution method.
Molecular model.
For modeling we used atomic coordinates of influenza virus A/Vietnam/1194/2003 (H5N1) HA complex with Neu5Acα2-3Galβ1-4GlcNAcβ (3′SLN; Protein Data Bank [PDB] accession number 4BGY) (54). Molecular models were generated using DS Viewer Pro, version 5.0 (Accelrys Inc.).
Statistical analyses.
Differences in the affinity constants (Kaffs) as well as in the ratios of Kaff(3′SLN)/Kaff[Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SiaLex)] between two groups of viruses, HPAIVs and LPAIVs, were determined using an unpaired two-tailed t test.
Nucleotide sequence accession numbers.
The HA gene nucleotide sequences have been deposited in the GenBank database under accession numbers CY107842 to CY107861 and KM190930 to KM190932.
RESULTS
HA receptor-binding and NA substrate specificities toward Neu5Acα2-3Gal-terminated SGPs were evaluated for 27 AIVs (13 HPAIVs and 14 LPAIVs) of different subtypes (Fig. 2). High-molecular-mass (∼1,000 kDa) SGPs were used in order to mimic the multivalent interactions between virus and receptors on the cell surface (43, 44). For receptor-binding assays, we used a set of SGPs containing different sialyloligosaccharides that allowed us to evaluate the affinities of AIVs for SGPs with β1-3- or β1-4-linkage in the carbohydrate core of Neu5Acα2-3Gal-terminated oligosaccharides (β1-3GlcNAcβ in SiaLec and β1-4GlcNAcβ in 3′SLN) as well as for their fucosylated analogs [β1-3(Fucα1-4)GlcNAcβ (SiaLea) and β1-4(Fucα1-3)GlcNAcβ (SiaLex), respectively]. The same monovalent sialyloligosaccharides labeled with the fluorescent dye BODIPY FL were used for evaluation of the substrate specificity of the influenza virus NAs.
HPAIVs possess enhanced receptor-binding specificity for 3′SLN and SiaLec compared to that of LPAIVs.
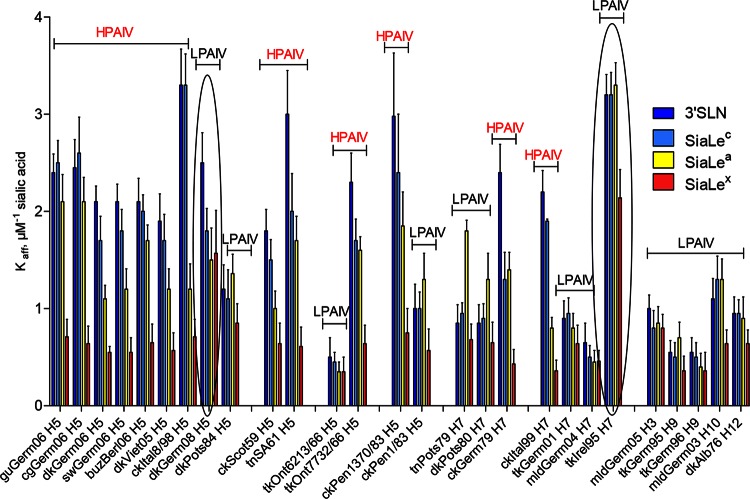
All HPAIVs had a high affinity for SGPs containing 3′SLN and SiaLec moieties (Fig. 3). In comparison with HPAIVs, all LPAIVs except tkIrel95 (H7N7) (explanations of virus abbreviations are given in Fig. 2) and dkGerm08 (H5N3) displayed substantially lower (from 1.5 to 9.4 times) receptor-binding affinity for both 3′SLN and SiaLec (Fig. 3). Two LPAIVs, tkIrel95 and dkGerm08, possessed receptor binding affinities for both SGPs similar to those of HPAIVs.
FIG 3.
Receptor-binding specificity of avian influenza viruses. The data are presented as affinity constants of virus in complexes with sialyloligosaccharides (Kaff ± SD μM−1 sialic acid). Viruses are grouped according to their subtypes and evolutionary relationships. The two circled LPAIVs were found to be exceptions as they revealed significantly higher binding affinities than other LPAIVs. Differences in the affinity constants (Kaffs) between two groups of viruses, HPAIVs and LPAIVs, were statistically significant (P < 0.0001) and were determined using an unpaired two-tailed t test. See Fig. 2 for virus abbreviations.
The affinities of 6 out of 13 HPAIVs (dkGerm06, swGerm06, dkViet05, ckItal8/98, ckScot59, and ckItal99) for the fucosylated SGP SiaLea declined compared with the affinities to the nonfucosylated counterpart, SiaLec (Fig. 3). Five LPAIVs (tnPots79, dkPots80, dkPots84, tkGerm95, and ckPen1/83) displayed higher (1.3 to 2.9 times) affinities for SiaLea than toward SiaLec. The affinities of the rest of the LPAIVs differed only slightly, if at all, compared with those for SiaLec (Fig. 3).
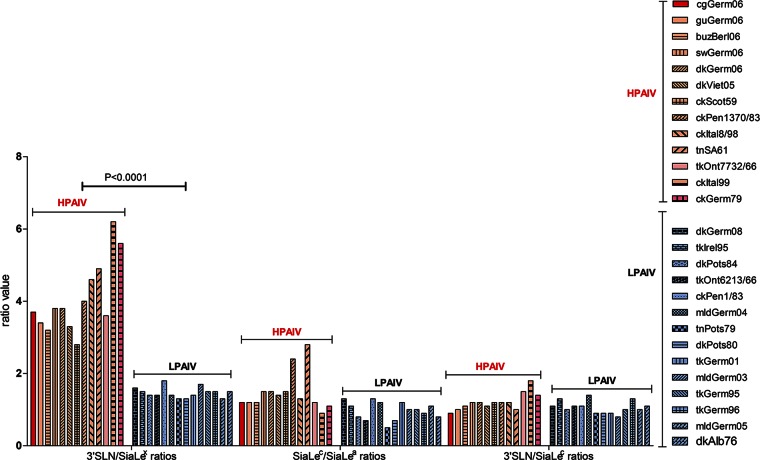
The affinity of all AIVs for fucosylated SGP SiaLex was lower (1.3 to 6.1 times) than that for the nonfucosylated counterpart 3′SLN (Fig. 3). Remarkably, despite the variation in absolute values of binding affinity, a strong correlation was observed between the virus pathogenic phenotype and the pattern of binding to 3′SLN and its fucosylated analog SiaLex. Indeed, when receptor-binding specificity was plotted as the Kaff(3′SLN)/Kaff(SiaLex) ratio, the values for all HPAIVs ranged between 3 and 6, whereas for LPAIVs these values were only about 1.5 (Fig. 4). In other words, HPAIVs increase binding to 3′SLN, compared to the values of LPAIVs, but the binding to SiaLex remains the same.
FIG 4.
Relative binding affinities of the viruses for 3′SLN versus SiaLex, SiaLec versus SiaLea, and 3′SLN versus SiaLec, determined Kaff(3′SLN)/Kaff(SiaLex), Kaff(SiaLec)/Kaff(SiaLea), and Kaff(3′SLN)/Kaff(SiaLec), respectively. Red indicates HPAIVs, and blue indicates LPAIVs. Differences in the ratio values of Kaff(3′SLN)/Kaff(SiaLex) between two groups of viruses, HPAIVs and LPAIVs, were statistically significant (P < 0.0001) and were determined using unpaired two-tailed t test. See Fig. 2 for virus abbreviations.
No correlation between pathogenicity and the ability to discriminate 3′SLN versus SiaLec or SiaLec versus SiaLea was found (Fig. 4).
Enhanced receptor-binding of HPAIVs for 3′SLN is provided by amino acid substitutions in the RBS.
Although the HA sequences of some of the viruses used in our study were available from the GenBank, we sequenced independently all HA genes of H5 and H7 subtype viruses to account for potential passaging-related distinctions (accession numbers are given in Fig. 2). Our focus was on HPAIVs and LPAIVs of these subtypes since only LPAIVs of H5 and H7 subtypes naturally evolve into HPAIVs. To understand the molecular basis of differences in the specific receptor-binding patterns of HPAIVs and LPAIVs, we analyzed their HA sequences at amino acid positions 110 to 240, which form the receptor-binding site (RBS), and the location of observed amino acid substitutions on the X-ray diffraction data model of H5 HA complex with 3′SLN (PDB accession number 4BGY) (54) (Fig. 5 and 6).
FIG 5.
Partial amino acid sequences of H5 and H7 in viruses (H3 numbering). Positions described in the text are highlighted in yellow. The RBS is marked in gray. HPAIVs are marked with asterisks. Additional glycosylation sites are marked with circles. The figure was generated using BioEdit. See Fig. 2 for virus abbreviations.
FIG 6.
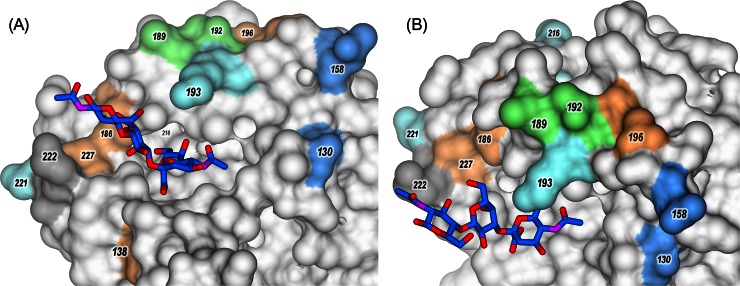
Molecular models of the H5 HA of the influenza virus A/Vietnam/1194/2004 complexed with 3′SLN (PDB accession number 4BGY). Panels A and B show different views of the same model. Sialosides are shown in stick representation, with carbon in blue, oxygen in red, and nitrogen in violet. HA is shown in surface representation; amino acids in positions discussed in the text (H3 HA numbering) are shown in color.
Comparison of H5 HA sequences of HPAIVs and LPAIVs.
Evolutionary relationships of the HA of AIVs included here were generated using amino acid sequences of the HA protein. Three groups of related H5 viruses were compared (Fig. 2 and 5A).
In the first group, ckPen1/83 appeared in chickens in Pennsylvania in April 1983 and subsequently became virulent in October 1983. Viruses isolated in April and October were very closely related, and genetic reassortment had not occurred during the development of virulence (55). The receptor-binding affinity of HPAIV ckPen1370/83 for 3′SLN was about 3-fold higher than that for LPAIV ckPen1/83, but both viruses displayed comparable affinities for fucosylated SGPs (Fig. 3). The HA sequence of the HPAIV differed from that of LPAIV by the substitutions Glu189Ala and Ala192Glu (Fig. 5A and 6). We conclude that these two substitutions in the RBS were responsible for the enhanced binding of ckPen1370/83 to 3′SLN.
Two viruses, tkOnt6213/66 and tkOnt7732/66, were isolated during similar disease episodes in turkeys occurring in a relatively restricted area in Ontario, Canada. The LPAIV tkOnt6213/66 was isolated in January 1966, and the HPAIV tkOnt7732/66 was isolated in March 1966. The HPAIV tkOnt7732/66 was serologically related to LPAIV tkOnt6213/66 (56, 57). Receptor-binding affinities of the HPAIV for 3′SLN and SiaLex were about 5- and 2-fold higher than those of the LPAIV, respectively. The HAs have six amino acid differences at positions 130, 138, 186, 193, 196, and 227 inside the RBS and in close proximity, which could account for the differences in receptor-binding specificity (Fig. 5A and 6). Moreover, two substitutions generate two additional glycosylation sites in the HPAIV HA in positions 130 to 133 and 158 to 160 (because of Asn130 and Ser160) (Fig. 5A and 6).
Last, we compared amino acid differences between HPAIVs and LPAIVs isolated in Germany in 2006, 2008, and 1984 (Fig. 5A). Two LPAIVs differed only in one position in the RBS, that is, Pro221 in dkPost84 and Ser221 in dkGerm08. LPAIV dkGerm08 was found to be a pronounced exception from LPAIVs as the exhibited HA affinity for 3′SLN was similar to that of HPAIV, but the virus bound SiaLex at a level 2-fold stronger than HPAIV levels. Thus, the presence of Ser221 with a hydrophilic and flexible back chain instead of the rigid Pro in HA of LPAIV dkPots84 (Fig. 6) is most likely responsible for the high affinity of LPAIV dkGerm08 and all HPAIVs toward nonfucosylated sialosides.
Comparison of the HA sequences of HPAIVs with the low-pathogenic virus dkGerm08 revealed three amino acid differences in the vicinity of the HA RBS (Fig. 5A). HAs of HPAIVs possess Glu130, Arg193, and Lys216, whereas the HA of dkGerm08 has Asp130, Lys193, and Glu216 (Fig. 5A and 6). These differences are thus responsible for the differences in the receptor specificities of HPAIVs and LPAIV dkGerm08.
Comparison of H7 HA sequences of HPAIVs and LPAIVs.
We compared HA sequences of HPAIVs and LPAIVs, namely, the viruses isolated in Germany in 1979 to 1980 and the European viruses of 1995 to 2004 (Fig. 5B).
Two HPAIVs, ckGerm79 and ckItal99, displayed the highest affinity for 3′SLN and the lowest affinity for SiaLex. One of the LPAIVs, namely, tkIrel95, possessed a high affinity for all SGPs investigated (Fig. 3). The receptor-binding affinity of HPAIV ckGerm79 for 3′SLN was about 3-fold higher than that for two LPAIVs, tnPots79 and dkPots80, but the HPAIV and both LPAIV viruses displayed comparable affinities for fucosylated SGPs (Fig. 3). The HPAIV ckGerm79 differed from both LPAIVs at two positions in the vicinity of the HA RBS (Fig. 5B). The HAs of LPAIVs possess amino acid residue Gly136, while the HA of the HPAIV has Asn136. The second feature of HPAIV HA is the presence of a negatively charged residue, Glu218, instead of Gly218 in the HAs of LPAIVs. This finding is consistent with previously reported data (58), in which the variant of X31 virus (H3N2) containing the single amino acid substitution Gly218Glu in the HA possessed the highest affinities for the α2-3 sialyloligosaccharides. Notably, the HA of LPAIV tkIrel95, which displayed high affinities for all SGPs, also has Glu218.
The receptor-binding affinity of HPAIV ckItal99 for 3′SLN was about 3-fold higher than the affinities of the LPAIVs tkGerm01 and mldGerm04, but the HPAIV and both LPAIV viruses displayed comparable affinities for fucosylated SGPs (Fig. 3). The HA of HPAIV ckItal99 has the following amino acid substitutions compared to the LPAIVs: Ala134Thr and Ala137Thr in the 130-loop and Ala219Glu in the 220-loop (Fig. 5B). Moreover, Ala134Thr in the HPAIV HA generates a potential glycosylation site at Asn132.
To predict the genetic and structural basis of different receptor-binding patterns of the H7 HPAIVs and LPAIVs (Fig. 3), we compared HAs of LPAIV tkIrel95 and HPAIV ckItal99 (Fig. 5B). The HA of tkIrel95 differed from that of ckItal99 by several amino acid substitutions in the region of the RBS (Fig. 5B); among them were Pro185Ser, Thr189Ala, and Gly218Glu. The tkIrel95 virus displayed significantly higher avidity for fucosylated SGPs than the HPAIV (as well as the other LPAIVs investigated), which possessed typically for H7 in positions 185 to 189 small amino acid residues (Ser-Gly-Ser-Thr-Thr). This observation is in good agreement with data reported earlier (40). It was suggested that the “left” side of the RBS of H7 HA (conserved amino acids in positions 185 to 189) is responsible for recognition of fucosylated receptors. Moreover, the rigid Pro185, which may radically change the RBS architecture, is an atypical substitution for H7 HAs. It was reported that the sequence analysis of about 70 HAs of H7 influenza viruses isolated in Eurasia and America from aquatic and terrestrial birds revealed Ser residues at position 185, without exception, in all viruses analyzed (40). These features of the tkIrel95 HA likely determine its enhanced receptor-binding affinity for α2-3 SGPs, including fucosylated ones (Fig. 3).
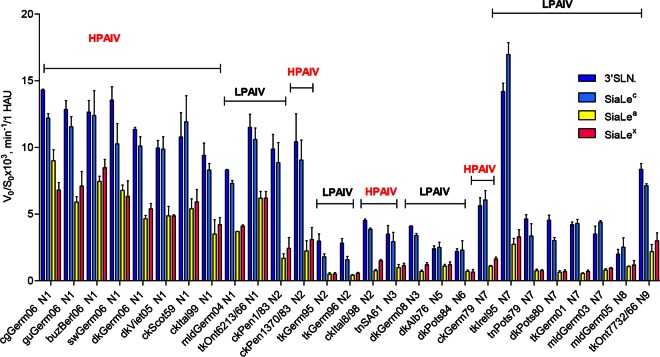
Substrate specificity of the NA does not correlate with pathogenic potential of the virus.
All AIVs hydrolyzed SiaLex and SiaLea much more slowly (1.4- to 8-fold) than their nonfucosylated counterparts 3′SLN and SiaLec, respectively. N1 subtype NAs of both HPAIVs and LPAIVs possessed high sialidase activity toward the nonfucosylated sialyloligosaccharides 3′SLN and SiaLec (Fig. 7). N2 NAs of HPAI virus ckPenn1370/83 and LPAI virus ckPenn1/83 showed identical and high sialidase activities toward 3′SLN and SiaLec (Fig. 7).
FIG 7.
NA substrate specificity of the avian influenza viruses was calculated for each sialoside as the slope of the linear region of the V0-versus-S0 kinetic curve (V0, initial rate of the desialylation; S0, initial substrate concentration). Substrate specificity of each virus NA was determined by comparison of V0/S0 values obtained for all substrates under the same conditions. Colors indicate the sialyloligosaccharide moiety of the SGP. Data are presented as indicated on the y axis, calculated for 1 HAU of virus. See Fig. 2 for virus abbreviations.
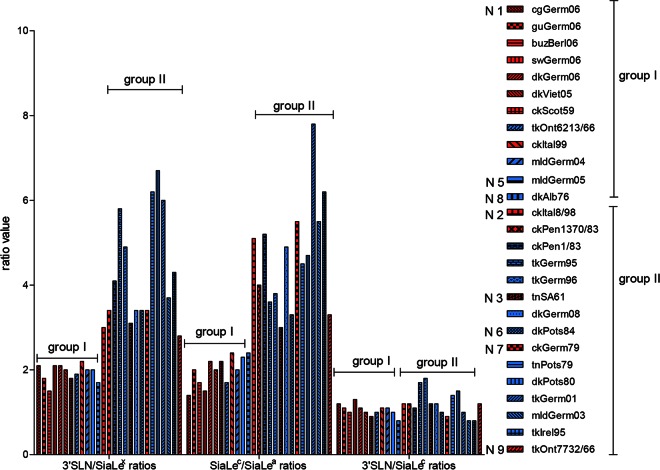
When substrate specificity was plotted as the 3′SLN/SiaLex ratio, all viruses were divided into two groups (Fig. 8). For AIVs (both HPAIVs and LPAIVs) with N1, N5, and N8 NAs, the ratio was between 1.5 and 2, while for those bearing N2, N3, N6, N7, and N9 NA subtypes, the ratio was significantly higher and varied between 3 and 7. Analysis of the SiaLec/SiaLea ratio revealed the same pattern (Fig. 8).
FIG 8.
Profiles of NA substrate specificities of the avian influenza viruses as 3′SLN/SiaLex, 3′SLN/SiaLec, and SiaLec/SiaLea substrate specificity ratios. Red indicates HPAIVs, and blue indicates LPAIVs.
It is known that NAs of the influenza type A viruses form two genetically and structurally distinct groups. The major structural difference between group I and group II is a large cavity adjacent to the active site in group I but not present in group II NAs (59). N1, N5, and N8 NAs belongs to group I, and NAs of the N2, N3, N6, N7, and N9 subtypes belong to group II. Thus, the profile of the substrate specificity of the NA correlated with the NA subtype and did not correlate with the pathogenic potential of the virus.
DISCUSSION
In this study, we compared receptor-binding specificities and sialidase substrate specificities of HPAIVs and LPAIVs of H5 and H7 subtypes and found that all HPAIVs investigated differed from LPAIVs by having higher HA receptor-binding affinities toward the trisaccharides 3′SLN and SiaLec.
Earlier it was reported that the most prominent feature of H5 poultry viruses is strong binding to 6-sulfo-3′SLN, and H7 viruses from all species were characterized by much stronger binding to 6-sulfo-3′SLN and 6-sulfo-SiaLex than to nonsulfated SGPs (36, 37, 39, 40). No obvious differences between HPAIVs and closely related LPAIVs were noticed. Here, we have focused on nonsulfated receptors and for the first time found correlations of receptor-binding properties of the HA with a highly pathogenic phenotype of poultry viruses. The analysis of the evolutionarily related groups of H5 and H7 viruses suggested that changes in the receptor-binding characteristics occurred in the course of the emergence of the HPAIVs from their low-pathogenic precursors. For example, low-pathogenic virus ckPen1/83 circulated for some months in poultry before evolving into the highly pathogenic variant ckPen1370/83 (60, 61). The latter virus differed from the ancestor virus by two amino acid substitutions in the RBS of the HA and by enhanced binding affinity for 3′SLN and SiaLec. Similarly, LPAIV tkOnt6213/66 and HPAIV tkOnt7732/66 isolated a few months later differed by amino acid substitutions in the vicinity of the RBS and by their receptor specificities. Thus, both HPAIVs differed from their low-pathogenic precursors by their receptor-binding properties.
Noteworthy, the HA of HPAIV ckItal99 possessed a potential glycosylation site at the Asn132, and its affinity for 3′SLN was higher than the affinity of HAs expressed by closely related LPAIVs tkGerm01 and mldGerm04. In previous studies it was postulated that a mutation in the H7 HA of an HPAIV isolated from a fatal human case (H7N7 virus A/Netherlands/219/2003) which introduces a potential glycosylation site at Asn132 (because of Thr134) contributed to the pathogenic properties of this virus (62, 63). Moreover, it was demonstrated later that the additional glycosylation site at this position of H7 HA significantly increases the binding affinity for 3′SLN (64). Data obtained are in a good agreement with these reports and support an assumption that the occurrence of a potential glycosylation site in the HA correlates with the receptor-binding properties of the HA and the highly pathogenic phenotype of the virus.
Comparison between fucosylated and nonfucosylated SGPs revealed that the affinities of all viruses tested for SiaLex were lower than their affinities for 3′SLN. These results are in accordance with data reported previously (38, 39). Namely, all duck viruses of the H1, H2, H3, H4, H5, H8, H9, H10, H11, and H14 subtypes demonstrated low affinity for SiaLex. Using molecular modeling authors of the previous studies suggested that partial overlap of the fucose moiety with a bulky amino acid (Arg, Lys, Thr, Leu, or Gln) in position 222 could be a general mechanism that reduces the capability of duck viruses to bind fucosylated receptors. Comparison of the HA sequences of all viruses investigated here showed that they possessed Lys222, Gln222, or Leu222. We assume that most AIVs with the bulky amino acid at position 222 have a reduced capability to bind fucosylated receptors.
In summary, our data show that all HPAIVs differed from LPAIVs by higher HA receptor-binding affinities toward the trisaccharides 3′SLN and SiaLec. The functional HA-NA balance is known to be essential for efficient replication of influenza viruses (20). We speculate therefore that enhanced binding of HPAIV to 3′SLN and SiaLec might be compensated by enhanced desialylation of these substrates by the NA. In this study, we determined substrate specificity of the NA as a ratio of enzymatic activities toward different substrates and found that substrate specificity did not correlate with the pathogenic potential of the virus. We did not determine absolute NA activities as this would require knowledge of the NA concentration in a reaction mixture, which is a difficult task. Further studies on the absolute activities of the NA of HPAIV and LPAIV are required to describe precisely the functional balance between NA and HA of these viruses.
Interestingly, two LPAIVs, dkGerm08 (H5N3) and tkIrel95 (H7N7), differed from other LPAIVs tested. These viruses revealed significantly higher HA receptor-binding affinities than other LPAIVs for 3′SLN and SiaLec, as well as for their fucosylated counterparts. HAs of each of these viruses have unique amino acids in the RBS. These features of the HA structure correlate with their high affinity for a wide range of α2-3 sialyloligosaccharides. We speculate that such a molecular characteristic and a high receptor-binding affinity of HA for most α2-3 sialosides might predispose this virus, in its low-pathogenicity phenotype, to act as a progenitor of a newly emerging HPAIV. Interestingly, both of these atypical LPAIVs were found to be nonpathogenic for chickens, with an intravenous pathogenicity index (IVPI) of 0.0 (data not shown). It will be of significant interest to test whether these viruses have an extended tissue tropism similar to that of HPAIVs and/or are more pathogenic for poultry than typical LPAIVs.
It is well known that HPAIVs and LPAIVs possess different tissue tropisms: LPAIVs cause a localized infection while HPAIVs cause generalized infection (reviewed in references 65 and 66). The extended tissue tropism of the HPAIVs (such as the ability to replicate in endothelial cells) compared to that of LPAIVs is provided by the acquisition of the multibasic cleavage site by the HPAIVs (67). We suppose that the ability of HPAIVs to replicate in a broader spectrum of cells may account for the gradual changes of the viral receptor-binding specificity. Alternatively, alterations of the receptor specificity could occur before the acquisition of the multibasic cleavage site and of the high cleavability of the HA. Our finding of two LPAIVs with receptor characteristics of HPAIVs would agree with the second scenario.
Thus, the findings allow us to conclude that acquisition of the highly pathogenic phenotype is accompanied by enhanced receptor-binding affinity first of all for 3′SLN, which is a typical poultry-like receptor. It was reported that the typical feature of poultry-adapted viruses is preferential binding to receptors having a β1-4 bond between the Neu5Acα2-3Gal moiety and the next sugar residue, such as Neu5Acα2-3Galβ1-4GlcNAc (3′SLN) (26, 36, 37, 39). We believe that enhanced binding to 3′SLN is provided by substitutions in the RBS of HA, which appeared in the HAs of LPAIVs in the course of transmission of LPAIVs from wild waterfowl into poultry flocks, with subsequent circulation and evolution/adaptation in poultry. The identification of LPAIVs with the receptor characteristics of HPAIVs described here argues that the sialic acid-binding specificity of the HA may be used as a novel phenotypic marker of HPAIVs.
ACKNOWLEDGMENTS
We thank Earl Brown (Ottawa, Canada) for providing the A/turkey/Ontario/6213/66 virus strain. We thank Orla Flynn, Central Veterinary Research Laboratory, Virology Division, Department of Agriculture, Food and the Marine Laboratories, Backweston, Celbridge, County Kildare, Ireland, for providing the IVPI for virus A/turkey/Ireland/PV8/95, H7N7. We thank Heidemarie Lehmann, Birgit Troschke, and Sven Tietze for excellent technical assistance.
The work was supported by a Molecular and Cell Biology grant of the Presidium of Russian Academy of Sciences and by the grant Immunogenicity and Protective Efficacy of Intranasal delNS1(H5N1) Influenza Vaccine SP5B-CT-2007-044512 (grant 01EZ0917) of the German Ministry for Education and Research and the European Union 7th Framework Programme (FP7/2007-2013) under grant agreement 278433-PREDEMICS.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus ADME. 2005. Characterization of a novel influenza a virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. 2014. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Organisation for Animal Health (OIE). 2005. Avian influenza, chapter 2.7.12. In OIE manual of diagnostic tests and vaccines for terrestrial animals, 5th ed World Organisation for Animal Health (OIE), Paris, France. [Google Scholar]
- 5.World Organisation for Animal Health (OIE). 2007. Avian influenza, chapter 2.7.12. In Terrestrial animal health code, 16th ed World Organisation for Animal Health, Paris, France: http://www.oie.int/doc/ged/d6430.pdf. [Google Scholar]
- 6.Fereidouni SR, Harder TC, Starick E. 2008. Rapid pathotyping of recent H5N1 highly pathogenic avian influenza viruses and of H5 viruses with low pathogenicity by RT-PCR and restriction enzyme cleavage pattern (RECP). J Virol Methods 154:14–19. doi: 10.1016/j.jviromet.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Gall A, Hoffmann B, Harder T, Grund C, Höper D, Beer M. 2009. Design and validation of a microarray for detection, hemagglutinin subtyping, and pathotyping of avian influenza viruses. J Clin Microbiol 47:327–334. doi: 10.1128/JCM.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinhauer DA. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1–201. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 9.Brown IH, Banks J, Manvell RJ, Essen SC, Shell W, Slomka M, Londt B, Alexander DJ. 2006. Recent epidemiology and ecology of influenza A viruses in avian species in Europe and the Middle East. Dev Biol (Basel) 124:45–50. [PubMed] [Google Scholar]
- 10.Capua I, Alexander DJ. 2009. Avian influenza infection in birds: a challenge and opportunity for the poultry veterinarian. Poult Sci 88:842–846. doi: 10.3382/ps.2008-00289. [DOI] [PubMed] [Google Scholar]
- 11.Capua I, Mutinelli F, Marangon S, Alexander DJ. 2000. H7N1 avian influenza viruses in Italy (1999 to 2000) in intensively reared chickens and turkeys. Avian Pathol 29:537–543. doi: 10.1080/03079450020016779. [DOI] [PubMed] [Google Scholar]
- 12.Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander DJ. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch Virol 146:963–973. doi: 10.1007/s007050170128. [DOI] [PubMed] [Google Scholar]
- 13.Harder TC, Werner O. 2006. Avian influenza. In Kamps B, Hoffmann C, Preiser W (ed), Influenza report 2006 Flying Publisher, Paris, France: http://influenzareport.com/ir/ai.htm. [Google Scholar]
- 14.Lee CW, Lee YJ, Senne DA, Suarez DL. 2006. Pathogenic potential of North American H7N2 avian influenza virus: a mutagenesis study using reverse genetics. Virology 353:388–395. doi: 10.1016/j.virol.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A 97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uyeki TM. 2009. Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin Infect Dis 49:279–290. doi: 10.1086/600035. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich M, Matrosovich T, Gray T, Roberts NA, Klenk H-D. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner R, Matrosovich M, Klenk H. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 21.Su B, Wurtzer S, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, Clavel F, Labrosse B. 2009. Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA). PLoS One 4:e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaverin N. 2010. Postreassortment amino acid substitutions in influenza A viruses. Future Microbiol 5:705–715. doi: 10.2217/fmb.10.43. [DOI] [PubMed] [Google Scholar]
- 23.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74:6015–6020. doi: 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baigent SJ, McCauley JW. 2003. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. Bioessays 25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- 25.Horimoto T, Kawaoka Y. 2001. Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev 14:129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matrosovich MN, Gambaryan AS, Klenk H-D. 2008. Receptor specificity of influenza viruses and its alteration during interspecies transmission, p 134–155. In Klenk H-D, Matrosovich MN, Stech J (ed), Avian influenza, vol 27 Karger, Basel, Switzerland. [Google Scholar]
- 27.Malik Peiris JS. 2009. Avian influenza viruses in humans. Rev Sci Tech 28:161–173. [DOI] [PubMed] [Google Scholar]
- 28.Capua I, Alexander DJ. 2006. The challenge of avian influenza to the veterinary community. Avian Pathol 35:189–205. doi: 10.1080/03079450600717174. [DOI] [PubMed] [Google Scholar]
- 29.Röhm C, Horimoto T, Kawaoka Y, Süss J, Webster RG. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664–670. doi: 10.1006/viro.1995.1301. [DOI] [PubMed] [Google Scholar]
- 30.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36. doi: 10.1016/j.virol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 31.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 32.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 33.Matrosovich M, Zhou N, Kawaoka Y, Webster R. 1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol 73:1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannecchini S, Clausi V, Di Trani L, Falcone E, Terregino C, Toffan A, Cilloni F, Matrosovich M, Gambaryan AS, Bovin NV, Delogu M, Capua I, Donatelli I, Azzi A. 2010. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology 408:167–173. doi: 10.1016/j.virol.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Gambaryan A, Webster R, Matrosovich M. 2002. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol 147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- 36.Gambaryan AS, Tuzikov AB, Pazynina GV, Webster RG, Matrosovich MN, Bovin NV. 2004. H5N1 chicken influenza viruses display a high binding affinity for Neu5Acα2-3Galβ1-4(6-HSO3)GlcNAc-containing receptors. Virology 326:310–316. doi: 10.1016/j.virol.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Gambaryan A, Yamnikova S, Lvov D, Tuzikov A, Chinarev A, Pazynina G, Webster R, Matrosovich M, Bovin N. 2005. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology 334:276–283. doi: 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, Klimov A. 2006. Evolution of the receptor-binding phenotype of influenza A (H5) viruses. Virology 344:432–438. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 39.Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, Klimov AI. 2008. 6-Sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J 5:85. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, Bovin NV, Klenk H-D, Matrosovich MN. 2012. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol 86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osterhaus ADME, Munster VJ, Fouchier RAM. 2008. Epidemiology of avian influenza, p 1–10. In Klenk H-D, Matrosovich MN, Stech J (ed), Avian influenza, vol 27 Karger, Basel, Switzerland. [Google Scholar]
- 42.Pantin-Jackwood MJ, Swayne DE. 2009. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev Sci Tech 28:113–136. [PubMed] [Google Scholar]
- 43.Matrosovich M, Klenk H-D. 2003. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor-binding. Rev Med Virol 13:85–97. doi: 10.1002/rmv.372. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki Y, Ito T, Suzuki T, Holland RE Jr, Chambers TM, Kiso M, Ishida H, Kawaoka Y. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 74:11825–11831. doi: 10.1128/JVI.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuzikov AB, Gambaryan AS, Juneja LR, Bovin NV. 2000. Conversion of complex sialooligosaccharides into polymeric conjugates and their anti-influenza virus inhibitory potency. J Carbohydr Chem 19:1191–1200. doi: 10.1080/07328300008544143. [DOI] [Google Scholar]
- 46.Pazynina GV, Sablina MA, Tuzikov AB, Chinarev AA, Bovin NV. 2003. Synthesis of complex 2-3 sialooligosaccharides, including sulphated and fucosylated ones, using Neu5Ac alfa 2-3Gal as building block. Mendeleev Commun 13:245–248. doi: 10.1070/MC2003v013n06ABEH001845. [DOI] [Google Scholar]
- 47.Shilova NV, Galanina OE, Pochechueva TV, Chinarev AA, Kadykov VA, Tuzikov AB, Bovin NV. 2005. High molecular weight neoglycoconjugates for solid phase assays. Glycoconj J 22:43–51. doi: 10.1007/s10719-005-0280-y. [DOI] [PubMed] [Google Scholar]
- 48.Mochalova LV, Korchagina EY, Kurova VS, Shtyria JA, Gambaryan AS, Bovin NV. 2005. Fluorescent assay for studying the substrate specificity of neuraminidase. Anal Biochem 34:190–193. doi: 10.1016/j.ab.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 49.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mochalova L, Gambaryan A, Romanova J, Tuzikov A, Chinarev A, Katinger H, Egorov A, Bovin N. 2003. Receptor-binding property of modern human influenza viruses primarily isolated in Vero and MDCK cells and chicken embryonated eggs. Virology 313:473–480. doi: 10.1016/S0042-6822(03)00377-5. [DOI] [PubMed] [Google Scholar]
- 51.Matrosovich MN, Gambaryan AS. 2012. Solid-phase assays of receptor-binding specificity. Methods Mol Biol 865:71–94. doi: 10.1007/978-1-61779-621-0_5. [DOI] [PubMed] [Google Scholar]
- 52.Mochalova L, Kurova V, Shtyrya Y, Korchagina E, Gambaryan A, Belyanchikov I, Bovin N. 2007. Oligosaccharide specificity of influenza H1N1 virus neuraminidases. Arch Virol 152:2047–2057. doi: 10.1007/s00705-007-1024-z. [DOI] [PubMed] [Google Scholar]
- 53.Shtyrya Y, Mochalova L, Voznova G, Rudneva I, Shilov A, Kaverin N, Bovin N. 2009. Adjustment of receptor-binding and neuraminidase substrate specificities in avian-human reassortant influenza viruses. Glycoconj J 26:99–109. doi: 10.1007/s10719-008-9169-x. [DOI] [PubMed] [Google Scholar]
- 54.Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ. 2013. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497:392. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 55.Bean WJ, Kawaoka Y, Wood JM, Pearson JE, Webster RG. 1985. Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J Virol 54:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang G, Rouse BT, Narayan O, Ferguson AE, Connell MC. 1968. A new influenza virus infection in turkeys. I. Isolation and characterization of virus 6213. Can Vet J 9:22–29. [PMC free article] [PubMed] [Google Scholar]
- 57.Lang G, Narayan O, Rouse BT, Ferguson AE, Connell MC. 1968. A new influenza A virus infection in turkeys. II. A highly pathogenic variant, A/Turkey/Ontario 7732/66. Can Vet J 9:151–160. [PMC free article] [PubMed] [Google Scholar]
- 58.Daniels PS, Jeffries S, Yates P, Schild GC, Rogers GN, Paulson JC, Wharton SA, Douglas AR, Skehel JJ, Wiley DC. 1987. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J 6:1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 60.Kawaoka Y, Naeve CW, Webster RG. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303–316. doi: 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 61.Londt BZ, Banks J, Alexander DJ. 2007. Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathol 36:347–350. doi: 10.1080/03079450701589134. [DOI] [PubMed] [Google Scholar]
- 62.Munster VJ, Wallensten A, Baas C, Rimmelzwaan GF, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2005. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg Infect Dis 11:1545–1551. doi: 10.3201/eid1110.050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol 84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H, Carney PJ, Donis RO, Stevens J. 2012. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol 86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alexander DJ. 2000. A review of avian influenza in different bird species. Vet Microbiol 74:3–13. doi: 10.1016/S0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 66.Alexander DJ. 2007. An overview of the epidemiology of avian influenza. Vaccine 25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 67.Feldmann A, Schafer MK, Garten W, Klenk H-D. 2000. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J Virol 74:8018–8027. doi: 10.1128/JVI.74.17.8018-8027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]