ABSTRACT
Acquisition of human cytomegalovirus (CMV) usually occurs by contact between contaminated bodily fluids, such as urine and saliva, and host mucosal cells. Langerhans-type dendritic cells (LC) are the only type of immune cells found in the outermost layers of the oral mucosae, where they not only provide a first line of defense against CMV but can easily be targeted by orally administered vaccines, while their bone marrow resident progenitors are important sites of virus latency. In this work, we tracked the progress of infection in CD34+ progenitor cells, immature LC (iLC), and mature LC (mLC) exposed to the clinical-like strain TB40-BAC4 or to the vaccine strain AD169varATCC, prior to their long-term maintenance under either immature or mature conditions. We show that the genomes of both strains are efficiently maintained in CD34+ cells during their differentiation into iLC, although this requires the presence of larger amounts of input AD169varATCC DNA. Lipopolysaccharide- and CD40 ligand-induced maturation of iLC derived from latently infected progenitors was not associated with robust viral genome replication and progeny production, while maturation of directly infected iLC increased and prolonged expression of the viral immediate early proteins. While effective replication of viral genomes from both strains occurred only in mLC, both iLC and mLC produced viral progeny, suggesting that both types of LC may contribute to CMV horizontal transmission in vivo.
IMPORTANCE Human CMV is usually acquired via the oral and nasal mucosae. Langerhans-type dendritic cells (LC) are the only type of immune cells found in the outermost layers of these tissues. Understanding how CMV interacts with LC and their hematopoietic progenitors is thus essential to develop innovative means of defense against this virus. Here we show that the genomes of a virulent and an attenuated strain of CMV are maintained in hematopoietic progenitor cells during their differentiation into immature LC and that maturation of these cells by exposure to lipopolysaccharide and CD40 ligand is not sufficient to trigger virus reactivation. While the extents of viral protein expression and genome replication were broadest in directly infected mature LC populations, similar amounts of viral progeny were detected in the supernatants of immature and mature LC, suggesting that these immune cells of the oral mucosa are likely to be important for CMV transmission within the human population.
INTRODUCTION
Human cytomegalovirus (CMV), a major cause of disease and death in immunocompromised and in congenitally infected individuals, is normally acquired by contact between infected bodily fluids such as urine and saliva and host mucosal surfaces, particularly those lining the oral and nasal cavities.
From these peripheral sites, CMV is thought to reach the circulation and hence the bone marrow, where latency is established in myeloid cells, including CD34+ hematopoietic progenitors (1–5). Reactivation from latency in the myeloid progeny of these cells is then believed to be the source of newly produced viral particles, which upon amplification in oral epithelial cells are released in the saliva and contribute to CMV horizontal transmission in vivo (6–8).
Immature Langerhans-type dendritic cells (iLC) are the only professional antigen-presenting cells located in the outermost layers of the oral mucosa (9–14). As such, iLC are bound to be the first immune cells to encounter invading pathogens that access their hosts via the oral cavity, such as CMV. Upon contact with “danger” signals, iLC migrate toward the draining lymph nodes and begin the process of maturation, which culminates with the activation of adaptive immune responses (15). Although mature Langerhans-type dendritic cells (mLC) usually reside within secondary lymphoid organs, their presence has been detected in the oral mucosae of individuals with inflammatory diseases such as gingivitis, periodontitis, and oral ulcers (16, 17), suggesting that under certain conditions, mLC may also get in direct contact with CMV.
As the number of latently infected mononuclear cells found in the blood of healthy donors and the amount of live LC that can be obtained from oral tissues are usually extremely low, CMV infection of CD34+ progenitor cells, iLC, and mLC has been studied predominantly by using in vitro culture systems.
In 1994, Kondo et al. first showed that viral genomes could be maintained in the absence of productive infection in fetal liver-derived granulocyte-macrophage progenitor cells (GM-Ps) exposed to the laboratory-adapted CMV strains Towne RC256 and Toledo (18). As cells proliferated over a time course of 28 days, viral DNA accumulated in parallel with the number of GM-Ps. Treatment with ganciclovir, an inhibitor of productive viral replication, did not affect this trend. In later studies using CD34+ cells purified from GM-Ps, establishment of latency without production or release of viral particles was also observed with the attenuated CMV strains TownevarRIT3 and AD169varATCC (19, 20).
In 2002, Goodrum et al. presented a new model to study CMV latency, consisting of bone marrow-derived CD34+ cells maintained on irradiated murine stromal cells in the presence of serum (21). While clinical-like strains underwent latency in this system, productive replication was observed in cells infected with Towne and AD169varATCC (21–23), leading to the conclusion that viral open reading frames (ORFs) missing from the genomes of attenuated strains were required to establish latency (23).
A year later, we introduced a new system to study CMV interactions with iLC and mLC differentiated in vitro from umbilical cord or peripheral blood CD34+ progenitor cells using a highly defined, serum-free cytokine cocktail (24), which was shown to generate LC displaying the same morphological and ultrastructural features of epithelial iLC (25–28) and expressing some of the markers of oral LC (24, 29, 30). Using this model, we showed that mLC are more permissive to infection onset than iLC and that this is due not to defects in viral entry but rather to the inefficient transcription of viral genes, particularly those encoding the immediate early proteins 1 and 2 (IE1/IE2), in iLC (9, 24). The fact that both clinical-like and laboratory-adapted strains could initiate their replication cycle in both LC types, moreover, implied that none of the proteins encoded by the ORFs lacking from the genomes of attenuated strains is absolutely required for CMV infection of LC (9, 24).
Using the same system, Reeves et al. then showed that differentiation of CD34+ cells latently infected with the clinical-like strain TB40/E was associated with the maintenance of viral genomes in a transcriptionally silent state, while iLC maturation triggered reactivation (31, 32).
In this work, we used our CD34+ cell-iLC-mLC differentiation/maturation model to (i) quantify changes in viral and cellular genome amounts occurring during the differentiation of latently infected CD34+ cells into iLC, (ii) compare the effects of maturation on CMV reactivation in LC derived from latently infected CD34+ cells and on CMV replication in directly infected LC, (iii) compare the kinetics of viral replication in CD34+ cells, iLC, and mLC to those in human foreskin fibroblasts (HFF), a cell type highly permissive to lytic infection, and (iv) compare the infection kinetics of the CMV vaccine strain AD169varATCC, possessing a shorter genome and a more restricted tropism, to that of the clinical-like strain TB40-BAC4, which is characterized by a more complete genome and a broader tropism.
Currently available data provide only qualitative evidence of viral genome carriage during the differentiation of CD34+ cells into iLC and of viral reactivation in mLC (31, 32). As the process of CD34+ cell differentiation is associated with substantial increases in cell numbers (up to 20- to 40-fold), the quantitative measurement of viral genome copy numbers present over time becomes crucial to understand whether viral DNA is lost, maintained, or replicated during latency. We show that, after an initial sharp drop, TB40-BAC4 genomes are maintained in proliferating/differentiating CD34+ cells in the absence of IE1/IE2 protein expression and progeny release, while the efficient carriage of AD169varATCC genomes requires the presence of high(er) amounts of viral genomes at the start of infection.
Thus far, the effects of LC maturation on CMV infection dynamics have been separately evaluated within the context of viral reactivation in lipopolysaccharide (LPS)-treated iLC differentiated from latently infected, bone marrow-derived CD34+ cells (31, 32) or of viral lytic replication in LPS- and/or CD40 ligand (CD40L)-treated iLC populations differentiated from umbilical cord or peripheral blood derived CD34+ cells (9, 24). Because these assays were conducted with LC populations differentiated from different sets of CD34+ cells donors, exposed to different maturation stimuli, and infected with different viral strains, results are difficult to compare. In this work, we asked whether maturation by exposure to CD40L and LPS, stimuli known to increase the susceptibility of iLC to infection onset, would also trigger CMV reactivation in iLC carrying latent viral genomes and whether the delivery of maturation signals after iLC exposure to viral particles would be as effective in supporting infection progress as in cells exposed to virus when already mature. We found that maturation of iLC differentiated from latently infected, umbilical cord blood-derived CD34+ cells was not accompanied by the robust amplification of viral DNA, although TB40-BAC4 genome amounts were larger in mature than in immature LC differentiated from latently infected CD34+ cells. The fact that no viral progeny was observed in culture supernatants, however, suggests that exposure to LPS and CD40L molecules is not sufficient to trigger vigorous CMV reactivation. Yet, maturation was associated with the enhanced and prolonged expression of the IE1/IE2 proteins in directly infected mLC and in iLC matured immediately after infection, suggesting that LPS/CD40L signaling can rapidly induce the establishment of an intranuclear environment supportive of viral gene transcription from genomes delivered by viral particles, but not as much from genomes already present in the nuclei of latently infected iLC. Interestingly, this enhancement in IE1/IE2 protein expression was not followed by a similar increase in viral genome amplification events. Instead, viral DNA replication was most efficient in cells infected when already mature, suggesting that the supportive milieu promoted by maturation must already be present at the time of infection onset to reach high genome replication rates.
Human fibroblasts have been traditionally used to study CMV pathogenesis on account of their high degree of permissiveness to CMV replication in vitro. Consequently, these cells have become a basis of comparison to assess CMV tropism for other cell types. Our evaluation of CMV infection progress in HFF, iLC, and mLC revealed that the extents of viral protein expression, genome amplification, and progeny production were dramatically lower in iLC and mLC than in HFF. This suggests that, irrespective of the susceptibility differences between iLC and mLC, both LC types are more resistant to infection than HFF, at least when exposed to the two CMV strains tested here.
Consistent with our previous results (24), we found that a larger proportion of mLC than iLC express the IE1/IE2 proteins at day two and that mLC produce and release viral progeny. Progress of infection in iLC, however, had not been previously assessed. Here we show that iLC too produce viral progeny, a quite surprising finding considering the absence of widespread IE1/IE2 protein synthesis and of large increases in viral genome amounts, as seen in mLC and in HFF cultures. Immature LC thus appear to be less resistant to CMV infection than previously believed, and they may contribute to CMV horizontal transmission in vivo by releasing newly produced virus in the oral cavity. Our finding that viral yields were reduced, rather than increased, upon maintenance of infected iLC under maturation conditions was also quite intriguing, as it may hint at the existence of cell defense mechanisms acting to restrict viral progeny production in infected iLC during their migration toward the draining lymph nodes in vivo, in order to prevent virus spread to other cell types within the host.
Finally, all studies of CMV latency making use of our CD34+ cell-iLC-mLC differentiation/maturation model (31, 32), as well as our initial work on LC susceptibility to CMV infection and immunomodulation (24, 33, 34), were conducted using the broadly tropic but genotypically heterogeneous strain TB40/E (35). Consequently, no data are currently available on the behavior of attenuated strains in this model, except for our findings that both Towne and AD169varATCC can initiate infection in iLC and mLC (9, 24). Studies using these strains in latency models have yielded contrasting results. While Towne and AD169varATCC were reported to establish latent infections in GM-Ps (18–20), both strains were found to initiate productive infections in CD34+ cells cultured on irradiated murine stromal cells (21–23). In this work, we compared the infection kinetics of TB40-BAC4, a genotypically uniform strain derived from the bacterial artificial chromosome cloning of a highly endotheliotropic subpopulation of TB40/E (35), to those of AD169varATCC. The progress of AD169varATCC infection in both iLC and mLC beyond the initial stages of infection was also tracked. No evidence of productive replication was found in AD169varATCC-infected CD34+ cell cultures, in accordance with results from GM-Ps (20) but not from CD34+ cells/murine stromal cells cultures (23). The only difference we observed between the infection kinetics of TB40-BAC4 and AD169varATCC was in the efficiency of viral genome maintenance during CD34+ cells differentiation, suggesting that no additional ORF beyond those contained in the genome of AD169varATCC is required to support entry and replication in directly infected iLC and mLC.
MATERIALS AND METHODS
Cells.
Umbilical cord blood CD34+ hematopoietic progenitor cells were purchased from Stemcell Technologies Inc., Vancouver, Canada. Immature LC populations were obtained by culturing CD34+ cells at a density of 1 × 104 to 2.5 × 104 cells per well in 48-well tissue culture plates using serum-free X-VIVO 15 medium (Lonza/BioWhittaker, Allendale, NJ) supplemented with 1,500 IU/ml of granulocyte-macrophage colony-stimulating factor (GM-CSF) (Leukine Sargramostim; Immunex, Seattle, WA), 2.5 ng/ml of tumor necrosis factor alpha, 20 ng/ml of stem cell factor, 100 ng/ml of Flt3 ligand, and 0.5 ng/ml of transforming growth factor β1 (all purchased from Peprotech, Rocky Hill, NJ), while maturation was induced by cell exposure to X-VIVO 15 medium containing 10% fetal bovine serum, 200 ng/ml of CD40 ligand (Immunex, Seattle, WA), 1,500 IU/ml of GM-CSF, and 250 ng/ml of lipopolysaccharide (Sigma-Aldrich, St. Louis, MO) as previously described (9, 24). Human foreskin fibroblasts were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal clone serum III (HyClone), 100 U/ml penicillin and 100 μg/ml streptomycin, 4 mM HEPES, and 1 mM sodium pyruvate (Gibco, Life Technologies, Grand Island, NY).
Virus strains and titrations.
AD169varATCC (36) and TB40-BAC4 (35), gifts from E. S. Mocarski (Emory University, Atlanta, GA) and C. Sinzger (University of Ulm, Ulm, Germany), respectively, were propagated on HFF and purified by ultracentrifugation as previously described (24). Titers of virus stocks and culture supernatants were determined by immunofluorescence staining analyses of HFF harvested at 24 h postinfection (hpi) and stained with the monoclonal antibody MAb810 (1:500; Chemicon, Temecula, CA) directed against the viral IE1/IE2 proteins.
Cell infection.
HFF were plated at a density of ∼ 2 × 104 cells/cm2 3 days before infection with TB40-BAC4 at a multiplicity of infection (MOI) of 0.05 PFU/cell. The virus inoculum was left in contact with cells at 37°C in 5% CO2 for 4 h, after which cells were washed twice prior to incubation in complete DMEM. Supernatant and cells were collected at 4 hpi and at days 2, 4, 6, 8, 10, and 14. Immediately after thawing, CD34+ cells from each donor were left uninfected or were exposed to AD169varATCC or TB40-BAC4 virions at a calculated MOI of 10 for 4 h prior to washing and plating in iLC medium. At day eight, iLC were harvested, counted, and replated in either immature or maturation medium. Differentiating CD34+ cells and supernatants were collected at 4 hpi and at days 2, 4, 6, and 8, while iLC derived from infected CD34+ cells and their supernatants were harvested at 4 hpi and at days 2, 4, 6, 8, 10, and 14. Immature LC differentiated from uninfected CD34+ cells were also harvested on day eight, counted, and exposed to AD169varATCC or TB40-BAC4 virions for 4 h, prior to washing and plating in either iLC or mLC medium, or were left uninfected and cultured under maturation conditions. These uninfected cells were then harvested at day two postmaturation, exposed to AD169varATCC or TB40-BAC4 virions for 4 h, washed, and plated again in mLC medium. Infected iLC and mLC were then collected at 4 hpi and at days 2, 4, 6, 8, 10, and 14. For foscarnet treatment, cells were exposed to medium containing 300 μg/ml (1 mM) of the viral polymerase inhibitor phosphonoformic acid, as in our previous work (37). All cells and supernatant samples were stored at −80°C prior to processing for titration and real-time quantitative PCR (qPCR) analyses, respectively.
Immunofluorescence staining analyses (IFA).
HFF grown on 12-mm-diameter glass coverslips and LC deposited on glass slides by centrifugation at 500 rpm for 3 min at room temperature (RT) using a Cytospin 4 (Thermo Shandon) were fixed in 1% formaldehyde for 30 min at RT, permeabilized in 0.5% Triton X-100 for 20 min on ice, treated with blocking buffer (20% fetal bovine serum in phosphate-buffered saline [PBS]) for 30 min at RT, and incubated with mouse monoclonal antibody MAb810 (1:400; EMD Millipore), directed against an epitope encoded by exon 2 of the UL122/123 ORF and common to both the IE1 and IE2 proteins (38) for 1 h at RT in a humidified chamber. After washing in blocking buffer, samples were incubated with Alexa Fluor 594-conjugated goat anti-mouse antibodies (1:200; Life Technologies, Carlsbad, CA) for 1 h at RT, followed by nuclear labeling with Hoechst 33342 (0.2 mg/ml; Molecular Probes, Eugene, OR) for 3 min at RT. For IE1/IE2 and ppUL44 double staining, fixed cells were first incubated with a monoclonal anti-ppUL44 antibody (1:500 dilution; Goodwin Institute, Plantation, FL) followed by an Alexa Fluor 594-conjugated goat anti-mouse antibody, blocked with normal mouse immunoglobulin G (1:100; Caltag, Burlingame, CA), and then stained with an Alexa Fluor 488-conjugated anti-IE1/IE2 monoclonal antibody. Slides were then mounted in 90% glycerol–10% PBS containing 2.5 g/liter of 1, 4-diazabicyclo-(2,2,2)-octane (DABCO) (Alfa Aesar, Pelham, NH) and viewed on a Nikon Eclipse E600 fluorescence microscope equipped with iVision-Mac imaging software. The percentage of IE1/IE2+ cells was then determined using the ImageJ 1.47v software.
Real-time quantitative genomic PCR.
Genomic DNA was extracted from infected and uninfected cells using the OmniGenX PureSpin gDNA miniprep kit (E&K Scientific, Santa Clara, CA). mRNA was obtained using the μMACS mRNA isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen, Life Technologies, Grand Island, NY). Real-time quantitative PCRs were performed using the iTaq SYBR green supermix with ROX (Bio-Rad, Hercules, CA) and an ABI7900 thermocycler (Applied Biosystems, Carlsbad, CA), with primers hybridizing to exon 2 of the viral UL122 and UL123 ORFs (forward primer, 5′-GGCCGAAGAATCCTCAAAA-3′; reverse primer, 5′-TCGTTGCAATCCTCGGTCA-3′) or to the cellular albumin gene (forward primer, 5′-GCTGTCATCTCTTGTGGGCTGT-3′; reverse primer. 5′-AAACTCATGGGAGCTGCTGGTT-3′) as in our previous work (37). The following cycling parameters were used: 95°C for 2 min to activate the iTaq polymerase, followed by 40 cycles of template denaturation at 95°C for 15 s, primer annealing at 51°C (UL122/123) or 57°C (albumin) for 30 s, and product extension at 72°C for 30 s. Absolute quantifications of viral and cellular genome amounts were obtained using a standard curve made by serial dilutions of plasmid pON303 (39) and of plasmid pTOPO-albumin (9). The number of viral genome copies per cell was calculated as number of viral DNA copies/(number of albumin DNA copies/2) (40).
Statistical analysis.
Student t tests (paired, two tailed) were conducted to compare the mean values of two data sets, while the nonparametric and distribution free Kolmogorov-Smirnov (KS) test was used to compare the cumulative distributions of any two data sets (http://www.wessa.net/rwasp_Reddy-Moores%20K-S%20Test.wasp/). Differences were considered significant at a P value of <0.05.
RESULTS
Hematopoietic cell culture systems used.
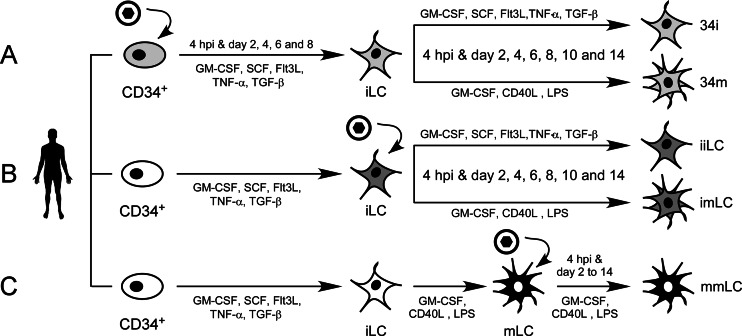
Umbilical cord blood-derived CD34+ cells from a total of six different donors were exposed to TB40-BAC4 or AD169varATCC virions for 4 h (Fig. 1A) or were left uninfected (Fig. 1B) prior to culture in iLC differentiation medium for 8 days. Infected CD34+ cells and their supernatants were collected at 4 hpi and at days 2, 4, 6, and 8. At day 8, differentiated iLC were counted and replated either in iLC medium (infected CD34 maintained as iLC [34i]) or in mLC maturation medium (infected CD34 maintained as mLC [34m]) for 14 days, with cells and supernatants collected again every 2 days (Fig. 1A).
FIG 1.
Schematic representation of the methods used to generate the five different types of dendritic cells analyzed in this work. Umbilical cord blood-derived CD34+ hematopoietic progenitor cells from single donors were partitioned into two groups immediately after thawing. One set was left uninfected (B, white oval), while the other was exposed to TB40-BAC4 or AD169varATCC virions for 4 h (A, light gray oval). Infected and uninfected CD34+ cells were then cultured in iLC differentiation medium for 8 days, with supernatants and cells collected every 2 days. (A) Immature LC derived from infected progenitors were harvested on day eight and replated in either fresh iLC differentiation medium (34i) or maturation medium (34m) for 14 days, with collection of cells and supernatants every 2 days. (B) Immature LC derived from uninfected progenitors were harvested on day eight and exposed to TB40-BAC4 or AD169varATCC virions for 4 h prior to culture in iLC differentiation medium (iiLC) or in maturation medium (imLC) for 14 days, with collection of cells and supernatants every 2 days. (C) A portion of the iLC differentiated from uninfected CD34+ cells was cultured in maturation medium for 2 days to generate mLC, which were then exposed to TB40-BAC4 or AD169varATCC virions for 4 h prior to culture in maturation medium (mmLC) for 14 days, with supernatants and cells collected every 2 days. White ovals and star-shaped outlines represent uninfected progenitor and dendritic cells, respectively, while light gray, dark gray, and black outlines represent infected cells. Black circles enclosing a black hexagon depict CMV virions. GM-CSF, granulocyte-macrophage-colony-stimulating factor; SCF, stem cell factor; Flt3L, Flt3 ligand; TNF-α, tumor necrosis factor alpha; TGF-β, transforming growth factor β1; CD40L, CD40 ligand; LPS, lipopolysaccharide.
Immature LC differentiated from uninfected CD34+ cells were also harvested at day eight, prior to exposure to TB40-BAC4 or AD169varATCC virions and culture either in iLC differentiation medium (infected iLC maintained as iLC [iiLC]) or in mLC maturation medium (infected iLC maintained as mLC [imLC]) for 14 days, with collections every 2 days (Fig. 1B).
A portion of the iLC harvested at day eight was kept uninfected and matured by exposure to LPS and CD40L for 2 days. Mature LC were then infected with TB40-BAC4 or AD169varATCC and harvested every 2 days until day 14 (infected mLC maintained as mLC [mmLC]) (Fig. 1C).
Real-time quantitative PCR with primers hybridizing to the viral UL122/123 open reading frame or to the cellular albumin gene was then used to determine the total number of cells per well and of viral genomes per well present at each time point and to calculate the proportion of cellular and viral genomes remaining over time relative to that at 4 hpi (input). The onset of lytic infection was tracked in each cell type by IFA using monoclonal antibodies directed against an N-terminal epitope shared by the IE1 (p72) and IE2 (p86) proteins expressed during lytic infections but absent from the smaller IE1x4 isoform expressed in CD34+ cells during latency (41). The amount of viral particles present in the collected supernatants was also quantified by IFA of HFF harvested at 24 h postexposure to each medium and stained for the IE1/IE2 proteins.
TB40-BAC4 genomes are robustly amplified in infected HFF with abundant progeny release.
To establish a basis of comparison for the kinetics of CMV infection in hematopoietic cells, the extent of cell proliferation, viral genome replication, IE1/IE2 protein expression, and viral progeny production were quantified in TB40-BAC4-infected HFF, as these cells are highly permissive to CMV lytic replication in vitro and are routinely used to study CMV pathogenesis (8, 42). Infections were conducted at an MOI of 0.05 PFU/ml to limit the number of cells receiving at least one viral particle to 5%, a percentage similar to that of iLC expressing the IE1/IE2 proteins after exposure to CMV (9). Upon removal of the inoculum at 4 hpi, HFF were washed and incubated in fresh medium supplemented or not with 300 μg/ml of the viral polymerase inhibitor foscarnet, as in our previous work (37). Cells and supernatants were collected at 4 hpi and at days 2, 4, 6, 8, 10, and 14.
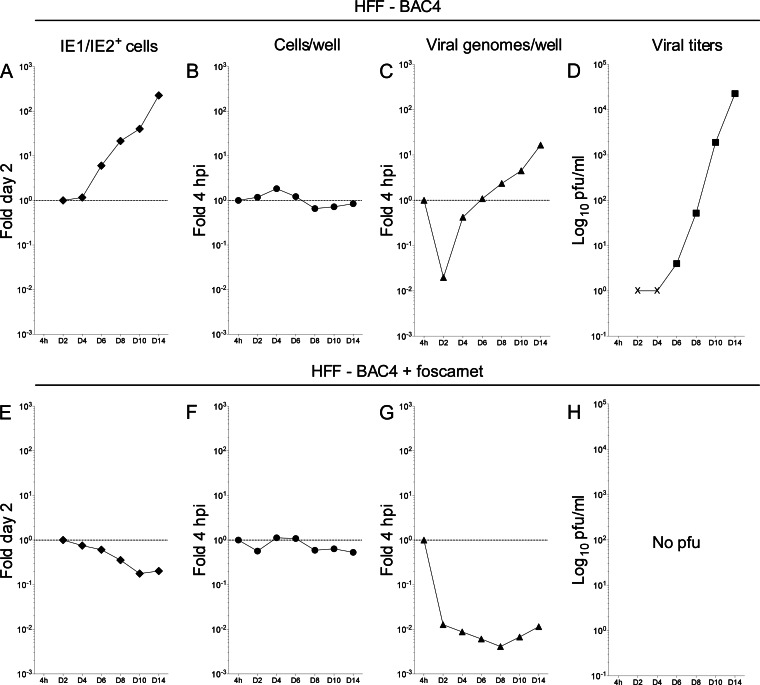
As expected for low-MOI infections, IE1/IE2 expression was initially detected in the nuclei of single cells scattered throughout the monolayer. From day four onwards, however, single positive cells became rare, while foci containing multiple IE1/IE2-expressing cells began to predominate. These foci progressively increased in size and in numbers, until 80 to 90% of the cells were IE1/IE2+ at day 14 (Fig. 2A). Although the initial number of antigen-expressing cells was identical in the presence and absence of foscarnet, the lack of polymerase activity prevented the development of plaques in foscarnet-treated cultures. Consequently, the total number of IE1/IE2+ cells decreased, instead of increasing, over time (Fig. 2E). Consistent with infections conducted at confluence and at low MOI, the total number of cells per well remained fairly constant over time, regardless of foscarnet treatment (Fig. 2B and F), indicating that addition of the inhibitor at this specific concentration did not induce substantial cell loss. After a dramatic drop at day two, the number of viral genomes per well rapidly increased, reaching values 17-fold higher than input on the last collection day (Fig. 2C). No genome amplification was observed in the presence of foscarnet. Instead, input DNA amounts sharply decreased to about 1% of the inoculum (Fig. 2G). While no viral progeny was detected at any time in the supernatants of foscarnet-treated cultures (Fig. 2H), viral particles remained undetectable in the medium of untreated cells only until day four (Fig. 2D, ×), after which yields rapidly and steadily increased, reaching values 300-fold higher than input at day 14 (Fig. 2D).
FIG 2.
TB40-BAC4 growth kinetics in confluent HFF infected at a low MOI. HFF were exposed to TB40-BAC4 virions at an MOI of 0.05 for 4 h prior to washing and further culture in the presence or absence of 300 μg/ml of the viral polymerase inhibitor foscarnet for 14 days. Cells and supernatants collected at days 2, 4, 6, 8, 10, and 14 were used to determine the proportion of cells expressing the IE1/IE2 proteins relative to that at day two (fold day 2) (A and E), the proportions of cells per well (B and F) and of viral genomes per well (C and G) present at each time point relative to the beginning of infection at 4 hpi (fold 4 hpi), and the amount of virus released into the supernatant (PFU/ml) (D and H). The dotted horizontal line marks the 1-fold point on the y axis.
In HFF infected at low MOI, thus, both the proportion of IE1/IE2-expressing cells and the total amounts of viral genomes per well increase linearly over time, while progeny production rates follow a sigmoidal distribution, as expected for multistep lytic replication.
TB40-BAC4 and AD169varATCC genomes are maintained in the absence of lytic gene expression and progeny production in proliferating/differentiating CD34+ cells.
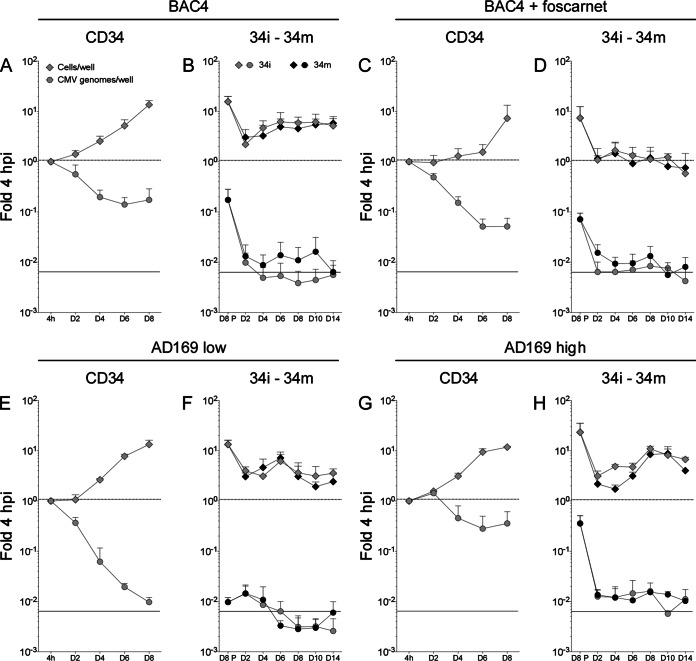
Consistent with the expected behavior for stem cells, the total number of infected CD34+ cells per well increased over time irrespective of the virus strain used and of the presence or absence of 300 μg/ml of foscarnet (Fig. 3A, C, E, and G, gray diamonds). The rates of cell growth were slightly different, however, with median values of 2-fold every 2 days for TB40-BAC4- and AD169varATCC-infected cells but of only 1.3-fold in the presence of foscarnet. This suggests that, in contrast to the case for HFF, foscarnet may be toxic to CD34+ cells at this specific concentration, possibly due to the reduced proliferation rates of HFF compared to CD34+ cells. No difference was observed in the total amplification extent (about 20-fold at day eight) of CMV- and mock-infected cells (not shown), indicating that infection had no negative effects on cell viability.
FIG 3.
Kinetics of TB40-BAC4 and AD169varATCC infection in proliferating/differentiating CD34+ cells and in derived LC maintained under immature (34i) or mature (34m) conditions. Immediately after thawing, CD34+ progenitor cells were exposed to TB40-BAC4 (A and C) or AD169varATCC (E and G) virions for 4 h prior to washing and culture in iLC differentiation medium containing (C) or not (A, E and G) 300 μg/ml of foscarnet. At the end of the iLC differentiation period on day eight, cells were harvested, counted, and replated in either iLC differentiation medium (34i) or maturation medium (34m) for 14 days, with (D) or without (B, F, and H) foscarnet. Real-time quantitative genomic PCR was used to determine the proportion of cells per well (light gray diamonds for CD34+ cells and 34i, black diamonds for 34m) and of viral genomes per well (light gray circles for CD34+ cells and 34i, black circles for 34m) present at each time point relative to the beginning of infection (fold 4 hpi). Panels A to F show results obtained from cultures containing 105 to 106 copies of viral DNA per well at 4 hpi, while panels G and H show results obtained from AD169varATCC-infected cells containing 106 to 107 copies of viral DNA per well at 4 hpi. Graphs show median and median absolute deviation values of data collected from six different CD34+ cell donors in six independent experiments (A to D), four different donors in three independent experiments (E and F), and three different donors in two independent experiments (G and H). The dotted line marks the 1-fold point, while the solid line marks the limit of detection point on the y axis. D8P refers to the proportion of cells per well or genomes per well replated on day eight relative to 4 hpi.
After an initial decrease, the total number of viral genomes per well in TB40-BAC4-infected cultures appeared to stabilize at values corresponding to 20% of the initial input in the absence of foscarnet (Fig. 3A, gray circles), while in the presence of the inhibitor, viral genome loss continued until only 5 to 7% of initial DNA amounts remained at day eight (Fig. 3C). These differences, however, did not reach statistical significance in donor paired two-tailed Student t tests (P = 0.233 at day six and P = 0.152 at day eight), and the two data distributions (Fig. 3A and C, day two to day eight) did not score as different in a two-sample KS test (P = 0.249).
A similar sharp decrease was also observed in AD169varATCC-infected cultures that contained amounts (105 to 106) of genomes per well at 4 hpi comparable to those for TB40-BAC4-infected cultures (Fig. 3E). In contrast, the loss of viral DNA in cultures derived from CD34+ cells harboring larger amounts (106 to 107) of AD169varATCC genomes per well at 4 hpi was substantially less profound, leveling at 20 to 36% of initial input at day six to eight (Fig. 3G), with the two data distributions (Fig. 3E and G) being substantially different according to a KS test (P = 0.014).
No IE1/IE2+ cells were observed by IFA analysis of cytospin preparations, and no expression of the UL122/123 genes was detected by real-time quantitative reverse transcription-PCR analyses of samples at each time postinfection (data not shown), while extremely small amounts of viral particles were detected in the supernatants of infected cultures, irrespective of the presence or absence of foscarnet (Table 1). As extracellular virions were not removed by proteolytic digestion at 4 hpi, these particles are more likely to represent unpenetrated virions than newly produced viral progeny.
TABLE 1.
Viral particle contents in the supernatants of CD34+, 34i, and 34m cells
| Cells | Viral particle content (PFU/ml) on daya: |
|||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 14 | |
| CD34+ BAC4 | 0.1 ± 0.5 | 0.2 ± 0.5 | 0.3 ± 0.8 | 0.6 ± 1.9 | ||
| CD34+ with foscarnet | 0.0 ± 0.0 | 0.2 ± 0.4 | 0.0 ± 0.0 | 0.2 ± 0.6 | ||
| CD34+ AD169 | NA | 0.3 ± 0.8 | 0.4 ± 0.5 | 0.0 ± 0.0 | ||
| 34i BAC4 | 0.2 ± 0.8 | 0.1 ± 0.4 | 0.3 ± 0.8 | 0.4 ± 1.7 | 0.1 ± 0.3 | 0.2 ± 0.6 |
| 34i with foscarnet | 0.4 ± 1.3 | 0.2 ± 0.7 | 0.2 ± 0.7 | 0.2 ± 0.7 | 0.2 ± 0.7 | 0.4 ± 0.9 |
| 34i AD169 | 0.5 ± 0.9 | 0.1 ± 0.4 | 0.4 ± 0.7 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 34m BAC4 | 0.2 ± 0.5 | 0.1 ± 0.7 | 0.1 ± 0.3 | 0.3 ± 1.7 | 0.0 ± 0.0 | 0.1 ± 0.7 |
| 34m with foscarnet | 0.2 ± 0.7 | 0.4 ± 1.3 | 0.0 ± 0.0 | 1.1 ± 3.3 | 0.0 ± 0.0 | 0.4 ± 1.3 |
| 34m AD169 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
Values are means ± standard deviations. NA, not available.
Together, these results indicate that both TB40-BAC4 and AD169varATCC establish nonlytic infections in proliferating and differentiating CD34+ cells, with their genomes being maintained over time in a process that may involve the activity of the viral polymerase, and that requires larger initial amounts of AD169varATCC genomes to attain the same degree of efficiency as in TB40-BAC4-infected cultures.
TB40-BAC4 and AD169varATCC genomes are maintained at low levels in LC derived from infected CD34+ cells irrespective of maturation.
At the end of the differentiation period at day eight, iLC derived from infected CD34+ cells were harvested and replated in either differentiation (34i) or maturation (34m) medium for 14 days (Fig. 1A). Cell replating was associated with a sharp 10-fold drop in both the total number of cells per well and viral genomes per well until day two postplating, after which the total number of cells per well started to rise again until day six to eight, irrespective of the medium used (Fig. 3B, F, and H, gray and black diamonds), and with the exception of TB40-BAC4-infected, foscarnet-treated cells (Fig. 3D). This increase was not mirrored by the total number of viral genomes per well, which did not increase again but remained instead at levels at or below background until day 14 (Fig. 3B, D, F, and H, gray and black circles). Viral genome loss was less severe in 34i and 34m cells derived from CD34+ cells containing large amounts of AD169varATCC genomes (Fig. 3H), and the cumulative distributions of data were considered to be different according to the KS comparisons of 34i data from populations containing low and high viral genome amounts (Fig. 3F and H, gray circles; P = 0.003) and to KS comparisons of 34m data from populations containing small and large viral genome amounts (Fig. 3F and H, black circles; P = 0.013).
Similarly, viral genome loss was less pronounced in 34m than in 34i cells derived from TB40-BAC4-infected progenitors (Fig. 3B), with the two distributions scoring as different (P = 0.013 for the donor paired KS comparison of 34i and 34m samples in Fig. 3B).
In contrast, treatment of 34i or 34m cells with foscarnet did not appear to have any major effect on their content in viral genomes over time. Accordingly, the distributions of data from treated and untreated samples were not statistically different (P = 0.139 for the KS comparison of treated to untreated 34i or 34m samples [Fig. 3B to D]).
No IE1/IE2+ cells were observed at any time point, and the amount of viral particles detected in the supernatants was again extremely small and very similar in cells treated or not with foscarnet (Table 1).
Together, these findings suggest that viral genomes are maintained at very low levels in both immature and mature LC derived from infected CD34+ progenitors and that this process is more efficient in 34i and 34m cells derived from progenitors containing large amounts of AD169varATCC DNA and in 34m cells derived from TB40-BAC4-infected CD34+ cells. Although some viral genome replication may be occurring, viral progeny is not produced in large quantities, if at all, by any of these cells. Thus, LPS and CD40L stimulation of iLC differentiated from infected progenitors does not appear to induce robust viral DNA replication and progeny production.
TB40-BAC4- or AD169varATCC-infected iLC maintained under immature conditions (iiLC) produce viral progeny in the absence of widespread IE1/IE2 gene expression and of robust genome amplification.
We next assessed the kinetics of viral replication in iLC directly exposed to TB40-BAC4 or AD169varATCC virions and subsequently maintained under immature (iiLC) or mature (imLC) conditions (Fig. 1B).
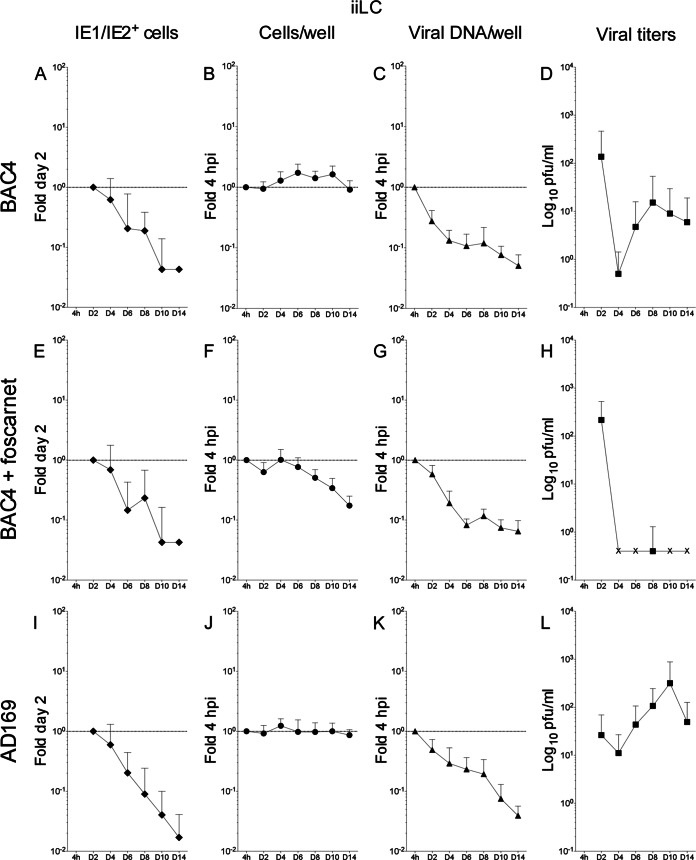
Consistent with previous results (9, 24), only a small fraction of iiLC expressed the IE1/IE2 proteins at day two postinfection with TB40-BAC4 (0.8% ± 1.5%) or AD169varATCC (1.7% ± 0.7%). These proportions rapidly decreased, plummeting to zero by day 14 (Fig. 4A, E, and I). The same trend was followed by the total number of viral genomes per well (Fig. 4C, G, and K) and by the total amount of cells per well in foscarnet-treated cultures (Fig. 4F). In contrast, the total amounts of TB40-BAC4- or AD169varATCC-infected cells remained fairly constant (Fig. 4B and J), indicating that the sharp decline in TB40-BAC4 and AD169varATCC genome copy numbers was not due to massive cell loss.
FIG 4.
Kinetics of TB40-BAC4 and AD169varATCC infection in iiLC. Immature LC differentiated from uninfected CD34+ progenitors were exposed to TB40-BAC4 (A to H) or AD169varATCC (I to L) virions for 4 h prior to washing and culture under immature conditions in the presence (E to H) or not of 300 μg/ml of foscarnet. Cells and supernatants were collected every 2 days. The proportion of cells expressing the viral IE1/IE2 proteins at each time postinfection relative to that at day two (A, E, and I) (fold day two) was determined by immunofluorescence staining analyses of cytospin preparations, while real-time, quantitative genomic PCR was used to determine the proportions of cells per well (B, F, and J) and of viral genomes per well (C, G, and K) present at each time point relative to the beginning of infection at 4 hpi (fold 4 hpi). The amount of virus present in the supernatants was quantified by immunofluorescence staining analyses of HFF collected at 24 h postexposure to each medium (D, H, and L). The “IE1/IE2+ cells” and “viral titers” graphs show mean and standard deviation values of data collected from five donors in four (A) and three (E) independent experiments and from three donors in three (I), four (D and L), and two (H) independent experiments. The “cells per well” and “viral DNA/well” graphs show median and median absolute deviation values of data collected from five donors in five independent experiments (B and C), five donors in three independent experiments (F and G), and three donors in four independent experiments (J and K). The dotted line marks the 1-fold point on the y axis.
Quite surprisingly, not only was viral progeny detected in the supernatants of both TB40-BAC4- and AD169varATCC-infected iiLC (Fig. 4D and L), but its amounts increased to values 30-fold higher than those at day four postinfection in both cases. No release of viral progeny was observed in the presence of foscarnet (Fig. 4H), suggesting that the activity of the viral polymerase is required for progeny production in TB40-BAC4-infected cultures.
Contrary to all expectations, thus, iiLC were productively infected by both TB40-BAC4 and AD169varATCC, despite the facts that the initial proportion of IE1/IE2+ cells was extremely small and that both the number of cells expressing the IE1/IE2 proteins and the total content of viral genomes rapidly plummeted during the course of infection.
As the CD34 molecule was reported to be present on the surface of endothelial cells (43), which are permissive to infection by CMV strains displaying the gH/gL/UL128-131A pentameric complex on their envelopes, we further assessed the identity of productively infected cells within iiLC populations, to rule out the possibility that small amounts of endothelial cells might have contributed to the observed viral yields. To this end, we determined the proportion of iiLC containing IE1/IE2+ or UL44+ polylobulated nuclei after infection with AD169varATCC, a pentamer-negative strain unable to infect endothelial cells. Polylobulated nuclei are a typical feature of LC but not of endothelial cells (44), while UL44, the processivity factor of the viral polymerase, accumulates at the sites of active viral DNA replication. We found that approximately 25% ± 6% of iiLC contained polylobulated nuclei at day two postinfection (Table 2). This proportion remained fairly stable until day six and slightly declining to about 17% from day 8 to 12. Of these polylobulated nuclei, 14% ± 7% displayed expression of the IE1/IE2 proteins at day two, but this percentage decreased to a steady 5% from day 4 to day 12. Quite interestingly, while none of the IE1/IE2+ polylobulated nuclei expressed the UL44 protein at day two, almost all became UL44+ from day four onwards. Together, these data show that iiLC cultures actually contain cells with polylobulated nuclei that are both IE1/IE2+ and UL44+, suggesting that at least a portion of the newly produced viral particles observed in the supernatant of AD169varATCC-infected iiLC cultures (Fig. 4L) are produced by bona fide LC. As the amounts of progeny detected after iiLC infection with TB40-BAC4 were not very different from those of AD169varATCC-infected cultures, we consider a potential contribution of contaminating endothelial cells to the observed viral titers to be highly unlikely.
TABLE 2.
Proportions of iiLC and mmLC with IE1/IE2+ and UL44+ polylobulated nuclei (pn)
| Cells or pn | % on daya: |
|||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 12 | |
| iiLC with pn | 25 ± 6 | 22 ± 8 | 23 ± 9 | 17 ± 7 | 15 ± 6 | 18 ± 5 |
| mmLC with pn | 37 ± 6 | 35 ± 14 | 24 ± 5 | 22 ± 6 | 16 ± 5 | 22 ± 7 |
| iiLC pn IE+ | 14 ± 7 | 5 ± 5 | 6 ± 6 | 4 ± 4 | 5 ± 7 | 4 ± 2 |
| mmLC pn IE+ | 23 ± 8 | 24 ± 12 | 22 ± 9 | 21 ± 10 | 17 ± 10 | 10 ± 8 |
| iiLC pn IE+ and UL44+ | 0 ± 0 | 100 ± 0 | 98 ± 7 | 93 ± 7 | 90 ± 30 | 20 ± 42 |
| mmLC pn IE+ and UL44+ | 78 ± 19 | 78 ± 25 | 66 ± 40 | 68 ± 40 | 55 ± 35 | 20 ± 33 |
Values are means ± standard deviations.
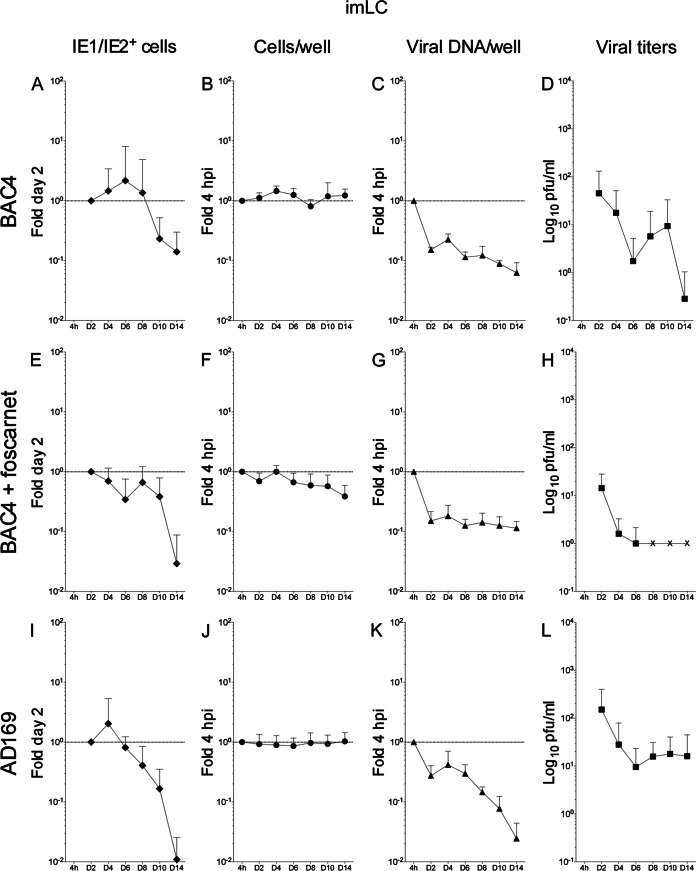
TB40-BAC4- or AD169varATCC-infected imLC display enhanced IE1/IE2 expression compared to iiLC but produce lower levels of viral progeny.
Exposure of iLC to LPS and CD40L immediately after TB40-BAC4 or AD169varATCC infection promoted expression of the IE1/IE2 proteins in a broader proportion of cells and for longer periods of time (Fig. 5A, E, and I) than for infected iLC maintained under immature conditions (Fig. 4A, E, and I), and populations displayed significantly different distributions (KS test P values of 7.293−06 for the comparison of TB40-BAC4-infected iiLC [see Fig. 7A, white diamonds] to imLC [see Fig. 7A, gray diamonds] and of 0.017 for the comparison of AD169varATCC-infected iiLC [see Fig. 7D, white diamonds] to imLC [see Fig. 7D, gray diamonds]).
FIG 5.
Kinetics of TB40-BAC4 and AD169varATCC infection in imLC. Immature LC differentiated from uninfected CD34+ progenitors were exposed to TB40-BAC4 (A to H) or AD169varATCC (I to L) virions for 4 h, prior to washing and culture in maturation medium in the presence (E to H) or not of 300 μg/ml of foscarnet. Cells and supernatants were collected every 2 days. Data collection and values shown in the graphs are as described in the legend to Fig. 4. The dotted line marks the 1-fold point on the y axis.
FIG 6.
Kinetics of TB40-BAC4 and AD169varATCC infection in mmLC. Mature LC obtained after 2 days of LPS and CD40L stimulation of iLC differentiated from uninfected CD34+ progenitors were exposed to TB40-BAC4 (A to H) or AD169varATCC (I to L) virions for 4 h prior to washing and further culture in maturation medium in the presence (E to H) or not of 300 μg/ml of foscarnet. Cells and supernatants were collected every 2 days. Data were collected as described in the legend to Fig. 4. The “IE1/IE2+ cells” and “viral titers” graphs show mean and standard deviation values of data collected from five donors in five (A), four (E), and six (D) independent experiments, from four donors in four independent experiments (L), and from three donors in three (I) and two (H) independent experiments. The “cells per well” and “viral DNA/well” graphs show median and median absolute deviation values of data collected from six donors in six independent experiments (B and C), five donors in four independent experiments (F and G), and three donors in four independent experiments (J and K). The dotted line marks the 1-fold point on the y axis.
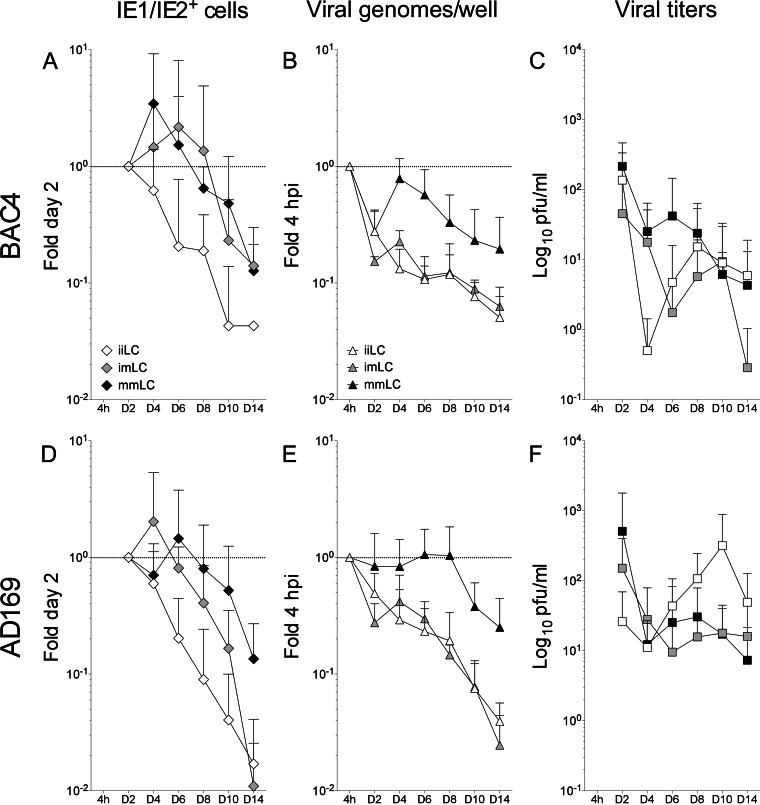
FIG 7.
Comparison of TB40-BAC4 and AD169varATCC infection kinetics in iiLC, imLC, and mmLC. The data shown in this figure are the same as in Fig. 4, 5, and 6 but rearranged for direct comparison of the proportions of IE1/IE2+ cells (A and D) and of viral genomes per well (B and E) present at each time point relative to those at day two and at 4 hpi, respectively, and of the amounts of viral particles released in the culture supernatants (C and F) after TB40-BAC4 or AD169varATCC infection of iiLC (white symbols), imLC (gray symbols), and mmLC (black symbols). The dotted line marks the 1-fold point on the y axis.
Similar to the case for iiLC cultures, the total number of cells per well did not change or increased slightly over time after AD169varATCC or TB40-BAC4 infection (Fig. 5B and J, respectively). In contrast, the amount of imLC per well in foscarnet-treated cultures (Fig. 5F) was larger than that of iiLC per well under the same conditions (Fig. 4F), suggesting a possible positive effect of maturation on cell survival.
Interestingly, the observed increase in the proportion of IE1/IE2-expressing cells was not followed by a similar intensification in the rates of viral genome amplification. After an initial drop at day two and a subsequent recovery at day four, the number of viral genomes per well in both TB40-BAC4- and AD169varATCC-infected imLC (Fig. 5C and K) rapidly declined in a fashion very similar to that of iiLC (Fig. 4C and K). Indeed, no difference in the distribution of data from infected iiLC and imLC was detected (KS test P values of 0.160 for the comparison of TB40-BAC4-infected iiLC [see Fig. 7B, white triangles] to imLC [see Fig. 7B, gray triangles] and of 0.644 for the comparison of AD169varATCC-infected iiLC [see Fig. 7E, white triangles] to imLC [see Fig. 7E, gray triangles]).
The kinetics of viral progeny production by infected imLC, in contrast, were different from those for iiLC. The initial decline in the viral particle content of supernatants was more protracted in imLC than in iiLC, lasting until day six to eight postinfection (Fig. 5D and L) instead of day four (Fig. 4D and L), and was then followed by a moderate boost in virus amounts, reaching levels 3- to 5-fold higher than those at day six or eight in TB40-BAC4-infected cultures (Fig. 5D), and by a milder but more protracted increase, peaking at 2-fold the amounts at day six in AD169varATCC-infected cells (Fig. 5L). These differences were, however, robust enough to reach statistical significance only for the KS comparison of AD169varATCC-infected iiLC (see Fig. 7F, white squares) to imLC (see Fig. 7F, gray squares) (P = 0.049) and not for the comparison of TB40-BAC4-infected iiLC (see Fig. 7C, white squares) to imLC (see Fig. 7C, gray squares) (P = 0.999). No viral particles were detected after day six in foscarnet-treated imLC (Fig. 5H).
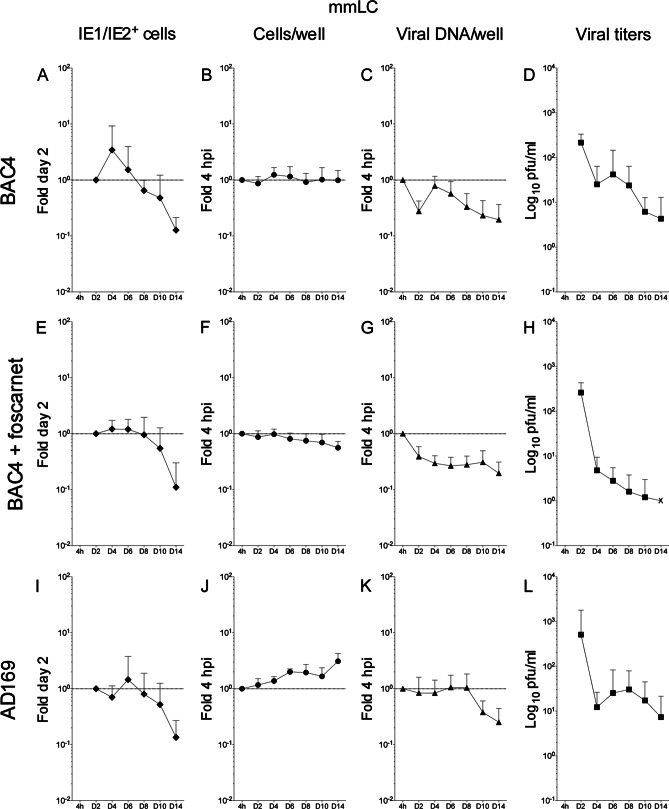
TB40-BAC4 and AD169varATCC genomes are maintained over time in infected mLC in the presence of prolonged IE1/IE2 gene expression and have modest progeny production.
As previously reported (9, 24), mLC infection with TB40-BAC4 or AD169varATCC was followed by the expression of the IE1/IE2 proteins in a higher proportion of cells than for iLC infection. Additionally, while IE1/IE2+ cells quickly became a rarity in infected iiLC cultures (Fig. 4A, E, and I), positive cells remained detectable in infected mmLC until the end of the time course and at proportions close to those of day two at least until day eight (Fig. 6A, E, and I). Accordingly, the data distributions were dramatically different, with P values of 8.294−07 for the KS comparison of TB40-BAC4-infected iiLC (Fig. 7A, white diamonds) to mmLC (Fig. 7A, black diamonds) and of 5.780−05 for the KS comparison of AD169varATCC-infected iiLC (Fig. 7D, white diamonds) to mmLC (Fig. 7D, black diamonds). In contrast, the pattern of IE expression was very similar in imLC and mmLC (Fig. 5A and D), as confirmed by the KS comparison of TB40-BAC4-infected imLC (Fig. 7A, gray diamonds) to mmLC (Fig. 7A, black diamonds) (P = 0.144) and of AD169varATCC-infected imLC (Fig. 7D, gray diamonds) to mmLC (Fig. 7D, black diamonds) (P = 0.079), indicating that the generation of an intracellular environment conducive to viral gene expression is rapidly achieved upon LC exposure to LPS and CD40L.
The distribution of TB40-BAC4 genomes per well in infected mmLC, by contrast, was rather different from that in both iiLC and imLC and consisted of an initial drop at day 2, a sharp rebound at day 4, and a slow decline from day 4 to 14 (Fig. 6C). Maturation also promoted the maintenance of AD169varATCC genomes at levels close to those of input (Fig. 6K), preventing their substantial loss as observed in iiLC and imLC (Fig. 4 and 5K). Correspondingly, KS comparisons yielded P values of 1.431−07 and 7.566−06 for the comparison of TB40-BAC4-infected mmLC (Fig. 7B, black triangles) to iiLC (white triangles) or to imLC (gray triangles), respectively, and of 7.504−07 and 2.224−06 for the comparison of AD169varATCC-infected mmLC (Fig. 7E, black triangles) to iiLC (white triangles) or to imLC (gray triangles), respectively.
Curiously, and in contrast to iiLC and imLC cultures, the total number of AD169varATCC-infected mmLC per well appeared to slightly increase over time (Fig. 6J), while no change in cell numbers was detected after TB40-BAC4 infection (Fig. 6B). The negative impact of foscarnet on cell survival was also significantly lessened by maturation, as the total number of mmLC per well did not decline over time in the presence of the inhibitor (Fig. 6F).
Even though IE1/IE2 gene expression and viral genome replication were enhanced and prolonged in mmLC compared to iiLC, viral progeny yields were somewhat modest. Akin to infected iiLC cultures, the total amount of virions present in the supernatant rapidly decreased until day four. Instead of rising sharply afterwards, only modest increases corresponding to 1.7-fold (TB40-BAC4, day six) (Fig. 6D) and 2.5-fold (AD169varATCC, day eight) (Fig. 6L) the amounts at day four postinfection were observed, so that the overall distribution of data from mmLC cultures was considered to be quite different from those of iiLC (KS test P values of 1.112−05 and 0.075 for the comparison of TB40-BAC4-infected mmLC [Fig. 7C, black squares] to iiLC [Fig. 7C, white squares] and of AD169varATCC-infected mmLC [Fig. 7F, black squares] to iiLC [Fig. 7F, white squares], respectively) and of imLC (KS test P values of 1.799−06 and 0.043 for the comparison of TB40-BAC4-infected mmLC [Fig. 7C, black squares] to imLC [Fig. 7C, gray squares] and of AD169varATCC-infected mmLC [Fig. 7F, black squares] to imLC [Fig. 7F, gray squares], respectively).
Interestingly, although mmLC cultures contained a higher proportion of cells with polylobulated nuclei than iiLC (Table 2) and although a larger percentage of these displayed expression of the IE1/IE2 proteins from day 2 to day 12, not all IE1/IE2+ nuclei also coexpressed the UL44 protein, as observed in iiLC populations. Instead, and consistent with what we previously reported (24), the proportion of IE1/IE2+ and UL44+ polylobulated nuclei averaged about 70% from day 2 to day 10 (Table 2), indicating that infection did not progress beyond the immediate early stages in a certain proportion of cells, possibly contributing to lower viral yields.
Consistent with previous data (24), thus, mmLC are productively infected by both TB40-BAC4 and AD169varATCC, but despite the facts that the IE1/IE2 proteins are expressed in a larger proportion of cells and for longer periods of time and that viral DNA is replicated and/or maintained better than in iiLC, mmLC produce lower levels of viral progeny over time.
Together, these data indicate that the intracellular signaling induced by exposure of iLC to LPS and CD40L promotes viral IE gene transcription, leading to a more widespread and prolonged expression of the IE1/IE2 proteins in infected cells. This permissive milieu is established quite rapidly after LC exposure to the maturation stimuli, as the pattern of IE1/IE2 protein expression was similar in LC infected when already mature (mmLC), and in LC infected when still immature but exposed to maturation stimuli immediately afterwards (imLC). This supportive environment is also maintained over time, preventing the loss of viral protein expression observed in iiLC (Fig. 7A and D).
In contrast to viral gene expression, maintenance of viral genomes over time occurs only when the supportive environment promoted by maturation is present at the moment of infection. Indeed, exposure of iLC to LPS and CD40L immediately after infection did not stimulate viral genome replication (Fig. 7B and E). Despite supporting viral gene expression and genome replication, maturation does not substantially stimulate viral progeny production, with iiLC, mmLC, and imLC all releasing virus over time (Fig. 7C and F).
DISCUSSION
CD34+ progenitor cells and their LC progeny are thought to play important roles in CMV pathogenesis in vivo as sites of viral latency, reactivation, and replication and as means of viral spread within the hosts and between hosts. Despite this, many aspects of CMV interactions with these cells remain unknown.
In this study, we sought to compare the efficiencies of infection onset, viral genome replication, and viral progeny production in CD34+ progenitor cells, iLC, and mLC derived from the same set of donors and exposed to the clinical-like CMV strain TB40-BAC4 or to the laboratory-adapted strain AD169varATCC prior to their maintenance under immature or mature conditions. Infection progress was tracked for up to 14 days and was compared to that in HFF, a highly permissive cell type customarily used in CMV pathogenesis studies.
Viral genomes are maintained in CD34+ cells during their differentiation into iLC.
This is the first report to quantify the numbers of viral and cellular genome copies present in CD34+ cell cultures during their differentiation into iLC. We show that while CD34+ cells proliferate over time, viral genome amounts remain fairly constant (Fig. 3A and G), suggesting either that viral DNA is maintained without amplification or degradation in cells that remain viable during the 8 days of differentiation or that it is replicated in a subset of cells and lost in another subset of similar size. Which one of these, and potentially other, hypotheses is correct cannot be definitely ascertained in the absence of quantitative PCR data from single cells.
Clearly, the number of viral DNA copies did not increase in parallel with the number of cellular genomes, as reported for Towne (20)- or AD169varATCC (18)-infected GM-Ps, suggesting that the behavior of viral genomes can change depending on the phenotype and differentiation state of the hosting cell. Consistent with data from GM-Ps (20), but in contrast to data from CD34+ cells cultured with irradiated murine stromal cells (23), no evidence of productive replication was found in AD169varATCC-infected CD34+ cell cultures (Table 1). Rather, AD169varATCC genome loads rapidly plummeted in cell populations containing the same starting amounts of viral DNA as TB40-BAC4-infected cells (Fig. 3E). The main difference we observed between clinical-like and laboratory-adapted strains, thus, was in the efficiency of viral genome maintenance during cell replication, rather than in the ability to establish nonlytic infections. Additionally, the defect in AD169varATCC genome maintenance was not absolute but could be rectified by increasing the amounts of input DNA by about 10-fold (Fig. 3G).
AD169varATCC virions do not contain the gH/gL/UL128-131A pentameric complex required for fusion of the viral envelope with endosomal membranes (45, 46), and they enter cells largely via the plasma membrane (47–49). Although the mechanisms used by CMV to access CD34+ cells have not been described, the ability to penetrate cells via both pathways may confer some advantage to pentamer-positive strains, akin to what we showed for iLC and mLC (9). Increasing the load of AD169varATCC input particles may thus augment the number of virions that reach the nucleus, improving their retention over time. Alternatively, the observed rapid loss of viral genomes may be due to the lack of expression of UL138 (and/or of other proteins), leading to the inefficient maintenance of viral DNA during latency, rather than to the onset of productive infections as reported for CD34+ cells cultured on irradiated murine stromal cells (21–23).
CMV infections in vivo are often associated with the development of thrombocytopenia (50). Whether this is due to some toxic effect of infection acting directly on hematopoietic progenitor cells or to some indirect negative influence on stromal cell functions remains unclear. Infection of CD34+ cell populations in vitro was reported to negatively impact their proliferation rates, but this was highly dependent on the CD34+ cell subtype, the virus strain, and the MOI used (4, 22, 51–56). We did not detect any difference in the total number of iLC obtained after differentiation of mock- or CMV-infected CD34+ cells in multiple experiments using TB40-BAC4 and AD169varATCC. Infected populations did not express the viral IE1/IE2 proteins and did not produce viral progeny (Table 1), suggesting that viral genome maintenance was not associated with extensive cytopathic effects leading to widespread cell death.
Maturation of LC differentiated from infected CD34+ cells does not substantially enhance the efficacy of viral genome maintenance and the frequency of reactivation events.
After an initial and unavoidable loss in cell and viral genome numbers following iLC harvest and replating procedures, cell proliferation resumed (Fig. 3B, F, and H), except in foscarnet-treated cultures (Fig. 3D), and was accompanied by the maintenance of viral genomes at relatively steady amounts until day 14 (Fig. 3B, D, and H). While no statistically significant difference was observed between the amounts of viral DNA present in 34i and 34m cultures derived from progenitors infected with AD169varATCC (Fig. 3F and H), TB40-BAC4-infected 34m cells contained, on average, 2.5-fold more viral genomes than 34i cells from day four onwards (Fig. 3B), and the two populations possessed statistically different distributions. None of the 34i or 34m cultures expressed the IE1/IE2 proteins throughout the time course, however, and the amounts of virus found in their respective supernatants were virtually identical (Table 1), indicating that maturation of iLC derived from infected progenitors was not associated with an increase in CMV reactivation events. These findings are in contrast to those of Reeves et al., who reported that mLC derived from the CD34+ cells of seropositive individuals contained 10-fold larger amounts of viral DNA than undifferentiated CD34+ cells and transcribed the UL122/123 genes encoding the viral IE1/IE2 proteins (57). IE gene expression was also observed at day one (32) or day three (31) postmaturation of iLC differentiated from progenitors experimentally infected with the clinical-like strain TB40/E, and virus was detected in mLC culture supernatants at day 10 or 14 postmaturation (31). Although the culture conditions used in these studies were more similar to the ones employed here, a number of key differences exist that can explain the observed discrepancies.
First, we used CD34+ progenitor cells harvested from the cord blood of different donors, as opposed to cells from the peripheral blood or the bone marrow of adults. Although all CD34+ progenitor cells, irrespective of their anatomical origin, can differentiate into iLC using the specific cytokine cocktail we employed in here, cord and peripheral blood CD34+ cells are not completely identical (58) and may thus react to CMV infection in different ways.
Second, viral genome amounts were quantified from Southern blot images of PCR-amplified genomic extracts obtained from the CD34+ cells and from the mLC of a single donor and at a single time point (57), as opposed to by real-time quantitative PCR analyses of genomic DNA derived from the cells of different donors, harvested at multiple time points, and in several independent experiments. While circulating CMV such as that found in naturally infected CD34+ cells is likely to have superior abilities to persist and reactivate in myeloid cells, the quantification method used may also have contributed to the observed differences in viral genome amounts. Additionally, both viral and cellular DNA copy numbers were simultaneously tracked in this study, as opposed to viral DNA amounts only (57).
Viral gene expression was also evaluated in a different way: while we determined the percentage of cells expressing the IE1/IE2 proteins at each time point, Reeves et al. used reverse transcription-PCR to assess the presence of UL122/123 transcripts (32, 57). On the one hand, we may have missed and/or underestimated the true extent of viral reactivation in 34i and 34m cells, particularly if IE1/IE2 protein expression occurred in only a very small percentage of cells. On the other hand, transcripts are not consistently translated into proteins, and no quantification of reverse transcription-PCR data was provided, making it difficult to gauge the actual extent of viral reactivation in mLC.
While naturally infected CD34+ cells were cultured under conditions very similar to those used in this study (57), experimentally infected cells were exposed to TB40/E for 24 h, incubated in the absence of differentiation cytokines for 3 days, and then differentiated into iLC (31, 32). The prolonged exposure to CMV virions (24 versus 4 h), coupled to the use of TB40/E, a genotypically and phenotypically heterogeneous strain (35) likely possessing a broader tropism than its bacterial artificial chromosome clone TB40-BAC4 used here, may have increased the proportion of infected CD34+ cells, as well as the load of viral particles per cell. The incubation of infected cells in cytokine-free medium prior to their differentiation into iLC, moreover, may have promoted the expansion of CD34+ cell subsets more permissive to the establishment of infection.
While we used a combination of LPS and CD40L molecules to trigger iLC maturation, Reeves et al. used LPS only (31, 32, 57). Our recent data indicate that iLC exposure to LPS alone, CD40L alone, or LPS plus CD40L can generate mLC with different susceptibilities to direct CMV infection (not shown), suggesting that signaling by these molecules can trigger very different responses in iLC, with potential influences on the ability of latent virus to reactivate. Interestingly, while reactivated virus was found (albeit not quantified) directly in the supernatant of mLC derived from experimentally infected CD34+ cells (31), CMV reactivation in mLC originating from naturally infected CD34+ cells was reported to occur after coculture with permissive fibroblasts (57). In our hands, maturation of latently infected iLC by exposure to LPS and CD40L was not sufficient to trigger robust viral reactivation, which we think may require additional stimulation beyond that provided by these molecules, which may be supplied by cytokines and other soluble factors secreted by HFF or produced by LC themselves upon contact with heterologous cell types. Indeed, coculture with HFF has historically been the most commonly used method to induce full reactivation (18–23, 59).
iLC maturation promotes and prolongs IE1/IE2 protein expression.
Stimulation of immature LC with LPS and CD40L for 2 days before infection (mmLC) or immediately after exposure to CMV virions (imLC) enhanced and prolonged viral IE protein expression compared to that by unstimulated cells (iiLC) (Fig. 7A and D). This implies that maturation promotes infection onset and does so relatively quickly, as the kinetics of viral protein expression in imLC and mmLC were almost identical. We previously showed that mLC contain substantially larger amounts of UL122/UL123 mRNA than iLC at 24, 48, and 72 h postinfection with different CMV strains (9). Maturation is thus likely to act by specifically boosting viral gene transcription, although changes in the efficiency of mRNA translation may also contribute to increase the proportion of IE1/IE2+ cells. This transcriptional enhancement may occur through a variety of possible mechanism, including the remodeling of repressive chromatin on the UL122/UL123 gene promoter, the disassembly of nuclear domain 10 bodies, and the replacement of transcriptional repressors with transcriptional activators on viral gene promoters. The major IE promoter was indeed reported to be associated with makers of transcriptionally active chromatin, such as acetylated H4 histones, but devoid of transcriptional repressors, such as the heterochromatin protein 1, in mLC (31, 32, 57), while removal of the cellular proteins hDaxx and ATRX from the nuclear domain 10 bodies of mLC was proposed to contribute to these cells' susceptibility to infection (60). In a recently completed microarray-based comparison of the transcriptional profile of iLC and mLC, we found that approximately 15% of the genes more highly expressed in iLC and 6% of those more highly expressed in mLC encode known transcription factors or chromatin-modifying enzymes. The potential effects of these proteins' activities on LC susceptibility to infection are under investigation.
iLC maturation prior to infection enhances viral genome replication.
Viral genome amounts were larger in cells infected when already mature (mmLC) than in cells matured after infection (imLC) (Fig. 7B and E), indicating that the intranuclear environment required for efficient viral DNA replication is not as quickly established as that promoting viral gene transcription and must already be present at the moment of infection onset to achieve effective genome amplification. Quite interestingly, expression of the viral IE1/IE2 proteins is not sufficient to generate this milieu, as the proportions of IE1/IE2+ in imLC and mmLC were very similar (Fig. 7A and D), yet viral genome numbers plummeted in imLC. A second block to the progress of viral infection may thus exist in iLC, acting after the one limiting IE gene expression and potentially shunting infection toward an abortive outcome. Removal of this second hindrance cannot be easily attained by signaling events ensuing exposure to LPS and CD40L or by the powerful activities of the IE1/IE2 proteins and may instead require the presence of specific cellular functions that need time to accumulate.
iLC maturation does not potently stimulate viral progeny production or release.
Quite unexpectedly, viral progeny was detected in the supernatant of iiLC cultures (Fig. 7C and F). This is rather remarkable considering that the already low proportion of IE1/IE2+ iiLC rapidly and dramatically decreased over time and was mirrored by viral genome amounts. These analyses were, however, not conducted at a single-cell level. It is thus possible that infection may have proceeded in the few cells initially expressing the IE1/IE2 proteins, leading to some low level of viral genome replication that may have escaped detection and culminating in the release of newly formed viral particles. The presence of the UL44 protein in virtually all IE1/IE2+ iiLC certainly lends support to this hypothesis, although this would require this subpopulation of permissive iLC to be particularly effective at producing viral progeny, considering the comparatively large amounts of virus detected in iiLC culture supernatants. Viral cycle progression in IE1/IE2+ iiLC may also have been boosted following some maturation or activation events induced by factors secreted by uninfected bystander cells and exerting more potent effects on viral replication than exposure to LPS and CD40L.
Also contrary to expectations, maturation did not substantially enhance viral yields. Considering that a larger proportion of mmLC expressed the IE1/IE2 proteins and that mmLC cultures contained larger amounts of viral genomes overall, we expected yields from these cells to exceed those of iiLC. Instead, values were similar (TB40-BAC4) (Fig. 7C) or lower (AD169varATCC) (Fig. 7F). As only virus contained in the culture medium was measured, however, mmLC may still be producing larger amounts of progeny than iiLC, retaining the majority of it intracellularly. Alternatively, infection may be blocked after IE1/IE2 protein expression in a certain proportion of cells, as our data pertaining UL44 expression seem to suggest (Table 2), leading to an overall decrease in viral yields from these cultures.
Interestingly, iLC maturation after infection reduced yields and delayed the time of appearance of new viral particles from day six to day eight (Fig. 7C and F, gray squares), so that imLC were the slowest and lowest producers of progeny overall. The occurrence of fewer viral genome replication events and the enhanced retention of newly produced virions within the cell may both have contributed to this effect.
Foscarnet's effects on cellular and viral genome copy numbers vary depending on the cell type.
Although inhibition of the viral DNA polymerase decreased the number of viral genomes present in CD34+ cells at late times postinfection (Fig. 3C), the observed differences did not reach statistical significance, and the rate of cell proliferation was also reduced, making it difficult to draw definitive conclusions as to whether the cellular or the viral enzyme is required for viral genome maintenance and, possibly, replication during latency. Treatment with ganciclovir, another viral polymerase inhibitor, was also reported to have no effect on the amplification of Towne genomes in latently infected GM-P cultures (18). In contrast to foscarnet, however, ganciclovir requires activation by the viral kinase UL97 (61, 62), which is not expressed during latency and may thus have been inactive in nonlytically infected cells.
Culture of infected iiLC in the presence of foscarnet also led to some considerable cell loss over time (Fig. 4F), while this effect was less pronounced in imLC cultures (Fig. 5F) and almost undetectable in mmLC populations (Fig. 6F), suggesting that maturation may be alleviating the negative impact of foscarnet on LC proliferation and/or viability. The specific bases of these toxic effects are difficult to pinpoint, as foscarnet was reported to only minimally affect the activity of host DNA polymerases at concentrations that completely abrogate viral DNA replication (63). Indeed, no cell loss was observed in treated HFF cultures when viral progeny production was completely blunted (Fig. 2F and H). The antiproliferative effects we observed may thus be specific to differentiating CD34+ cells and to iiLC, perhaps on account of their higher proliferation rates than those of confluent HFF populations. Exposure of exponentially growing human embryo cells to 1 mM foscarnet, as in the present study, was indeed reported to reduce their division rates by 50% (64).
Aside from its effect on cell numbers, foscarnet treatment substantially reduced the content of viral genomes in mmLC (KS test P value of 0.016 for the comparison of untreated to treated mmLC samples) (Fig. 6C to G) but had little effect on iiLC (KS test P value of 0.727 for the comparison of untreated to treated iiLC samples) (Fig. 4C to G) and imLC (KS test P value of 0.076 for the comparison of untreated to treated iiLC samples) (Fig. 5C to G). Of note, viral genome amounts in treated mmLC cultures were larger than those in treated iiLC (KS test P value of 6.38−05) and imLC (KS test P value of 1.207−08), suggesting that maturation may promote maintenance of viral DNA even in the absence of replication, possibly by protecting viral genomes from degradation.
LC are substantially less permissive to infection than HFF.
A direct comparison of CMV replication kinetics in HFF and LC is difficult to perform. Population-wise, LC are much less susceptible to CMV infection than HFF. When infected at an MOI of 10, only about 2% ± 0.7% of iLC and 10% ± 5% of mLC (9), but between 80% and 100% of HFF (65), express the IE1/IE2 proteins. While the percentage of IE1/IE2+ HFF can be increased by raising the MOI, the total numbers of LC expressing the IE1/IE2 proteins after infection at MOIs of 10 and 100 remain virtually identical. Finally, whereas the proportion of IE1/IE2+ HFF progressively increases over time until all cells become positive (Fig. 2A), only imLC and mmLC support prolonged IE protein expression (Fig. 7A and D), but even in these populations, it never becomes universal. Some of these features of LC cultures can be explained by their being substantially less phenotypically homogeneous than HFF. At least two subpopulations of LC are likely to exist, one highly resistant and one more permissive to infection, so that when all cells from the latter have been colonized, no new infections can take place. We also speculate that particle spread may be somewhat restricted in LC, particularly so in iiLC. New virions begin to appear in the supernatants of both HFF and iiLC cultures between day four and day six (Fig. 2D and 6D and L). In HFF populations, a good portion of these particles proceed to start a new cycle in neighboring uninfected cells, as attested by the marked surge in the number of IE1/IE2+ cells from day four to day six and afterwards (Fig. 2A), ensuing the development and enlargement of plaques. No such increase is observed in iiLC populations (Fig. 4A and I), suggesting that the released progeny does not initiate new replication cycles in uninfected cells. This may be due to the fact that no additional permissive cells are available within the population, either because all susceptible iiLC have already been infected or because bystander cells have acquired resistance in response to soluble mediators released by infected iiLC. Maturation appears to somewhat mitigate this effect and to render iLC more susceptible to infection onset (Fig. 5 and 6A and I). At the same time, however, viral particle production and/or release is negatively affected in imLC and mmLC, leading to lower titers in their supernatants than in those of iiLC (Fig. 7C and F). Although to much lower levels than HFF, both iiLC and mmLC nevertheless produce viral progeny and may theoretically contribute to CMV spread from the oral mucosae to other organs within the host, as well as to CMV transmission to new hosts via the saliva in vivo. Quite interestingly, no major differences in the replication kinetics of TB40-BAC4 and AD169varATCC were detected. This suggests that use of AD169varATCC in orally delivered vaccines could potently trigger LC responses, potentially inducing the production of protective antibodies directly at the sites of CMV entry. The production of viral progeny within the oral cavity following vaccination is, however, likely to be of concern, even if viral spread from infected LC to neighboring epithelial cells is likely to remain limited due to the absence of the pentameric complex from the surface of AD169varATCC's virions. The most important future direction in this area of research, therefore, is to determine whether these findings, obtained using in vitro-differentiated LC, also hold true in vivo.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI099372 and by a subgrant from the Canadian Institutes of Health Research.
REFERENCES
- 1.Mendelson M, Monard S, Sissons P, Sinclair J. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol 77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 2.Khaiboullina SF, Maciejewski JP, Crapnell K, Spallone PA, Dean Stock A, Pari GS, Zanjani ED, Jeor SS. 2004. Human cytomegalovirus persists in myeloid progenitors and is passed to the myeloid progeny in a latent form. Br J Haematol 126:410–417. doi: 10.1111/j.1365-2141.2004.05056.x. [DOI] [PubMed] [Google Scholar]
- 3.Minton EJ, Tysoe C, Sinclair JH, Sissons JG. 1994. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol 68:4017–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhuravskaya T, Maciejewski JP, Netski DM, Bruening E, Mackintosh FR, St Jeor S. 1997. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood 90:2482–2491. [PubMed] [Google Scholar]
- 5.Slobedman B, Mocarski ES. 1999. Quantitative analysis of latent human cytomegalovirus. J Virol 73:4806–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 7.Crough T, Khanna R. 2009. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev 22:76–98. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Lauron EJ, Yu D, Fehr AR, Hertel L. 2014. Human cytomegalovirus infection of Langerhans-type dendritic cells does not require the presence of the gH/gL/UL128-131A complex and is blocked after nuclear deposition of viral genomes in immature cells. J Virol 88:403–416. doi: 10.1128/JVI.03062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allam JP, Novak N, Fuchs C, Asen S, Berge S, Appel T, Geiger E, Kochan JP, Bieber T. 2003. Characterization of dendritic cells from human oral mucosa: a new Langerhans' cell type with high constitutive FcepsilonRI expression. J Allergy Clin Immunol 112:141–148. doi: 10.1067/mai.2003.1607. [DOI] [PubMed] [Google Scholar]
- 11.Allam JP, Niederhagen B, Bucheler M, Appel T, Betten H, Bieber T, Berge S, Novak N. 2006. Comparative analysis of nasal and oral mucosa dendritic cells. Allergy 61:166–172. doi: 10.1111/j.1398-9995.2005.00965.x. [DOI] [PubMed] [Google Scholar]
- 12.Barrett AW, Cruchley AT, Williams DM. 1996. Oral mucosal Langerhans' cells. Crit Rev Oral Biol Med 7:36–58. doi: 10.1177/10454411960070010301. [DOI] [PubMed] [Google Scholar]
- 13.Cutler CW, Jotwani R. 2006. Dendritic cells at the oral mucosal interface. J Dent Res 85:678–689. doi: 10.1177/154405910608500801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novak N, Haberstok J, Bieber T, Allam JP. 2008. The immune privilege of the oral mucosa. Trends Mol Med 14:191–198. doi: 10.1016/j.molmed.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Steinman RM. 2007. Dendritic cells: understanding immunogenicity. Eur J Immunol 37(Suppl 1):S53–S60. doi: 10.1002/eji.200737400. [DOI] [PubMed] [Google Scholar]
- 16.Jotwani R, Palucka AK, Al-Quotub M, Nouri-Shirazi M, Kim J, Bell D, Banchereau J, Cutler CW. 2001. Mature dendritic cells infiltrate the T cell-rich region of oral mucosa in chronic periodontitis: in situ, in vivo, and in vitro studies. J Immunol 167:4693–4700. doi: 10.4049/jimmunol.167.8.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farthing PM, Matear P, Cruchley AT. 1990. The activation of Langerhans cells in oral lichen planus. J Oral Pathol Med 19:81–85. doi: 10.1111/j.1600-0714.1990.tb00801.x. [DOI] [PubMed] [Google Scholar]
- 18.Kondo K, Kaneshima H, Mocarski ES. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci U S A 91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung AK, Abendroth A, Cunningham AL, Slobedman B. 2006. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 108:3691–3699. doi: 10.1182/blood-2005-12-026682. [DOI] [PubMed] [Google Scholar]
- 20.Cheung AK, Gottlieb DJ, Plachter B, Pepperl-Klindworth S, Avdic S, Cunningham AL, Abendroth A, Slobedman B. 2009. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: implications for virus elimination during latency. Blood 114:4128–4137. doi: 10.1182/blood-2008-12-197111. [DOI] [PubMed] [Google Scholar]
- 21.Goodrum FD, Jordan CT, High K, Shenk T. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc Natl Acad Sci U S A 99:16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrum F, Jordan CT, Terhune SS, High K, Shenk T. 2004. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood 104:687–695. doi: 10.1182/blood-2003-12-4344. [DOI] [PubMed] [Google Scholar]
- 23.Goodrum F, Reeves M, Sinclair J, High K, Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937–945. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertel L, Lacaille VG, Strobl H, Mellins ED, Mocarski ES. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J Virol 77:7563–7574. doi: 10.1128/JVI.77.13.7563-7574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. 1992. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature 360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- 26.Strobl H, Riedl E, Scheinecker C, Bello-Fernandez C, Pickl WF, Rappersberger K, Majdic O, Knapp W. 1996. TGF-beta 1 promotes in vitro development of dendritic cells from CD34+ hemopoietic progenitors. J Immunol 157:1499–1507. [PubMed] [Google Scholar]
- 27.Strobl H, Bello-Fernandez C, Riedl E, Pickl WF, Majdic O, Lyman SD, Knapp W. 1997. flt3 ligand in cooperation with transforming growth factor-beta1 potentiates in vitro development of Langerhans-type dendritic cells and allows single-cell dendritic cell cluster formation under serum-free conditions. Blood 90:1425–1434. [PubMed] [Google Scholar]
- 28.Gatti E, Velleca MA, Biedermann BC, Ma W, Unternaehrer J, Ebersold MW, Medzhitov R, Pober JS, Mellman I. 2000. Large-scale culture and selective maturation of human Langerhans cells from granulocyte colony-stimulating factor-mobilized CD34+ progenitors. J Immunol 164:3600–3607. doi: 10.4049/jimmunol.164.7.3600. [DOI] [PubMed] [Google Scholar]
- 29.Strunk D, Rappersberger K, Egger C, Strobl H, Kromer E, Elbe A, Maurer D, Stingl G. 1996. Generation of human dendritic cells/Langerhans cells from circulating CD34+ hematopoietic progenitor cells. Blood 87:1292–1302. [PubMed] [Google Scholar]
- 30.Rozis G, Benlahrech A, Duraisingham S, Gotch F, Patterson S. 2008. Human Langerhans' cells and dermal-type dendritic cells generated from CD34 stem cells express different toll-like receptors and secrete different cytokines in response to toll-like receptor ligands. Immunology 124:329–338. doi: 10.1111/j.1365-2567.2007.02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves MB, Lehner PJ, Sissons JG, Sinclair JH. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol 86:2949–2954. doi: 10.1099/vir.0.81161-0. [DOI] [PubMed] [Google Scholar]
- 32.Reeves MB, Sinclair JH. 2010. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J Gen Virol 91:599–604. doi: 10.1099/vir.0.015602-0. [DOI] [PubMed] [Google Scholar]
- 33.Lee AW, Hertel L, Louie RK, Burster T, Lacaille V, Pashine A, Abate DA, Mocarski ES, Mellins ED. 2006. Human cytomegalovirus alters localization of MHC class II and dendrite morphology in mature Langerhans cells. J Immunol 177:3960–3971. doi: 10.4049/jimmunol.177.6.3960. [DOI] [PubMed] [Google Scholar]
- 34.Lee AW, Wang N, Hornell TM, Harding JJ, Deshpande C, Hertel L, Lacaille V, Pashine A, Macaubas C, Mocarski ES, Mellins ED. 2011. Human cytomegalovirus decreases constitutive transcription of MHC class II genes in mature Langerhans cells by reducing CIITA transcript levels. Mol Immunol 48:1160–1167. doi: 10.1016/j.molimm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 36.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA III, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol 154:125–169. [DOI] [PubMed] [Google Scholar]
- 37.Hertel L, Mocarski ES. 2004. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. J Virol 78:11988–12011. doi: 10.1128/JVI.78.21.11988-12011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazeron MC, Jahn G, Plachter B. 1992. Monoclonal antibody E-13 (M-810) to human cytomegalovirus recognizes an epitope encoded by exon 2 of the major immediate early gene. J Gen Virol 73:2699–2703. doi: 10.1099/0022-1317-73-10-2699. [DOI] [PubMed] [Google Scholar]
- 39.Spaete RR, Mocarski ES. 1985. Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J Virol 56:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bressollette-Bodin C, Coste-Burel M, Besse B, Andre-Garnier E, Ferre V, Imbert-Marcille BM. 2009. Cellular normalization of viral DNA loads on whole blood improves the clinical management of cytomegalovirus or Epstein Barr virus infections in the setting of pre-emptive therapy. J Med Virol 81:90–98. doi: 10.1002/jmv.21334. [DOI] [PubMed] [Google Scholar]
- 41.Tarrant-Elorza M, Rossetto CC, Pari GS. 2014. Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe 16:43–54. doi: 10.1016/j.chom.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Fortunato EA. 2014. Use of diploid human fibroblasts as a model system to culture, grow, and study human cytomegalovirus infection. Methods Mol Biol 1119:47–57. doi: 10.1007/978-1-62703-788-4_4. [DOI] [PubMed] [Google Scholar]
- 43.Fina L, Molgaard HV, Robertson D, Bradley NJ, Monaghan P, Delia D, Sutherland DR, Baker MA, Greaves MF. 1990. Expression of the CD34 gene in vascular endothelial cells. Blood 75:2417–2426. [PubMed] [Google Scholar]
- 44.Zelickson AS. 1965. The Langerhans cell. J Invest Dermatol 44:201–212. [DOI] [PubMed] [Google Scholar]
- 45.Huber MT, Compton T. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol 72:8191–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol 87:2451–2460. doi: 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 47.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol 74:7720–7729. doi: 10.1128/JVI.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanarsdall AL, Chase MC, Johnson DC. 2011. Human cytomegalovirus glycoprotein gO complexes with gH/gL, promoting interference with viral entry into human fibroblasts but not entry into epithelial cells. J Virol 85:11638–11645. doi: 10.1128/JVI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isaacson MK, Compton T. 2009. Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J Virol 83:3891–3903. doi: 10.1128/JVI.01251-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almeida-Porada GD, Ascensao JL. 1996. Cytomegalovirus as a cause of pancytopenia. Leuk Lymphoma 21:217–223. [DOI] [PubMed] [Google Scholar]
- 51.Simmons P, Kaushansky K, Torok-Storb B. 1990. Mechanisms of cytomegalovirus-mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc Natl Acad Sci U S A 87:1386–1390. doi: 10.1073/pnas.87.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sing GK, Ruscetti FW. 1990. Preferential suppression of myelopoiesis in normal human bone marrow cells after in vitro challenge with human cytomegalovirus. Blood 75:1965–1973. [PubMed] [Google Scholar]
- 53.Movassagh M, Gozlan J, Senechal B, Baillou C, Petit JC, Lemoine FM. 1996. Direct infection of CD34+ progenitor cells by human cytomegalovirus: evidence for inhibition of hematopoiesis and viral replication. Blood 88:1277–1283. [PubMed] [Google Scholar]
- 54.Sindre H, Tjoonnfjord GE, Rollag H, Ranneberg-Nilsen T, Veiby OP, Beck S, Degre M, Hestdal K. 1996. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood 88:4526–4533. [PubMed] [Google Scholar]
- 55.Maciejewski JP, St Jeor SC. 1999. Human cytomegalovirus infection of human hematopoietic progenitor cells. Leuk Lymphoma 33:1–13. [DOI] [PubMed] [Google Scholar]
- 56.Crapnell K, Zanjani ED, Chaudhuri A, Ascensao JL, St Jeor S, Maciejewski JP. 2000. In vitro infection of megakaryocytes and their precursors by human cytomegalovirus. Blood 95:487–493. [PubMed] [Google Scholar]
- 57.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci U S A 102:4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawley RG, Ramezani A, Hawley TS. 2006. Hematopoietic stem cells. Methods Enzymol 419:149–179. doi: 10.1016/S0076-6879(06)19007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn G, Jores R, Mocarski ES. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci U S A 95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saffert RT, Penkert RR, Kalejta RF. 2010. Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J Virol 84:5594–5604. doi: 10.1128/JVI.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Littler E, Stuart AD, Chee MS. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 62.Sullivan V, Talarico CL, Stanat SC, Davis M, Coen DM, Biron KK. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162–164. doi: 10.1038/358162a0. [DOI] [PubMed] [Google Scholar]
- 63.McVoy MA, Nixon DE. 2005. Impact of 2-bromo-5,6-dichloro-1-beta-d-ribofuranosyl benzimidazole riboside and inhibitors of DNA, RNA, and protein synthesis on human cytomegalovirus genome maturation. J Virol 79:11115–11127. doi: 10.1128/JVI.79.17.11115-11127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stenberg K, Skog S, Tribukait B. 1985. Concentration-dependent effects of foscarnet on the cell cycle. Antimicrob Agents Chemother 28:802–806. doi: 10.1128/AAC.28.6.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller MS, Hertel L. 2009. Onset of human cytomegalovirus replication in fibroblasts requires the presence of an intact vimentin cytoskeleton. J Virol 83:7015–7028. doi: 10.1128/JVI.00398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]