ABSTRACT
Antisera raised against the avian hepatitis E virus (HEV) capsid protein are cross-reactive with human and swine HEV capsid proteins. In this study, two monoclonal antibodies (MAbs) against the avian HEV capsid protein, namely, 3E8 and 1B5, were shown to cross-react with the swine HEV capsid protein. The motifs involved in binding both MAbs were identified and characterized using phage display biopanning, peptide synthesis, and truncated or mutated protein expression, along with indirect enzyme-linked immunosorbent assay (ELISA) and Western blotting. The results showed that the I/VPHD motif is a necessary core sequence and that P and H are two key amino acids for recognition by MAb 3E8. The VKLYM/TS motif is the minimal amino acid sequence necessary for recognition by MAb 1B5. Cross-reactivity between the two epitopes and antibodies against avian, swine, and human HEVs in sera showed that both epitopes are common to avian, swine, and human HEVs. In addition, amino acid sequence alignment of the capsid proteins revealed that the key motifs of both novel epitopes are the same in HEVs from different animal species, predicting that they may be common to HEV isolates from boars, rabbits, rats, ferrets, mongooses, deer, and camels as well. Protein modeling analysis showed that both epitopes are at least partially exposed on the surface of the HEV capsid protein. Protective capacity analysis demonstrated that the two epitopes are nonprotective against avian HEV infection in chickens. Collectively, these studies characterize two novel linear B-cell epitopes common to avian, swine, and human HEVs, which furthers the understanding of HEV capsid protein antigenic structure.
IMPORTANCE More and more evidence indicates that the host range diversity of hepatitis E virus (HEV) is a global public health concern. A better understanding of the antigenic structure of the HEV capsid protein may improve disease diagnosis and prevention. In this study, binding site mapping and localization as well as the antigenic biology of two novel linear B-cell epitopes common to several different species of HEV were characterized. These findings partially reveal the antigenic structure of the HEV capsid protein and provide potential applications for the development of diagnostics and interventions for HEV infection.
INTRODUCTION
Hepatitis E is usually an acute and self-limiting disease in humans, is an epidemic in many developing countries in Asia and Africa (1–3), and occurs sporadically in some industrialized countries (4–6). The causative agent, hepatitis E virus (HEV), belongs to the Hepeviridae family and is a single-stranded, positive-sense RNA virus. The viral genome contains three open reading frames (ORFs), ORF1, -2, and -3, encoding a viral nonstructural protein, a capsid protein, and a small multifunctional phosphoprotein (2, 7), respectively. The proteins encoded by ORF2 and ORF3 are produced from a bicistronic subgenomic (SG) mRNA, and the coding region of ORF2 overlaps ORF3, but neither overlaps ORF1 (8, 9). To date, complete genomes of many animal HEVs, including HEVs from pigs (10), chickens (11, 12), rabbits (13), deer (14), mongooses (15), rats (16, 17), bats (18), and ferrets (19), have been characterized, and the host range and molecular diversity of HEVs are continually expanding (20). Swine HEV, the first identified animal strain of HEV, shares 80% to 90% nucleotide sequence identity with human HEV and can infect nonhuman primates (10). Hepatitis E is now regarded as a zoonotic disease, and pigs are the main reservoir animal for human infection (21–24).
Avian HEV, the second known animal strain of HEV, was isolated from chickens with big liver and spleen disease or hepatitis-splenomegaly syndrome. Although its genome of 6.6 kb is only about 48% identical to those of human and swine HEVs, avian HEV is antigenically related to human and swine HEVs (25, 26).
The capsid protein of HEV contains the major antigenic epitopes of the viral particle (27, 28) and is responsible for induction of the protective humoral immune response (29). Thus, the antigenic structure of the protein must be finely characterized for vaccine design and selection of serodiagnostic antigens. At present, four antigenic domains (I to IV) from the C-terminal 268 amino acids (aa) (aa 339 to 606) of the avian HEV capsid protein have been documented (26). One epitope in antigenic domain I (aa 389 to 410) is common to avian, human, and swine HEVs, and one or more epitopes in domain IV (aa 583 to 600) are common between avian and human HEVs (30). However, some swine and human antisera to HEVs have been shown to cross-react only with the C-terminal 268 aa residues, not the isolated domain I antigen (30). This suggests that other epitopes common to avian, human, and swine HEVs exist in the C-terminal 268-aa region of the avian HEV capsid protein, in regions other than the domain I region.
In the present study, to identify additional epitopes in the C-terminal 268-aa region of the avian HEV capsid protein common to avian, human, and swine HEVs, six monoclonal antibodies (MAbs) against the avian HEV capsid protein were generated using the C-terminal 268 aa of the avian HEV capsid protein as the immunizing antigen. Two of these MAbs, 3E8 and 1B5, cross-reacted with the swine HEV capsid protein. Binding motifs of the two reactive MAbs were finely mapped using a phage display random peptide library, peptide synthesis, and truncated or mutated protein expression. Cross-reactions between the two epitopes and serum antibodies against avian, swine, and human HEVs revealed that both epitopes are common to avian, swine, and human HEVs and induce a host immune response postinfection. Using aa sequence alignment and protein modeling, the two epitopes were shown to be highly conserved in different animal HEV isolates and located on the surface of the capsid protein. In addition, protective capacity analysis showed that they are nonprotective against avian HEV infection in chickens. These findings shed light on the antigenic structure of the HEV capsid protein and are important for the application of serodiagnosis and vaccine design for HEV.
MATERIALS AND METHODS
Serum samples.
Antisera against avian, human, and swine HEVs were separately collected from specific-pathogen-free (SPF) chickens infected experimentally with genotype 3 avian HEV isolated from China (CaHEV) (31), from human beings infected naturally by genotype 4 human HEVs, which were identified by reverse transcription-PCR (RT-PCR) as previously described (32) in the Affiliated Hospital of Shandong Agricultural University (33), and from swine herds infected naturally by genotype 4 swine HEV in Shandong Province (34), respectively. These serum samples were analyzed by indirect enzyme-linked immunosorbent assays (ELISAs) using the C-terminal 268 aa of avian and swine HEV capsid proteins (aORF2-C and sORF2-C, respectively), peptide 1 (389GNQHVDDRPSPAPAPKRALGTL410), or peptide 2 (461SVDWTKATVDGVQVKTVDASSGSNRFAALPAF492) as the coating antigen, as described in previous studies (30, 31, 34). Peptide 1 and peptide 2 included full-length antigenic domains I (common to avian, swine, and human HEVs) and II (unique to avian HEV), respectively (26, 30). sORF2-C was obtained from a swine HEV isolate from Shandong Province, China (CHN-SD-sHEV; genotype 4), which was described previously (34). Serum samples were also used to react with the peptides containing the novel epitopes identified in the study.
Monoclonal antibodies.
Six MAbs (3E8, 1B5, 7H12, 1H5, 3A1, and 6G8) were generated by immunization of BALB/c mice with aORF2-C as documented previously (35). These MAbs were used to cross-react with sORF2-C by indirect ELISA and Western blotting as described below. MAb 2C9 against the CaHEV ORF3 protein was used as the negative control (36). The seven MAbs were purified from the tissue culture medium by use of a goat anti-mouse IgG affinity column according to the manufacturer's instructions (JinSiTe Biotech Co., Nanjing, China), and the antibody concentrations were determined using a spectrophotometer (UV-2450; Shimadzu Corporation, Kyoto, Japan) with an absorption coefficient of the optical density at 280 nm (OD280)/1.35 (mg/ml).
Biopanning of a phage display random library.
To identify the MAb motifs that cross-react with sORF2-C, three rounds of biopanning were carried out based on the manufacturer's instructions for a PhD-C7C phage display peptide library kit (New England BioLabs Inc., Beverly, MA). Briefly, ELISA plates (Nunc immunoplate I; Nunc, Roskilde, Denmark) were coated with the purified MAbs (1.5 μg/well) in 0.1 M NaHCO3 (pH 8.6) and incubated overnight at 4°C. The plates were then blocked with blocking buffer (0.1 M NaHCO3, pH 8.6, 0.02% NaN3, and 5 mg/ml bovine serum albumin [BSA]) for 2 h at 4°C. After washing the plates with washing buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% Tween 20, pH 7.5), approximately 1.5 × 1011 PFU (4 × 1010 phage; 10 μl of the original library) was added to the wells and incubated for 1 h at room temperature. After washing again, the bound phage was eluted with elution buffer (0.2 M glycine-HCl, pH 2.2) containing 1 mg/ml BSA and subsequently amplified in early-log-phase Escherichia coli ER2738 cells. After three rounds of biopanning, 9 or 10 individual blue phage clones were selected and assayed for target binding by phage ELISA as described previously (37). The positive phage clones were then sequenced in an ABI3730 genetic analyzer (JinSiTe Biotech Co., Nanjing, China), using the 96 gIII sequencing primer (5′-TGAGCGGATAACAATTTCAC-3′). The amino acid sequences were determined using the inserted nucleotide sequences and were aligned with the amino acids of the CaHEV and CHN-SD-sHEV capsid proteins to find consensus sequences by using the MegAlign program of Lasergene 7.1 (DNASTAR Inc., Madison, WI).
Peptide synthesis.
To finely map the motifs where the two MAbs had successfully bound, a series of truncated and mutated peptides were commercially synthesized based on the sequencing results for positive phage clones and the corresponding amino acids of the CaHEV and CHN-SD-sHEV capsid proteins (Invitrogen Bio Co., Shanghai, China). All peptides were conjugated to keyhole limpet hemocyanin (KLH). Amino acid sequences of the peptides are shown in Table 1.
TABLE 1.
Synthetic peptides conjugated to KLH to test the specificity of reaction with MAb 3E8 or 1B5
| Short peptide or capsid protein | No. of amino acids | Purity (%) | Sequence (N terminal to C terminal)a |
|---|---|---|---|
| Pep3E8 | 7 | 98.0 | CMIPHDEN |
| Pep3E8-PH | 7 | 98.2 | CMIERDEN |
| Pep3E8-P | 6 | 97.3 | CAGIEHD |
| Pep3E8-H | 6 | 96.8 | CAGIPRD |
| Pep3E8-2 | 7 | 97.9 | VHALVPHC |
| Pep3E8-3 | 7 | 99 | CVHALNPH |
| Pep1B5-1 | 6 | 95.1 | CVKLYMS |
| Pep1B5-2 | 4 | 95.4 | CVKLY |
| Pep1B5-3 | 5 | 97.2 | CKLYMS |
| Avian HEV capsid | 355VKLYMSVEDAVNDKPIMVPHDIDLG379b | ||
| Swine HEV capsid | 410VKLYTSVENAQQDKGIAIPHDIDLG434c |
The C (cysteine) was added according to the hydrophobicity of the peptide. Bold letters indicate common motifs, and underlined letters indicate mutated amino acids.
Corresponding amino acid sequence of the CaHEV capsid protein.
Corresponding amino acid sequence of the CHN-SD-sHEV capsid protein.
Expression and purification of truncated capsid proteins from avian and swine HEVs.
To test whether the six MAbs would cross-react with avian and swine HEV capsid proteins, aORF2-C and sORF2-C were expressed from a bacterial system and purified as previously described (31, 34).
To further confirm the mapping of the binding sites for the MAbs, a series of wild-type, truncated, and mutated polypeptides from the CaHEV capsid protein were expressed using a bacterial system. The N protein from porcine reproductive and respiratory syndrome virus (PRRSV) (strain SD-16; GenBank accession number JX087437) was also expressed with the same system to serve as a negative-control protein (38). The genes of these polypeptides were amplified using the primer pairs shown in Table 2 and then cloned into the pET-28a(+) vector with a His tag (Novagen, Madison, WI), using the BamHI and XhoI restriction sites. The positive recombinant plasmids were transformed into Escherichia coli Rosetta (DE3)/pLysS and induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C to produce the recombinant proteins. The recombinant proteins were purified using a high-affinity Ni-nitrilotriacetic acid (Ni-NTA) resin column according to the manufacturer's instructions (Jinsite Biotechnology Corp., Nanjing, China).
TABLE 2.
Synthetic oligonucleotide primers used for cloning and expression of mutated and truncated ORF2 proteins of the CaHEV strain and the N protein of the PRRSV SD16 strain
| Oligonucleotide | Sequenced | Motif |
|---|---|---|
| aORF2 355-570-Sa | CGGGATCCGTGAAATTGTATATGT | 355VKLYM |
| aORF2 360-570-S | CGGATCCTCGGTGGAGGATGCTGTC | 360SVEDAV |
| aORF2 365-570-S | CGGATCCGTCAACGACAAGCCCATT | 365VNDKPI |
| aORF2 371-570-S | CAAGGATCCATGGTCCCTCACGACATTGA | 371MVPHD |
| aORF2 380-570-S | GAAGGATCCACTAGCACTGTTACCTGC | 380TSTVTC |
| aORF2 PH373-374RR-Sb | AAAGGATCCATGGTCCGTCGCGACATTGAC | 371MVRRD |
| aORF2 P373R-S | AAAGGATCCATGGTCCGTCACGACATTGAC | 371MVRHD |
| aORF2 H374R-S | CAAGGATCCATGGTCCCTCGCGACATTGAC | 371MVPRD |
| aORF2 V372A-S | CAAGGATCCATGGCCCCTCACGACATT | 371MAPHD |
| aORF2 D375E-S | CAAGGATCCATGGTCCCTCACGAGATTGACCT | 371MVPHE |
| aORF2-Rc | GAACTCGAGGGTGGTGCAATCATCAC | |
| N Protein-S | CGGGATCCATGCCAAATAACAACGGCAAG | |
| N Protein-R | AACTCGAGTCATGCTGAGGGTGATGCTGT |
Forward primer encoding a truncated protein for epitope 1B5.
Forward primer encoding a recombinant protein (aa 371 to 570) for epitope 3E8, with aa P373 and H374 mutated to R.
Reverse primer for 10 mutation and truncation proteins from avian HEV ORF2.
Letters in italics indicate BamHI or XhoI sequences.
Indirect ELISA.
To test the immune reactions of the synthesized peptides and expressed polypeptides with the MAbs, an indirect ELISA was performed using procedures described by Guo et al. (30). Briefly, a 96-well ELISA plate was coated with peptides (Table 1) and polypeptides (Table 2) (200 ng/well) in phosphate-buffered saline (PBS; pH 7.2) and incubated overnight at 4°C, followed by blocking with blocking buffer (2.5% dry milk in 0.05% Tween 20 in PBS [PBST]) for 1 h at room temperature. After the plate was incubated with the MAbs for 1 h at 37°C and washed, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Jackson Immunoresearch, West Grove, PA) was added to the wells at a dilution of 1:10,000. The color was developed with tetramethylbenzidine (TMB; Sigma Chemical Co., St. Louis, MO) for 15 min and was stopped with 3 M H2SO4. The OD450 was read by an automated ELISA plate reader (Bio-Rad, Hercules, CA).
Western blotting.
The purified truncated proteins and polypeptides with a His tag (Table 2) were loaded at 1.0 × 10−3 nmol per lane, separated by SDS-PAGE on a 12% acrylamide gel, and transferred electrophoretically to a polyvinylidene difluoride (PVDF) membrane by use of a minitransblot apparatus (Bio-Rad Co.). After blocking with blocking buffer at 4°C overnight, the membrane was washed and incubated with the MAbs for 1 h at room temperature. After washing, the membrane was incubated with HRP-conjugated goat anti-mouse IgG (1:5,000) in blocking buffer for 1 h. Membranes were washed three times and then visualized using a diaminobenzidine (DAB) Enhance liquid substrate system (TianGen Co., Beijing, China). The reaction was stopped with deionized water, and membranes were subsequently dried for analysis.
Cross-reactivities of antisera against avian, swine, and human HEVs.
The cross-reactivities of antisera against avian, swine, and human HEVs with various binding motifs were evaluated using indirect ELISA. Indirect ELISAs were performed as described above, with the following modifications: plates were separately coated with aORF2-C, sORF2-C, peptide 1, peptide 2, Pep3E8, 3E8-PH, Pep1B5-1, and Pep1B5-2; primary antibodies were replaced with chicken (challenged experimentally by genotype 3 avian HEV), swine (infected naturally by genotype 4 swine HEV), and human (infected naturally by genotype 4 human HEV) serum samples; and the secondary antibodies were replaced with HRP-conjugated goat anti-chicken, anti-swine, and anti-human IgGs, respectively (Jackson ImmunoResearch Laboratories, West Grove, PA).
Computer analysis.
To analyze the conservation of the common epitopes between avian and mammalian HEVs, the aa residues of capsid proteins from different avian and mammalian HEV isolates were aligned using the Clustal W method in Lasergene 7.1 (DNASTAR Inc., Madison, WI).
To determine the spatial locations of common epitopes, the protein crystal structure of the HEV capsid protein was obtained from the RCSB Protein Data Bank (PDB) (entry 2ZTN). Areas of interest on the protein were highlighted using the PyMOL molecular graphics system (www.pymol.org) (39).
Protective capacity analysis.
To assess the protective capacity of the synthesized peptides containing the common epitopes, an animal immunization study was performed. Seven-week-old SPF chickens (n = 30) were purchased from the Beijing Meiliyaweitong Experimental Animal Technology Company and confirmed to be negative for avian HEV RNA and antibodies by using RT-PCR and indirect ELISA (12, 31). Chickens were divided into 5 groups (6/group) and housed at 2 chickens per cage. For groups 1 to 4, Pep3E8 and Pep1B5-1 (peptides containing common epitopes), aORF2-C, and KLH (mock treatment; control peptide), respectively, were used as immunizing antigens. In group 5, nonimmunized and unchallenged chickens served as controls. The antigen (200 μg) for the first immunization was emulsified with Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO) and injected intramuscularly. One booster injection was given 2 weeks later, with an equal amount of antigen in Freund's incomplete adjuvant. Two weeks after the second immunization, serum samples were collected from chickens in groups 1 to 3 and tested separately for titrations of anti-Pep3E8, anti-Pep1B5-1, and anti-aORF2-C antibodies, respectively, using indirect ELISA as described previously (30). The chickens in groups 1 to 4 were then challenged intravenously with 800 μl of the CaHEV infectious stock at a virus dose of 104 genomic equivalents (GE)/ml or 500 50% chicken infectious doses (CID50)/ml. Serum and fecal samples were collected from all chickens at 0, 1, 2, 3, 4, 5, 6, and 7 weeks postinoculation and were tested qualitatively and quantitatively for avian HEV RNA by using reverse transcription nested PCR (RT-nPCR) as described by Dong et al. (35) and quantitative RT-PCR (qRT-PCR) as described by Troxler et al. (40), respectively. In addition, serum samples were used to detect anti-avian HEV antibodies by indirect ELISA, using peptide 10 as the coating antigen, as described by Guo et al. (41). Peptide 10, which is located in the N terminus of the avian HEV capsid protein, was used to differentiate anti-avian HEV antibodies induced by the avian HEV challenge from the antibody response induced by immunization. All chickens were euthanized at 7 weeks postinoculation. At necropsy, livers were observed for gross lesions and were photographed digitally. Animal experiments were conducted under the guidelines of the Institutional Animal Care and Use Committee of Northwest A&F University.
HEV capsid protein aa sequence accession numbers.
Forty-two HEV capsid protein aa sequences were used in this study and were downloaded from GenBank in February 2015. The accession numbers of these isolates are as follows: avian HEV sequences, accession numbers JN997392, JN597006, AM943647, AY535004, GU954430, and AM943646; human HEV sequences, accession numbers D11093, X98292, M74506, AB074918, AB089824, HM439284, and AJ272108; pig HEV sequences, accession numbers KJ507956, AF082843, KF176351, and AY594199; wild boar HEV sequences, accession numbers AB573435, FJ705359, and AB602441; rabbit HEV sequences, accession numbers AB301710, AF082843, FJ906895, GU937805, and JX121233; rat HEV sequences, accession numbers AB847305, JN167537, JN167538, GU345042, JX120573, and AB847306; ferret HEV sequences, accession numbers JN998607, AB890001, AB890374, and JN998606; camel HEV sequences, accession numbers KJ496143 and KJ496144; deer HEV sequence, accession number AB189071; mongoose HEV sequence, accession number AB591733; bat HEV sequences, accession numbers JQ001749 and NC_018382; and cutthroat trout HEV sequence, accession number HQ731075.
RESULTS
Cross-reactivities of six MAbs with the swine HEV capsid protein.
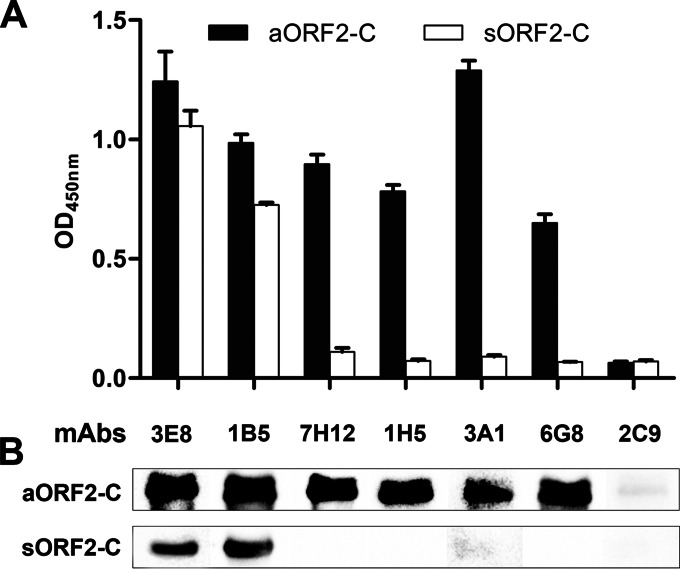
Indirect ELISA (Fig. 1A) and Western blot (Fig. 1B) experiments demonstrated that two MAbs, 3E8 and 1B5, reacted with both sORF2-C from genotype 4 swine HEV and aORF2-C from genotype 3 avian HEV. These findings suggest that the two epitopes recognized by MAbs 3E8 and 1B5 may be common between genotype 3 avian and genotype 4 swine HEVs.
FIG 1.
Anti-avian HEV capsid MAbs 3E8 and 1B5 cross-react with swine HEV capsid protein. Data are shown for indirect ELISA (A) and Western blot (B) analyses of cross-reactivity between MAbs 3E8, 1B5, 7H12, 1H5, 3A1, and 6G8 and the genotype 3 avian HEV capsid protein (aORF2-C) or the genotype 4 swine HEV capsid protein (sORF2-C). The MAb 2C9 (anti-avian HEV ORF3 protein) was used as a negative control.
Epitope prediction using phage biopanning.
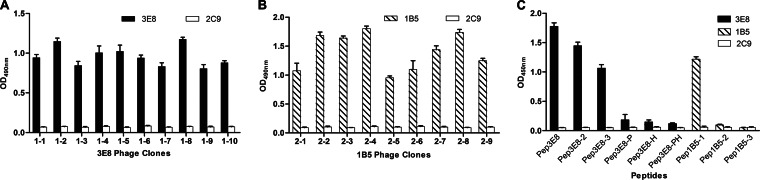
After three rounds of biopanning, 10 MAb 3E8- and 9 MAb 1B5-reactive phage clones were acquired from the phage display library. Phage ELISA results showed that all 10 3E8 phage clones (Fig. 2A) and all 9 1B5 clones (Fig. 2B) showed specific reactivities with MAbs 3E8 and 1B5, respectively (OD450 > 0.74), but not with MAb 2C9 (OD450 < 0.16). All positive phage clones were amplified for DNA sequencing. The deduced aa sequences from insert DNA sequences were aligned with aa sequences from both the CaHEV and CHN-SD-sHEV capsid proteins. The alignments showed that nine 3E8-reactive clones displayed a consensus sequence of I/VPHD, which is highly homologous to the 372VPHD375 motif (found in CaHEV) and the 427IPHD430 motif (found in CHN-SD-sHEV) (Table 3). Of the nine 1B5-reactive clones, seven displayed a consensus sequence of VKLYT/MS, which is identical to the 355VKLYMS360 motif (found in CaHEV) and the 410VKLYTS415 motif (found in CHN-SD-sHEV) (Table 3). Thus, the core motifs of the novel epitopes recognized by MAbs 3E8 and 1B5 are I/VPHD and VKLYT/MS, respectively.
FIG 2.
Identification of binding motifs of MAbs 3E8 and 1B5 by biopanning of a phage display random library and by Pepscan assay. Ten phage clones of MAb 3E8 (1-1 to 1-10) (A) and nine clones of MAb 1B5 (2-1 to 2-9) (B), selected after three rounds of biopanning with a PhD-C7C phage display peptide library kit, were tested for binding to the respective MAbs by phage ELISA. The 2C9 MAb served as a negative control. (C) Reactivity of 3E8 or 1B5 with a series of synthetic peptides by indirect ELISA. The IPHD (reactive with MAb 3E8) and VKLYMS (reactive with MAb 1B5) motifs were the targets for mutation or truncation. Sequences of these synthetic peptides can be found in Table 1. Error bars indicate the standard ranges of individual values from two independent experiments. The anti-HEV ORF3 MAb 2C9 served as a negative control.
TABLE 3.
Identification of consensus sequences of epitopes recognized by MAbs 3E8 and 1B5, based on an alignment of 7-mer peptide sequences from ELISA-positive phage clonesa
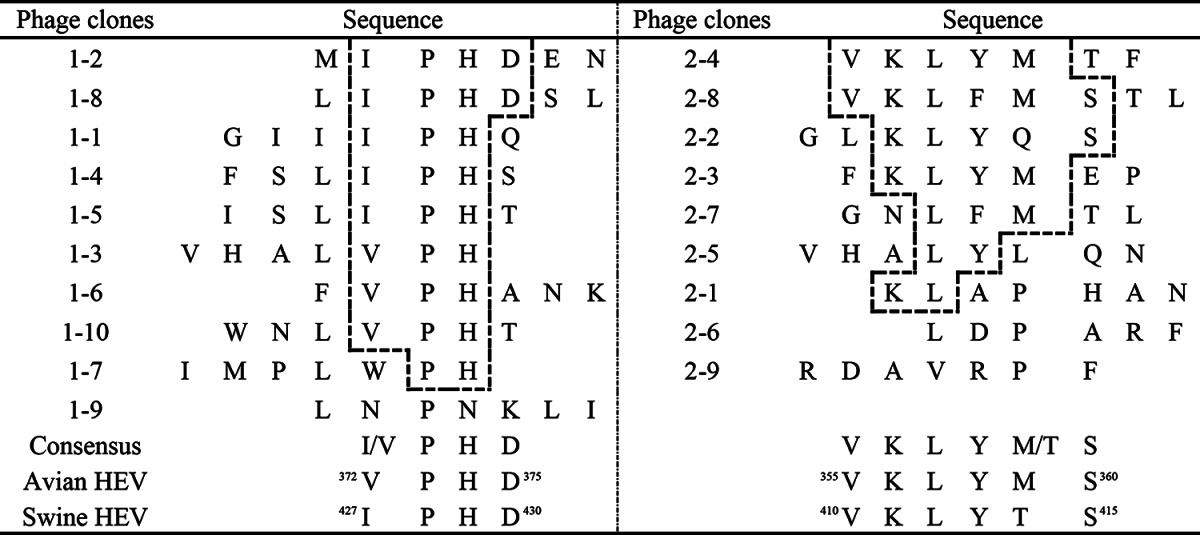
The consensus amino acid residues from phage clones are shown in boxes. The corresponding matching sequences of 372VPHD375 and 355VKLYMS360 for CaHEV and 427IPHD430 and 410VKLYTS415 for CHN-SD-sHEV are shown at the bottom of the alignment for comparison.
Precise definition of the motifs for the two novel epitopes.
To precisely define the minimal motifs required for binding by 3E8 and 1B5, a panel of truncated and mutated peptides was synthesized based on the core amino acid sequence of I/VPHD for 3E8 and VKLYT/MS for 1B5 (Table 1). These peptides were used as coating antigens to react with MAbs 3E8 and 1B5 in an indirect ELISA. For 3E8, the following three peptides reacted: Pep3E8 (containing IPHD), Pep3E8-2 (containing VPH), and Pep3E8-3 (containing PH). Pep3E8 showed higher reactivity than Pep3E8-2 and Pep3E8-3 (Fig. 2C). The peptides Pep3E8-PH (PH mutated to ER), Pep3E8-P (P mutated to E), and Pep3E8-H (H mutated to R) did not react (Fig. 2C). These results confirmed that the minimal motifs in the linear common epitope which can be recognized by 3E8 are 372VPHD375 (CaHEV) and 427IPHD430 (CHN-SD-sHEV), in which P and H are the two critical amino acids. Mutations of the V (CaHEV) or I (CHN-SD-sHEV) and D amino acids reduced the reactive ability. For 1B5, peptide Pep1B5-1 (containing VKLYMS) showed reactivity, while Pep1B5-2 (containing VKLY) and Pep1B5-3 (containing KLYMS) were not recognized. These results confirmed that the minimal amino acid sequences required for epitope recognition by 1B5 are 355VKLYMS360 (CaHEV) and 410VKLYTS415 (CHN-SD-sHEV).
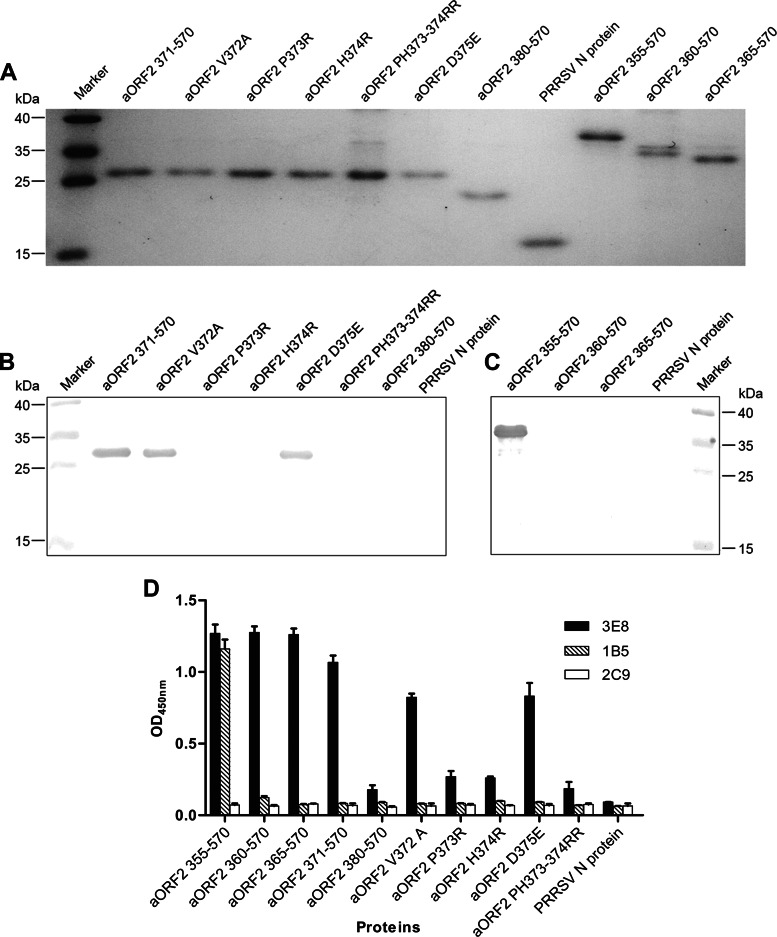
To further confirm the analysis of the Pepscan assay, 10 mutated or truncated His-fusion recombinant avian HEV capsid proteins were designed, expressed, and purified based on the consensus sequence I/VPHD for 3E8 and VKLYT/MS for 1B5 (Table 2). SDS-PAGE analysis showed that the 10 recombinant proteins were each expressed and purified with the expected sizes (Fig. 3A). The cross-reactivities of the 10 proteins with MAbs 3E8 and 1B5 were then tested using indirect ELISA and Western blotting. Indirect ELISAs showed that 3E8 reacted only with the recombinant proteins containing the specific amino acid sequence VPHD or with proteins where the sequence contained only a mutated V or D residue (aORF2 355–570, 360–570, 365–570, 371–570, V372A, and D375E). 3E8 did not react with the recombinant proteins containing P or H mutations (aORF2 P373R, H374R, and PH373–374RR) (Fig. 3D). Western blotting confirmed that 3E8 recognized the recombinant proteins containing the amino acids P and H but not the truncated P and H proteins (Fig. 3B). The results from indirect ELISA and Western blot experiments showed that the proteins containing the core amino acid sequence VKLYMS (aORF2 355–570) reacted with MAb 1B5, but the truncated proteins did not (Fig. 3C and D). The control MAb 2C9 did not react with any proteins, and the control antigen (PRRSV N protein) did not react with either MAb 3E8 or 1B5 in indirect ELISA or Western blots (Fig. 3B to D).
FIG 3.
Fine mapping of the epitopes recognized by MAbs 3E8 and 1B5, using His-fusion polypeptides containing truncated or mutated motifs derived from the sequence IPHD for 3E8 and VKLYMS for 1B5. The design of the truncated and mutated polypeptides is shown in Table 2. (A) Analysis of expression and purification of His-fusion polypeptides by SDS-PAGE. (B) Western blot analysis of MAb 3E8 reacting with eight truncated and mutated polypeptides targeting the IPHD motif. (C) Western blot analysis of MAb 1B5 reacting with four truncated polypeptides designed to target the VKLYMS motif. (D) Reactivities of all truncated and mutated polypeptides with MAbs 3E8 and 1B5 by indirect ELISA. The anti-HEV ORF3 MAb 2C9 and the N protein of PRRSV served as negative controls. Data shown are based on the means for two replicates.
Cross-reactivities of anti-avian, anti-swine, and anti-human HEV-positive serum samples.
To determine the reactivities of the two epitopes with anti-avian, anti-swine, and anti-human HEV sera, positive serum samples from chickens challenged by genotype 3 avian HEV, pigs infected by genotype 4 swine HEV, and humans infected by genotype 4 human HEV were separately used to react with four peptides (Pep3E8, Pep3E8-PH, Pep1B5-1, and Pep1B5-2) in an indirect ELISA. These serum samples were also used to react with the antigens aORF2-C, sORF2-C, peptide 1, and peptide 2 by indirect ELISA. In these experiments, aORF2-C, sORF2-C, Pep3E8, and Pep1B5-1 (containing the core motifs of the newly identified common epitopes) reacted with the positive serum samples, but the Pep3E8-PH and Pep1B5-2 peptides did not react (Table 4). These results suggested that the two newly identified epitopes that are common to avian, swine, and human HEVs exist in the native viral particles and can induce an immune response in the host. Furthermore, peptide 1, which contains another common epitope, also reacted with the positive serum samples, consistent with previous work (30). However, peptide 2, which contains an epitope unique to avian HEV, reacted only with anti-avian HEV sera (Table 4).
TABLE 4.
Antigenic cross-reactivities of antisera from chickens infected experimentally with CaHEV (genotype 3), pigs infected naturally with swine HEV (genotype 4), and humans infected naturally with HEV (genotype 4) with six synthetic peptides and truncated avian and swine HEV capsid proteinsb
| Source of antisera | Indirect ELISA result (OD450) against six peptides and two proteinsa |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pep3E8 | 3E8-PH | Pep1B5-1 | Pep1B5-2 | Peptide 1 | Peptide 2 | ORF2-C | ORF2-C | |
| Pig | 0.932 ± 0.242 | 0.215 ± 0.074 | 0.668 ± 0.206 | 0.250 ± 0.095 | 0.592 ± 0.163 | 0.275 ± 0.086 | 1.287 ± 0.338 | 1.438 ± 0.357 |
| Human | 0.624 ± 0.191 | 0.212 ± 0.056 | 0.529 ± 0.127 | 0.275 ± 0.076 | 0.859 ± 0.232 | 0.306 ± 0.065 | 0.904 ± 0.292 | 1.258 ± 0.333 |
| Chicken | 0.913 ± 0.227 | 0.245 ± 0.090 | 1.077 ± 0.383 | 0.292 ± 0.043 | 0.739 ± 0.218 | 0.692 ± 0.224 | 1.732 ± 0.363 | 0.943 ± 0.297 |
The data are means ± standard deviations. Values in bold are considered positive.
Each five antisera against swine, human, and avian HEVs were separately detected against peptides and truncated capsid proteins by using indirect ELISA. The sera were diluted 1/100, and peptides/proteins were used at 100/200 ng per well.
Amino acid sequence alignment of different animal species HEV capsid proteins.
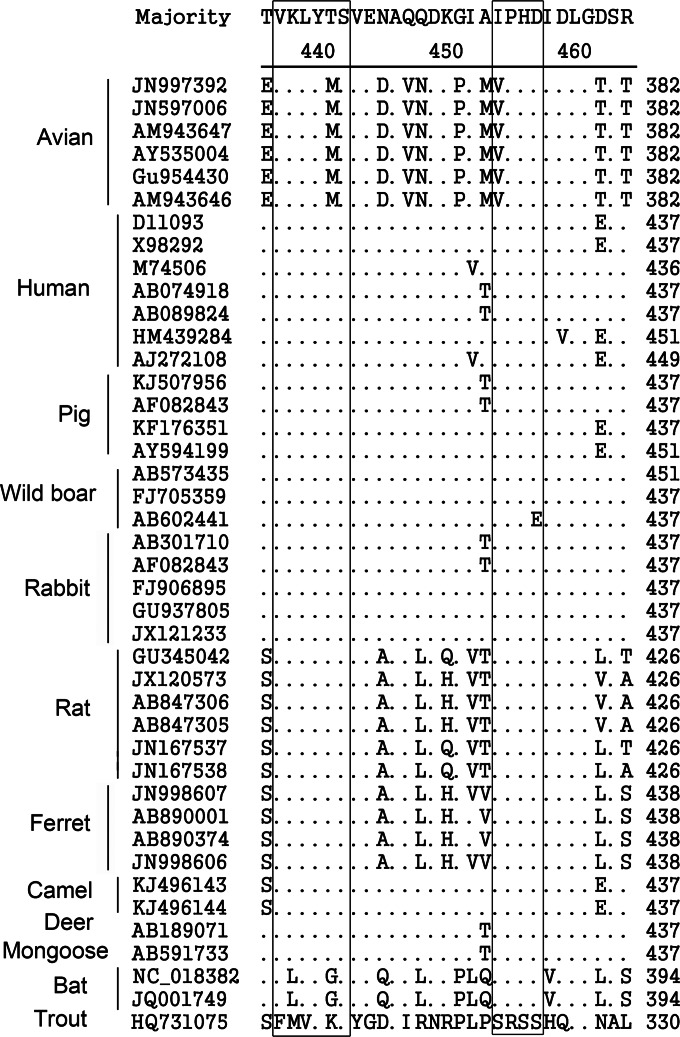
To analyze the conservation of the novel common epitopes among HEV strains from different animal species, the aa sequences of capsid proteins from 42 HEV isolates, including human, swine, avian, boar, rabbit, rat, ferret, mongoose, deer, camel, bat, and cutthroat trout HEV isolates, were BLAST searched using the public database in GenBank. Alignment analysis revealed that the epitopes recognized by 3E8 and 1B5 are highly conserved in many HEV isolates (Fig. 4), predicting that both of the novel linear B-cell epitopes may be common to HEVs in other species, excluding bat and cutthroat trout HEVs. The genomes of bat HEVs contain the I/VPHD motif of the 3E8 epitope, but the VKLYT/MS motif of the 1B5 epitope is mutated to VLLYGS. The cutthroat trout virus, a HEV-like virus, shares neither the 1B5 nor the 3E8 epitope.
FIG 4.
Conservation of the common epitopes among different HEV strains from different species. Amino acid alignments of capsid proteins from 42 HEV isolates from different species, including avian, swine, human, boar, bat, ferret, rat, deer, camel, mongoose, and cutthroat trout HEVs, were performed using the Clustal W method of the MegAlign program (version 7.1; Lasergene). The key amino acid residues of the epitopes recognized by 3E8 and 1B5 are shown in boxes, and dots indicate sequence identity. The key motifs for both epitopes are highly conserved among many HEV isolates, excluding bat and cutthroat trout HEVs.
Protein modeling.
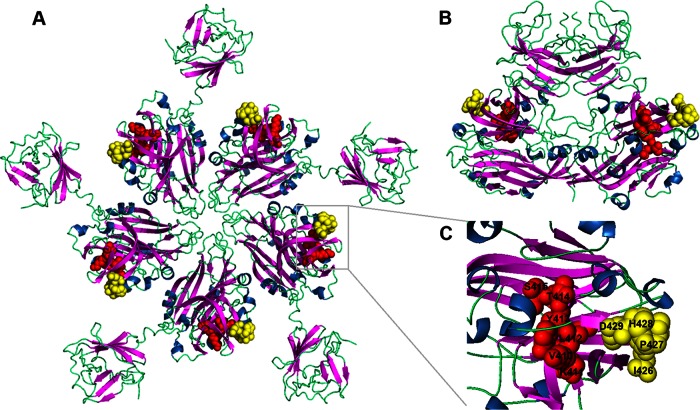
Protein modeling studies of the HEV capsid protein were undertaken to spatially localize the motifs for 3E8 and 1B5 binding. Using the protein modeling program PyMol, the two common epitopes were mapped to the 2ZTN HEV capsid protein structure obtained from the RCSB PDB (39). The epitope recognized by MAb 3E8, spanning aa 426 to 429 of human and swine HEV capsid proteins, was located in a region protruding from the molecule in both the pentamer (Fig. 5A) and dimer (Fig. 5B) formations. In contrast, the epitope recognized by 1B5, spanning aa 411 to 415, only partly protruded from the surface of the molecule in the pentamer (Fig. 5A) and dimer (Fig. 5B) formations. Both common epitopes are located in the loop region between two beta folding sheets and on the outer edge of the HEV capsid protein (Fig. 5C).
FIG 5.
Spatial locations on the HEV capsid protein of the novel epitopes recognized by the 3E8 and 1B5 MAbs. The overall structures of the HEV capsid protein pentamer (A) and dimer (B) and a detailed assessment of epitopes 3E8 and 1B5 (C) are shown. The spatial locations of the MAb 3E8 binding epitope (yellow) (aa 426 to 429) and the MAb 1B5 binding epitope (red) (aa 410 to 415) protruding from the HEV capsid protein are highlighted. The site of the epitope recognized by 3E8 is situated completely on the outer surface of the pentamer and dimer forms of the HEV capsid protein, but the epitope recognized by 1B5 is situated only partially on the outer surface. The three-dimensional capsid protein structure 2ZTN was obtained from the RCSB PDB (39).
Assessment of protection against avian HEV infection via novel epitope immunization.
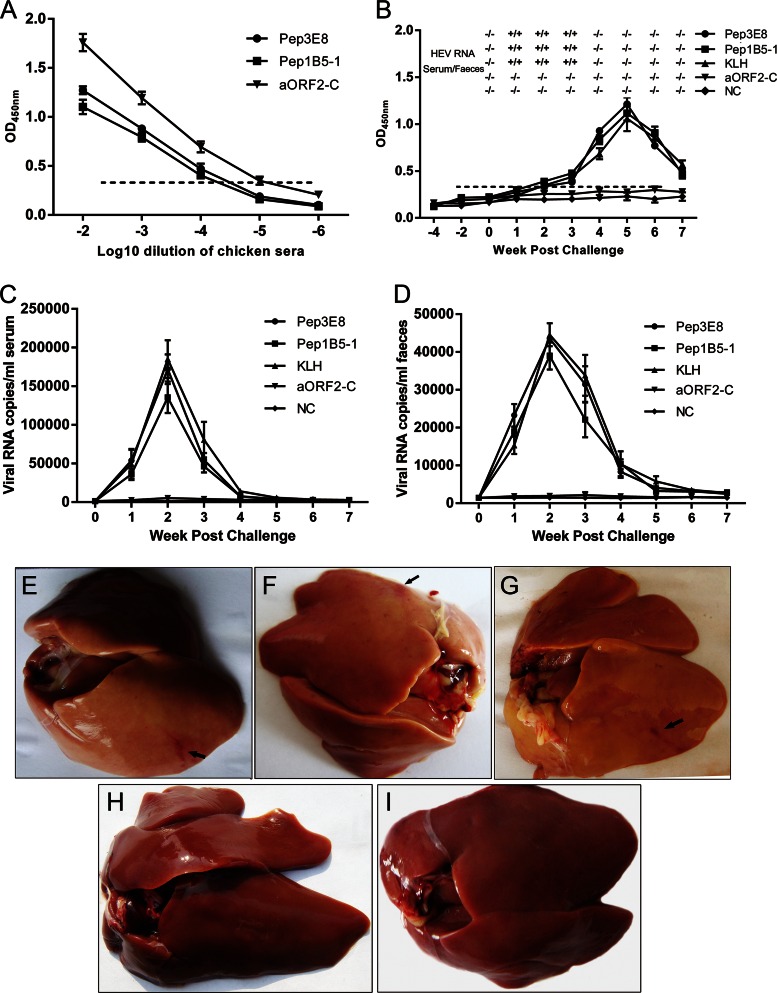
To assess whether the two novel epitopes are protective against avian HEV infection, an avian immunization study was performed. Thirty-seven-week-old SPF chickens were divided into five groups, and six SPF chickens each in groups 1 to 4 were immunized separately with Pep3E8 and Pep1B5-1 (containing common epitopes), aORF2-C, and KLH, respectively, followed by challenge with CaHEV. Group 5 was used as a control. After a second round of immunizations, the titers of anti-Pep3E8, anti-Pep1B5-1, and anti-aORF2-C antibodies in chicken serum samples from groups 1 to 3 were each determined separately; the titer was defined as the last serum dilution to give an OD450 of >0.368 (10). The results showed that the immunized chickens had serum titers of approximately 1:10,000 for the Pep3E8 and Pep1B5-1 groups and 1:100,000 for the aORF2-C group (Fig. 6A). Serum and fecal samples collected at 0 weeks postinoculation from all chickens were all negative for avian HEV RNA (Fig. 6B to D). After the CaHEV challenge, both serum and fecal viral RNAs were detected by RT-nPCR in chickens immunized with Pep3E8, Pep1B5-1, and KLH starting at 1 week postinoculation, and they remained positive for up to 3 weeks (Fig. 6B). By qRT-PCR detection, there was no significant difference in the amounts of avian HEV RNAs in serum and fecal samples from chickens immunized with Pep3E8, Pep1B5-1, and KLH following challenge (Fig. 6C and D). However, the serum and fecal samples from the group immunized with aORF2-C and from the control group were always negative by RT-nPCR and qRT-PCR testing (Fig. 6B to D). To differentiate the antibodies induced by immunization with peptides or aORF2-C versus avian HEV infection, peptide 10 (aa 27 to 50 of the avian HEV capsid protein) was selected as the antigen to assess avian HEV infection. As shown in Fig. 6B, the challenged chickens from the groups immunized with Pep3E8, Pep1B5-1, and KLH seroconverted to anti-peptide 10 antibodies at 3 weeks postinoculation, and at 7 weeks postinoculation the antibody was still detectable. However, the chickens in the group immunized with aORF2-C and in the control group were negative for antibodies against peptide 10 (Fig. 6B). These results showed that the chickens immunized separately with Pep3E8, Pep1B5-1, or KLH developed active avian HEV infections, as evidenced by seroconversion to avian HEV antibodies, viremia, and fecal virus shedding. These features were not observed in the group immunized with aORF2-C or in the control group. In addition, when the chickens were assessed at 7 weeks postinoculation, moderate to severe subcapsular hemorrhages were observed in the livers of the immunized and subsequently challenged chickens. Four of 6 animals in the Pep3E8 group (Fig. 6E), 3 of 6 in the Pep1B5-1 group (Fig. 6F), and 4 of 6 in the KLH group (Fig. 6G) exhibited hemorrhaging. In contrast, the chickens from the aORF2-C-immunized and control groups displayed no notable subcapsular hemorrhages in their livers (Fig. 6H and I). These results indicate that the two novel linear common epitopes are nonprotective against avian HEV infection in chickens.
FIG 6.
Protective capacity analysis of the novel common epitopes in chickens immunized against CaHEV infection. (A) ELISA titration of chicken sera collected 2 weeks after the second immunization with Pep3E8, Pep1B5-1, or aORF2-C. The titers of MAbs 3E8 and 1B5 were approximately 1:10,000, and the titer of aORF2-C was approximately 1:100,000. (B) Kinetics of anti-peptide 10 antibodies and qualitative detection of avian HEV RNA in Pep3E8-, Pep1B5-1-, aORF2-C-, or KLH-immunized animals and the control groups following vaccination. Serum samples (diluted 1:100) collected from all chickens were tested for the presence of anti-peptide 10 antibodies. Each point represents the mean value (± standard error of the mean [SEM]) obtained for six chickens immunized with the same antigen. The copy numbers of virus RNA in serum (C) and feces (D) in the different weeks postchallenge were also determined by qRT-PCR. (E to I) Gross lesions in livers of chickens from five groups are shown, with subcapsular hemorrhages indicated with arrows. (E) Pep3E8 group; (F) Pep1B5-1 group; (G) KLH group; (H) aORF2-C group; (I) control group.
DISCUSSION
Avian HEV, which belongs to the genus Orthohepevirus (42), is classified as a separate, floating species in the Hepeviridae family (43) and is related genetically and antigenically to swine and human HEVs (22). In previous studies, it was found that capsid proteins of avian, swine, and human HEVs share similar antigenic structures and common epitopes (26). One epitope in domain I (aa 389 to 410) of the avian HEV capsid protein that is also common to swine and human HEVs was identified previously (30). However, some clinical sera positive for swine and human HEVs were shown to react with the avian HEV capsid protein but not with isolated domain I, indicating that other common epitopes exist. In the present study, two novel linear B-cell epitopes common to avian, swine, and human HEVs, located at aa 427 to 430 and aa 411 to 415, were identified, supporting the notion that binding can occur outside domain I.
In this study, the motifs of these two linear epitopes were finely mapped using phage display, peptide synthesis, and expression of truncated and mutated proteins. The results showed that for the epitope to be recognized by MAb 3E8, the motif must contain the contiguous amino acids P and H. This initially suggested that the specificity of MAb 3E8 binding may be poor. However, when 3E8 was tested using the aORF2 380–570 and PRRSV N proteins, which also contain the contiguous amino acids P and H (data not shown), it did not react with either (Fig. 3B and D). These results indicate that the contiguous amino acids P and H can react with MAb 3E8 only if they are located within aa residues 372 to 375 of the avian HEV capsid protein or aa residues 442 to 445 of the swine HEV capsid protein. For the epitope recognized by MAb 1B5, only the mutated peptides and recombinant proteins which contained an unabridged VKLYT/MS sequence were shown to react (Fig. 3C and D). According to the current results, VKLYT/MS is believed to be the minimum essential residue sequence for MAb 1B5, but further research is needed to elucidate the function of these specific amino acids.
In previous studies, the antisera raised against the capsid protein of avian HEV were cross-reactive with human and swine HEV capsid proteins (26). In the present study, clinical sera positive for human or swine HEV reacted with peptides containing both newly identified common epitopes, further confirming the interaction (Table 4). In addition, aa sequence alignment of the HEV capsid proteins from different species revealed that the binding motifs of both common epitopes are highly conserved in human, swine, avian, boar, rabbit, deer, ferret, rat, camel, and mongoose HEV strains (Fig. 4). This predicts that the two linear B-cell epitopes recognized by MAbs 3E8 and 1B5 may also be common to boar, rabbit, deer, ferret, rat, camel, and mongoose HEVs. Experiments testing the cross-reactivity of the two novel epitopes with HEV-positive serum samples from each of these species will be needed to confirm this finding. Based on crystal structure analysis, the capsid protein of HEV-like particles folds into three major domains: the shell (S) domain (aa 129 to 319), which forms an internal scaffold structure of the particle; and the middle (M) domain (aa 320 to 455) and protruding (P) domain (aa 456 to 604), which are located on the surface of the particle and act as the predominant antigenic domains (39). According to the locations of both linear epitopes (aa 410 to 415 and aa 426 to 429 of the swine HEV capsid protein), they are located in the M domain and should be at the surface of HEV-like particles. Protein modeling of the capsid protein confirmed that the two common epitopes are at least partially exposed on the surface (Fig. 5A). More experiments are needed to determine the biological function of both epitopes in this region of the capsid protein.
In a previous study, it was found that the immunodominant linear B-cell epitopes on the avian HEV capsid protein are not protective against HEV infection and that the protective neutralizing epitope may be conformational instead of linear (41). In the current study, immunization with peptides containing the newly identified epitopes did not provide protection against avian HEV infection (Fig. 6). In addition, as shown in Fig. 6B, six chickens in the group immunized with aORF2-C were able to induce protective immunity against avian HEV infection, which supports previous findings (44). These results show that both of the novel B-cell epitopes are nonprotective, which further supports the idea that neutralization epitopes in the C terminus of the avian HEV capsid protein may be conformational, like the neutralization epitopes identified on the human HEV capsid protein.
In conclusion, two novel linear B-cell epitopes that are common to avian, swine, and human HEVs were identified and characterized. For the epitope to be recognized by MAb 3E8, specific localization of the amino acids P and H is necessary, and for epitope recognition by MAb 1B5, the minimal core amino acid sequence needed is VKLYMS. Both epitopes are highly conserved among strains from different species, excluding bat and cutthroat trout HEVs, and are nonprotective against avian HEV infection in chickens. These findings improve our understanding and interpretation of the antigenic structure of the HEV capsid protein, which is essential for future advancements in the diagnosis, treatment, and prevention of HEV.
ACKNOWLEDGMENTS
This study was supported by grants from the Natural Science Foundation of China (grant 31372464 to E.-M.Z. and grant 31402233 to Q.Z.), the Open Fund of the Key Laboratory of Veterinary Etiologic Biology (grant SKLVEB2013KFKT008), and the Specialized Fund for the Basic Research Operating Expenses Program of Central College (grant Z109021307 to Q.Z.).
REFERENCES
- 1.Arankalle VA, Chadha MS, Tsarev SA, Emerson SU, Risbud AR, Banerjee K, Purcell RH. 1994. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc Natl Acad Sci U S A 91:3428–3432. doi: 10.1073/pnas.91.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Zhang H, Ling R, Li H, Harrison TJ. 2000. The complete sequence of hepatitis E virus genotype 4 reveals an alternative strategy for translation of open reading frames 2 and 3. J Gen Virol 81:1675–1686. [DOI] [PubMed] [Google Scholar]
- 4.Amon JJ, Drobeniuc J, Bower WA, Magana JC, Escobedo MA, Williams IT, Bell BP, Armstrong GL. 2006. Locally acquired hepatitis E virus infection, El Paso, Texas. J Med Virol 78:741–746. doi: 10.1002/jmv.20617. [DOI] [PubMed] [Google Scholar]
- 5.Erker JC, Desai SM, Schlauder GG, Dawson GJ, Mushahwar IK. 1999. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J Gen Virol 80:681–690. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Kang JH, Ohnishi S, Hino K, Miyakawa H, Miyakawa Y, Maekubo H, Mishiro S. 2003. Full-length sequences of six hepatitis E virus isolates of genotypes III and IV from patients with sporadic acute or fulminant hepatitis in Japan. Intervirology 46:308–318. doi: 10.1159/000073210. [DOI] [PubMed] [Google Scholar]
- 7.Emerson SU, Purcell RH. 2003. Hepatitis E virus. Rev Med Virol 13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 8.Huang YW, Opriessnig T, Halbur PG, Meng XJ. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol 81:3018–3026. doi: 10.1128/JVI.02259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graff J, Torian U, Nguyen H, Emerson SU. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol 80:5919–5926. doi: 10.1128/JVI.00046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, Haynes JS, Thacker BJ, Emerson SU. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J Gen Virol 82:2449–2462. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Q, Zhou EM, Dong SW, Qiu HK, Zhang L, Hu SB, Zhao FF, Jiang SJ, Sun YN. 2010. Analysis of avian hepatitis E virus from chickens, China. Emerg Infect Dis 16:1469–1472. doi: 10.3201/eid1609.100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, Wang Y. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Kitajima N, Abe N, Mishiro S. 2004. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330:501–505. doi: 10.1016/j.virol.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M, Takahashi K, Taira K, Taira M, Ohno A, Sakugawa H, Arai M, Mishiro S. 2006. Hepatitis E virus infection in wild mongooses of Okinawa, Japan: demonstration of anti-HEV antibodies and a full-genome nucleotide sequence. Hepatol Res 34:137–140. doi: 10.1016/j.hepres.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich RG. 2010. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16:1452–1455. doi: 10.3201/eid1609.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell RH, Engle RE, Rood MP, Kabrane-Lazizi Y, Nguyen HT, Govindarajan S, St Claire M, Emerson SU. 2011. Hepatitis E virus in rats, Los Angeles, California, USA. Emerg Infect Dis 17:2216–2222. doi: 10.3201/eid1712.110482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose SM, Adu-Sarkodie Y, Oppong SK, Kalko EK, Osterman A, Rasche A, Adam A, Muller MA, Ulrich RG, Leroy EM, Lukashev AN, Drosten C. 2012. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86:9134–9147. doi: 10.1128/JVI.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj VS, Smits SL, Pas SD, Provacia LB, Moorman-Roest H, Osterhaus AD, Haagmans BL. 2012. Novel hepatitis E virus in ferrets, the Netherlands. Emerg Infect Dis 18:1369–1370. doi: 10.3201/eid1808.111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng XJ. 2006. Hepatitis E as a zoonotic disease, p 611–623. In Thomas H, Zuckermann A, Lemon S (ed), Viral hepatitis, 3rd ed Blackwell Publishing Ltd, Oxford, United Kingdom. [Google Scholar]
- 21.Hirano M, Ding X, Li TC, Takeda N, Kawabata H, Koizumi N, Kadosaka T, Goto I, Masuzawa T, Nakamura M, Taira K, Kuroki T, Tanikawa T, Watanabe H, Abe K. 2003. Evidence for widespread infection of hepatitis E virus among wild rats in Japan. Hepatol Res 27:1–5. doi: 10.1016/S1386-6346(03)00192-X. [DOI] [PubMed] [Google Scholar]
- 22.Meng XJ. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol 140:256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng XJ. 2011. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res 161:23–30. doi: 10.1016/j.virusres.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuo H, Yazaki Y, Sugawara K, Tsuda F, Takahashi M, Nishizawa T, Okamoto H. 2005. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J Med Virol 76:341–349. doi: 10.1002/jmv.20364. [DOI] [PubMed] [Google Scholar]
- 25.Huang FF, Sun ZF, Emerson SU, Purcell RH, Shivaprasad HL, Pierson FW, Toth TE, Meng XJ. 2004. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J Gen Virol 85:1609–1618. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- 26.Haqshenas G, Huang FF, Fenaux M, Guenette DK, Pierson FW, Larsen CT, Shivaprasad HL, Toth TE, Meng XJ. 2002. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J Gen Virol 83:2201–2209. [DOI] [PubMed] [Google Scholar]
- 27.Purdy MA, McCaustland KA, Krawczynski K, Spelbring J, Reyes GR, Bradley DW. 1993. Preliminary evidence that a trpE-HEV fusion protein protects cynomolgus macaques against challenge with wild-type hepatitis E virus (HEV). J Med Virol 41:90–94. doi: 10.1002/jmv.1890410118. [DOI] [PubMed] [Google Scholar]
- 28.Tsarev SA, Tsareva TS, Emerson SU, Govindarajan S, Shapiro M, Gerin JL, Purcell RH. 1994. Successful passive and active immunization of cynomolgus monkeys against hepatitis E. Proc Natl Acad Sci U S A 91:10198–10202. doi: 10.1073/pnas.91.21.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krawczynski K, Meng XJ, Rybczynska J. 2011. Pathogenetic elements of hepatitis E and animal models of HEV infection. Virus Res 161:78–83. doi: 10.1016/j.virusres.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Zhou EM, Sun ZF, Meng XJ, Halbur PG. 2006. Identification of B-cell epitopes in the capsid protein of avian hepatitis E virus (avian HEV) that are common to human and swine HEVs or unique to avian HEV. J Gen Virol 87:217–223. doi: 10.1099/vir.0.81393-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Q, Sun Y, Zhao J, Hu S, Zhao F, Chen F, Clavijo A, Zhou EM, Xiao Y. 2013. Development and application of an indirect ELISA for detection of antibodies against avian hepatitis E virus. J Virol Methods 187:32–36. doi: 10.1016/j.jviromet.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 32.Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, Toth TE, Meng XJ. 2002. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol 40:1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Jing S, Zhou EM, Sun P, Dong S, Li Y. 2009. Detection and characterization of human serum antibodies against avian hepatitis E virus. Chin J Health Lab Technol 19:1741–1744. [Google Scholar]
- 34.Wang XJ, Zhao Q, Jiang FL, Liu BY, Zhao JN, Dang L, Sun YN, Mu Y, Xiao SQ, Wang CB, Hsu WH, Liu L, Widen F, Zhou EM. 2014. Genetic characterization and serological prevalence of swine hepatitis E virus in Shandong Province, China. Vet Microbiol 172:415–424. doi: 10.1016/j.vetmic.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Dong S, Zhao Q, Lu M, Sun P, Qiu H, Zhang L, Lv J, Zhou EM. 2011. Analysis of epitopes in the capsid protein of avian hepatitis E virus by using monoclonal antibodies. J Virol Methods 171:374–380. doi: 10.1016/j.jviromet.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Q, Sun YN, Hu SB, Wang XJ, Xiao YH, Hsu WH, Xiao SQ, Wang CB, Mu Y, Hiscox JA, Zhou EM. 2013. Characterization of antigenic domains and epitopes in the ORF3 protein of a Chinese isolate of avian hepatitis E virus. Vet Microbiol 167:242–249. doi: 10.1016/j.vetmic.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Sun EC, Ma JN, Liu NH, Yang T, Zhao J, Geng HW, Wang LF, Qin YL, Bu ZG, Yang YH, Lunt RA, Wang LF, Wu DL. 2011. Identification of two linear B-cell epitopes from West Nile virus NS1 by screening a phage-displayed random peptide library. BMC Microbiol 11:160. doi: 10.1186/1471-2180-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, Zhao Q, Liang C, Dang L, Ma Y, Gao J, Li Q, Huang B, Kong N, Zhang C, Zhou EM. 2012. Complete genome sequence of a highly pathogenic porcine reproductive and respiratory syndrome virus variant. J Virol 86:8906. doi: 10.1128/JVI.01281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, Shima R, Moriishi K, Tsukihara T, Li TC, Takeda N, Miyamura T, Matsuura Y. 2009. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci U S A 106:12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troxler S, Marek A, Prokofieva I, Bilic I, Hess M. 2011. TaqMan real-time reverse transcription-PCR assay for universal detection and quantification of avian hepatitis E virus from clinical samples in the presence of a heterologous internal control RNA. J Clin Microbiol 49:1339–1346. doi: 10.1128/JCM.01626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo H, Zhou EM, Sun ZF, Meng XJ. 2008. Immunodominant epitopes mapped by synthetic peptides on the capsid protein of avian hepatitis E virus are non-protective. Viral Immunol 21:61–67. doi: 10.1089/vim.2007.0082. [DOI] [PubMed] [Google Scholar]
- 42.Smith DB, Simmonds P, International Committee on Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng XJ, Anderson DA, Arankalle VA, Emerson SU, Harrison TJ, Jameel S, Okamoto H. 2012. Hepeviridae, p 1021–1028. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom. [Google Scholar]
- 44.Guo H, Zhou EM, Sun ZF, Meng XJ. 2007. Protection of chickens against avian hepatitis E virus (avian HEV) infection by immunization with recombinant avian HEV capsid protein. Vaccine 25:2892–2899. doi: 10.1016/j.vaccine.2006.09.038. [DOI] [PubMed] [Google Scholar]