ABSTRACT
Bluetongue virus (BTV) causes bluetongue, a major hemorrhagic disease of ruminants. In order to investigate the molecular determinants of BTV virulence, we used a BTV8 strain minimally passaged in tissue culture (termed BTV8L in this study) and a derivative strain passaged extensively in tissue culture (BTV8H) in in vitro and in vivo studies. BTV8L was pathogenic in both IFNAR−/− mice and in sheep, while BTV8H was attenuated in both species. To identify genetic changes which led to BTV8H attenuation, we generated 34 reassortants between BTV8L and BTV8H. We found that partial attenuation of BTV8L in IFNAR−/− mice was achieved by simply replacing genomic segment 2 (Seg2, encoding VP2) or Seg10 (encoding NS3) with the BTV8H homologous segments. Fully attenuated viruses required at least two genome segments from BTV8H, including Seg2 with either Seg1 (encoding VP1), Seg6 (encoding VP6 and NS4), or Seg10 (encoding NS3). Conversely, full reversion of virulence of BTV8H required at least five genomic segments of BTV8L. We also demonstrated that BTV8H acquired an increased affinity for glycosaminoglycan receptors during passaging in cell culture due to mutations in its VP2 protein. Replication of BTV8H was relatively poor in interferon (IFN)-competent primary ovine endothelial cells compared to replication of BTV8L, and this phenotype was determined by several viral genomic segments, including Seg4 and Seg9. This study demonstrated that multiple viral proteins contribute to BTV8 virulence. VP2 and NS3 are primary determinants of BTV pathogenesis, but VP1, VP5, VP4, VP6, and VP7 also contribute to virulence.
IMPORTANCE Bluetongue is one of the major infectious diseases of ruminants, and it is listed as a notifiable disease by the World Organization for Animal Health (OIE). The clinical outcome of BTV infection varies considerably and depends on environmental and host- and virus-specific factors. Over the years, BTV serotypes/strains with various degrees of virulence (including nonpathogenic strains) have been described in different geographical locations. However, no data are available to correlate the BTV genotype to virulence. This study shows that BTV virulence is determined by different viral genomic segments. The data obtained will help to characterize thoroughly the pathogenesis of bluetongue. The possibility to determine the pathogenicity of virus isolates on the basis of their genome sequences will help in the design of control strategies that fit the risk posed by new emerging BTV strains.
INTRODUCTION
Bluetongue is one of the major infectious diseases of ruminants and is caused by bluetongue virus (BTV), an arbovirus transmitted by Culicoides spp. (1–3). BTV is a member of the Orbivirus genus within the Reoviridae. The double-stranded RNA (dsRNA) genome of BTV is composed of 10 segments encoding seven structural (VP1 to VP7) and four nonstructural (NS1 to NS4) proteins (4, 5). The BTV virion is an icosahedral particle assembled in a triple layer of capsid shells (6, 7). The viral core contains linear dsRNA genomic segments associated with replication complexes formed by three minor proteins, VP1 (RNA-dependent RNA polymerase), VP4 (capping enzyme including methyltransferase), and VP6 (RNA-dependent ATPase and helicase) enclosed by layers of VP3 (subcore) and VP7 (core) (4, 6, 8, 9). The outer capsid is formed by two structural proteins, VP2 and VP5, that are responsible for cell attachment and entry (10–12). NS1, the largest of the nonstructural proteins, forms tubules in the cytoplasm of BTV-infected cells and is a positive regulator of viral protein synthesis (13–15). NS2 is a major component of viral inclusion bodies (VIB) which are the sites of BTV morphogenesis and RNA replication (16–18). NS3, the only BTV glycoprotein, is involved in virus exit from infected cells and is thought to play a role in counteracting the innate immune response of the host cell (19–21). The recently discovered NS4 is the smallest of the BTV proteins, and it also confers a replication advantage in cells in an antiviral state (5). At present, there are 26 different BTV serotypes circulating worldwide. The serotype is determined predominantly by VP2, which is the most variable of BTV proteins and a main target of neutralizing antibodies (22–25).
Bluetongue is remarkably variable in its clinical manifestations, which can range from asymptomatic infection to a lethal hemorrhagic fever (2, 26–28). This variability is due to several factors related both to the infected host and the virus (2, 29–33). Over the years, BTV has been used extensively as a model to study the orbivirus replication cycle, structural biology, and interaction with the host cell. However, we have little understanding of the molecular determinants of BTV virulence.
The northern European strain of BTV8 was the cause of one of the largest outbreaks in the history of bluetongue. The virus emerged in 2006 in an area between the Netherlands, Belgium, and northern Germany and then spread throughout the continent, causing high mortality in naive sheep flocks and, occasionally, also clinical disease in cattle (34–36). Interestingly, no clinical signs were detected in sentinel sheep when the virus reached northern Italy and Sardinia in 2008. Experimental infections with the northern European strain of BTV8 (BTV8NET2006) and the Italian strain of BTV8 (BTV8IT2008) confirmed a marked reduction in the virulence of the latter virus (33). A comparison of the consensus sequences of both viruses showed 24 nucleotide differences between the two strains, resulting in eight amino acid residue substitutions in VP1, VP2, VP4, VP6, NS1, and NS2.
Genetic drift occurring during natural transmission cycles plays a significant role in the diversification of BTV strains and their pathogenicity. Similarly, in vitro passage of BTV was shown to have an impact on virulence in vivo (37–39). In particular, strains isolated from severe clinical cases and consequently adapted to mammalian tissue culture have been reported to have reduced virulence in experimentally infected animals (33, 38). Interestingly, a decrease in pathogenicity was shown to occur even in minimally passaged strains and was attributed to a genetic bottleneck that occurs after a single passage in mammalian cells rather than to mutations in the consensus sequence (33). Conversely, live attenuated vaccines and tissue culture-adapted strains of BTV with a history of multiple passages in vitro show increased accumulation of nucleotide substitutions correlating with increasing numbers of passages in mammalian cells (39). Genomic segments 1, 2, and 8 (Seg1, Seg2, and Seg8, respectively, encoding VP1, VP2, and NS2) were shown to be consistently mutated in high-passage-number strains of BTV2, BTV4, and BTV9 viruses and to be attenuated in murine models of bluetongue (39).
Here, in order to determine the roles played by specific BTV genomic segments in virus adaptation to tissue culture and attenuation in vivo, we compared genetic and phenotypic differences between BTV8NET2006 minimally passaged in tissue culture, a derivative of this strain extensively passaged in cell culture, and 34 reassortants between the two viruses.
MATERIALS AND METHODS
Cell lines.
All cell cultures were grown at 37°C in 5% CO2 humidified atmosphere. BSR cells (a variant of BHK-21 cells; kindly provided by Karl-Claus Conzelmann) were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P-S). Transfections in BSR cells were performed in DMEM with reduced FBS and no antibiotics. CPT-Tert cells (40) are sheep choroid plexus cells immortalized with simian virus 40 (SV40) T antigen and human telomerase reverse transcriptase. CPT-Tert cells were propagated in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS and 1% P-S. The CHO cell line is derived from adult Chinese hamster ovary, and pgsA-745 (ATCC number CRL-2242) is a CHO-derived cell line deficient in xylotransferase that does not produce glycosaminoglycans (GAGs). Both cell lines were propagated in Ham's F-12 medium supplemented with 10% FBS and 1% P-S.
Primary endothelial cell cultures.
Primary ovine aortic endothelial cells (OvEC) were isolated directly from aortas harvested from euthanized animals as recently described (41). Cells were maintained at 37°C in 5% CO2 and 3% O2 for a maximum of three passages.
Virus strains.
BTV8NET2006 (Pirbright reference collection number NET2006/04) was originally isolated from a naturally infected sheep during the 2006 outbreak in northern Europe and has been previously described (5, 33). In this study, we refer to this virus as BTV8L. The subscript “L” is used to indicate that this virus has a low number of passages in cultures. BTV8H (H, high passage number) was obtained by 65 serial passages of the BTV8L strain in BSR cells, followed by plaque purification.
Reverse genetics.
Plasmids used for the rescue of BTV8L (resulting in rgBTV8L) were described previously (5, 42). Plasmids used for the rescue of BTV8H by reverse genetics (resulting in rgBTV8H) were derived by site-directed mutagenesis of nonsynonymous sites in BTV8L. Reassortants between rgBTV8L and rgBTV8H are denoted with the name of virus forming the backbone (either BTV8L or BTV8H) followed by the substituted segment marked with the “L” or “H” subscript to indicate the virus of origin. For example, BTV8L+S2H is a reassortant containing the rgBTV8L backbone with Seg2 from rgBTV8H. Rescued versions of both BTV8L and BTV8H (rgBTV8L and rgBTV8H, respectively) and reassortants between both parental viruses were obtained using established procedures for BTV reverse genetics (5, 42, 43).
Virus titrations and replication curves.
Titers of viral stocks were determined by standard plaque assays in CPT-Tert cells as previously described (25, 43). Virus replication in CPT-Tert cells and OvEC was assessed by infecting cells at a multiplicity of infection (MOI) of 0.01 and collecting supernatants at 2, 24, 48, and 72 h postinfection (p.i.). To compare replication kinetics of selected viruses in CHO and pgsA-745 cells, infections were performed at an MOI of 0.01, and supernatants were collected only at 72 h p.i. Supernatants were centrifuged for 5 min at 500 × g to remove cell debris and then titrated by endpoint dilution assays. Titers were expressed as the log10 50% tissue culture infective doses/ml (TCID50/ml). All virus titration experiments were performed at least three times independently. Statistical calculations were carried out using GraphPad Prism.
BTV genome sequencing.
The full genome sequence of BTV8H was obtained using the Illumina platform. BSR cells were infected, and total RNA was extracted using TRIzol reagent (Invitrogen). Single-stranded RNA was precipitated using lithium chloride, and double-stranded RNA was harvested from the supernatant by precipitation with isopropanol in the presence of sodium acetate. Double-stranded RNA was used as a template for full-length amplification of cDNA (FLAC) by reverse transcription-PCR (RT-PCR) using established methods (44). Samples were analyzed using an Illumina Genome Analyser. The libraries were constructed from the PCR samples using a TruSeq DNA sample preparation kit (Illumina) according to the manufacturer's instructions. Briefly, the DNA samples were fragmented, the fragment was end repaired, and the 3′ ends were adenylated. After adaptor ligation steps, the fragments were purified by size selection on agarose gel, and the fragments containing adaptors on the 3′ and 5′ ends were enriched by PCR. Sequencing was performed on a GAIIX sequencer (Illumina) according to the manufacturer's protocol. Genomes were assembled using Maq software (45) with BTV8L used as a reference sequence. The assemblies were manually curated using Tablet for sequence visualization (46), and consensus sequences were generated as Fasta files.
In vivo experiments.
Animal experiments were carried out at the Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise G. Caporale (Teramo, Italy) in accordance with locally and nationally approved protocols regulating animal experimental use (protocol no. 11427/2012). Experiments in sheep were conducted in an insect-proof isolation unit with veterinary care. Prior to the experiments the animals were confirmed to be seronegative for BTV using a blocking enzyme-linked immunosorbent assay (ELISA), as described previously (33). Groups (n = 5 animals per group) of Dorset Poll sheep were infected intradermally with a total of 2 × 106 PFU (in 5 ml) of BTV8L and BTV8H by multiple inoculations in the inner leg and in the prescapular areas. Control animals were inoculated with 5 ml of mock-infected cell supernatant. Blood samples were collected from all animals daily for the first 15 days and at 17, 19, 21, and 28 days p.i. and then examined for the presence of viremia by quantitative RT-PCR (qRT-PCR). Sera were tested by virus neutralization assay for the presence of BTV-specific antibodies in the samples collected at 28 days p.i. All sheep were examined for the presence of clinical signs and fever, starting 1 week prior to inoculation and continuing for 15 days p.i., with further observations on days 17, 19, 21, and 28 p.i. Clinical signs were scored using a clinical reaction index (CRI) as previously described (33).
Transgenic mice deficient in type I interferon (IFN) receptor (129sv IFNAR−/−) were maintained at biosafety level 3. For each experiment, groups of adult mice matched for sex and age (n = 5 per group) were infected intraperitoneally or subcutaneously with 300 PFU or 3,000 PFU of virus or mock-infected cell culture medium as a control. Mice were examined for clinical signs daily until the experiment was concluded at 14 days p.i.
Serum neutralization assays.
The presence of neutralizing antibodies was assessed by neutralization assays as previously described (33, 43).
qRT-PCR.
Levels of viremia in infected sheep were assessed by qRT-PCR as described before (33). Red blood cells were lysed with water for 10 min on ice and centrifuged at 4°C for 10 min at 13,000 × g. Armored West Nile RNA (Asurage, USA) was spiked into each sample as an internal control for nucleic acid extraction. Total RNA was extracted using a High Pure nucleic acid extraction kit (Roche, Nutley, NJ) according to the manufacturer's instructions. For all samples, 250 ng of total RNA was amplified by one-step qRT-PCR using primers and probes for BTV Seg5. Armored RNA and β-actin were amplified as control reactions.
Levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, IFN-β, Mx1, and RSAD2 expression in infected OvEC were measured by qRT-PCR. Briefly, cells were seeded in 24-well plates and infected 48 h afterwards with rgBTV8L, rgBTV8H, or selected reassortants (MOI of 1). The medium was replaced after 1 h, and the cells were incubated for a further 17 h at 37°C. Next, supernatants were collected, and monolayers were directly lysed in 0.5 ml of TRIzol (Life Technologies). Phase separation was performed according to the manufacturer's instructions, whereupon the aqueous phase was mixed with ethanol and purified using an RNeasy minikit (Qiagen), including an RNase Free/DNase Set on-column DNase treatment step. Residual contaminating genomic DNA was removed using a Turbo DNA-Free kit (Ambion) according to the manufacturer's conditions. Reverse transcription was performed using 100 ng of RNA using random hexamers and SuperScript III (Life Technologies) for 1 h at 45°C. Quantitative PCR (qPCR) was performed using Brilliant III Ultra-Fast QPCR mastermix reagents (Agilent) and in-house designed primers/probes (sequences available upon request) targeting ovine GAPDH, β-actin, IFN-β, Mx1, and RSAD2. Samples were run on an Mx3005P PCR machine with rgBTV8L-infected cells set as a calibrator. GAPDH was used as normalizing gene against which fold induction was determined for IFN-β, Mx1, RSAD2, and β-actin.
IFN protection assays.
Measurement of IFN levels in cell supernatants was based on methods described previously (5, 41). Briefly, OvEC were seeded in 24-well plates and infected after 2 days at an MOI of 1 with selected viruses. Medium was replaced after 1 h of incubation, and supernatants were collected 18 h after infection and treated for 20 min with UV light to inactivate infectious virus. CPT-Tert cells were seeded in 96-well plates, and 24 h later 2-fold serial dilutions of UV-treated supernatants or serial dilutions of a known concentration of universal interferon (UIFN) were added to the cells. After 24 h of incubation, supernatants were removed, and cells were infected with encephalomyocarditis virus (EMCV) and incubated for further 48 h. Cells were then inspected for EMCV-induced cytopathic effect. The levels of IFN in supernatants collected from BTV-infected OvEC were calculated based on the number of wells protected from EMCV-induced cell death compared to those of the UIFN control wells.
Cell protection by pretreatment with UIFN was performed using CPT-Tert cells. Cells were seeded in 24-well plates and 24 h later treated with 1,000 units of UIFN. After 18 h of incubation, UIFN was removed, and cells were infected with selected viruses at an MOI of 0.01. In parallel, untreated CPT-Tert cells were infected with the same set of viruses. Inocula were replaced by fresh tissue culture medium 1.5 h later. At 48 h p.i. supernatants were collected, and viral titers were determined by endpoint dilution assay and expressed as the log10 TCID50/ml.
RESULTS
Phenotype of tissue culture-adapted BTV8.
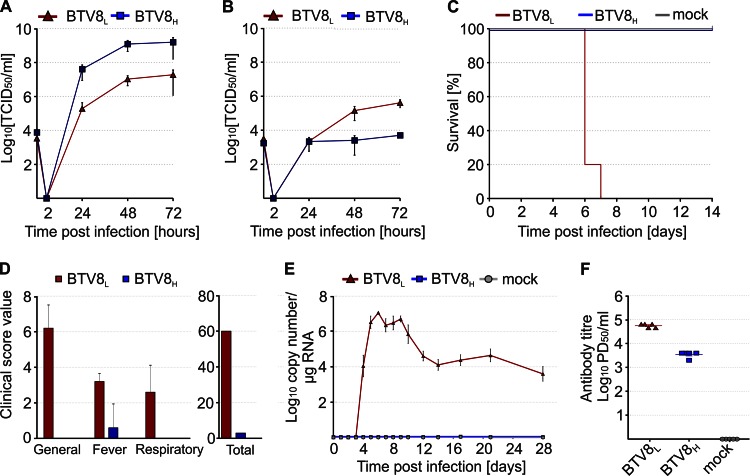
BTV8L was serially passaged 65 times in BSR cells and plaque purified. The resulting virus, BTV8H, and the parental BTV8L were then titrated in sheep CPT-Tert cells and in primary ovine endothelial cells (OvEC). CPT-Tert cells are unable to initiate an IFN response following virus infection (5, 40), while OvEC are IFN competent (41). Both viruses replicated very efficiently in IFN-deficient CPT-Tert cells, but BTV8H reached titers approximately 100-fold higher than its parental virus (Fig. 1A). Both viruses produced lower yields in OvEC than in CPT-Tert cells, and BTV8L replicated better than BTV8H in these cells (Fig. 1B).
FIG 1.
In vivo and in vitro phenotype of minimally passaged BTV8L and tissue culture-adapted BTV8H. (A and B) BTV8L and BTV8H replication kinetics in ovine CPT-Tert cells (A) and primary ovine endothelial cells (OvEC) (B). Cells were infected with BTV8L or BTV8H at an MOI of 0.01. Supernatants were collected at 2, 24, 48, and 72 h p.i. and then titrated in BSR cells by limiting dilution analysis. Virus titers are expressed as log10(TCID50/ml). (C) Survival plot of IFNAR−/− mice (n = 5 per group) infected with 300 PFU of BTV8L and BTV8H or mock infected. (D to F) Virulence of BTV8L and BTV8H in sheep. Clinical scores (D), viremia (E), and neutralizing antibodies (F) at 28 days p.i. were measured in infected and mock-infected sheep (n = 5 per group) as described in Materials and Methods. PD50, 50% protective dose.
Next, we determined the pathogenicity of BTV8L and BTV8H in IFNAR−/− mice. BTV8L was highly virulent in this mouse model, inducing 100% mortality within 7 days p.i., whereas BTV8H was completely attenuated (Fig. 1C). Next, we assessed the virulence of BTV8L and BTV8H in sheep, the natural host of BTV infection, in order to characterize further the phenotype of these viruses in vivo. Sheep infected with BTV8L showed fever and general clinical signs, including depression and mild respiratory distress. Viremia was detected in all BTV8L-infected animals (n = 5) from approximately day 4 p.i. until the end of the experiment at day 28 p.i. On the other hand, sheep infected with BTV8H did not show any clinical signs of bluetongue or viremia (Fig. 1D and E). As expected, both BTV8L- and BTV8H-infected sheep developed neutralizing antibodies although the antibody titers were lower in the BTV8H-infected animals (Fig. 1F).
Genotype and phenotype of rgBTV8L and rgBTV8H.
The data obtained so far indicate that BTV8H had accumulated mutations that affected virus replication in IFN-competent cells and resulted in attenuation in vivo both in IFNAR−/− mice and in sheep. Full-genome sequencing of BTV8H revealed 31 nucleotide mismatches compared to the sequence of BTV8L, out of which 16 resulted in amino acid residue substitutions (Fig. 2A). Nonsynonymous mutations were present in each of the 10 genomic segments and affected all characterized BTV proteins with the exception of NS4.
FIG 2.
Genetic differences between BTV8L and BTV8H and virulence of rescued viruses in IFNAR−/− mice. (A) Schematic representation of the 10 genomic segments of BTV8L and BTV8H. Mutations in BTV8H compared to the sequence of the minimally passaged BTV8L are indicated with red dots. Nonsynonymous mutations are marked with asterisks, and the numbers relative to the mutated amino acid residues in the corresponding viral proteins are shown. The plus sign indicates a nucleotide insertion. The length of the schematic genome segments and the relative position of mutations are indicative only. (B) Survival plots of IFNAR−/− mice (n = 5 per group) infected intraperitoneally with 300 PFU of rgBTV8L or rgBTV8H.
In order to identify BTV8 genomic segments affecting virulence, we rescued both BTV8L and BTV8H by reverse genetics. We reconstructed BTV8H by substituting the 16 nucleotide mutations of BTV8H into the BTV8L vectors by site-directed mutagenesis. Hence, we rescued rgBTV8L with the same nucleotide sequence as the original virus (BTV8L) while rgBTV8H was identical to the original high-passage-number virus at the amino acid level but did not contain the silent mutations present in the genome of BTV8H. We then inoculated IFNAR−/− mice with rgBTV8L and rgBTV8H in order to test whether the phenotypes of the rescued viruses were identical to those of the parental BTV8L and BTV8H. Groups of five mice were inoculated intraperitoneally with 300 PFU of each virus and monitored over a 14-day period (Fig. 2B). All mice infected with rgBTV8H survived the challenge while rgBTV8L caused 100% mortality. These data showed that both rescued viruses retained the phenotypes of the original viruses.
Pathogenicity of reassortants between rgBTV8L and rgBTV8H.
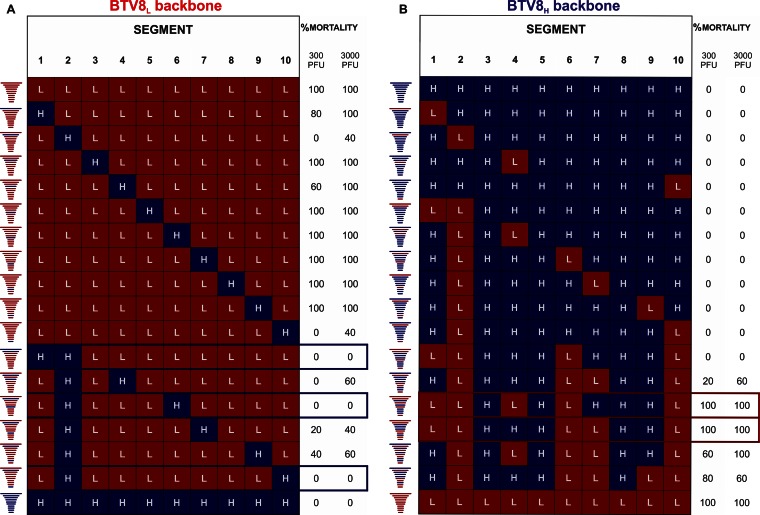
In order to define the genomic segments carrying attenuating mutations, we rescued in total 34 reassortants. A set of reassortants was based upon the BTV8L backbone with a single genomic segment or a combination of segments from BTV8H. Four monoreassortants showed reduced mortality in IFNAR−/− mice (Fig. 3A). In particular, BTV8L+S2H and BTV8L+S10H caused no mortality in mice at an inoculation dose of 300 PFU and caused 40% mortality with 3,000 PFU. Full attenuation using both inoculation doses was achieved with double reassortants BTV8L+S1/2H (which contains Seg1 and Seg2 of BTV8H), BTV8L+S2/6H, and BTV8L+S2/10H. These data suggest that VP2 and NS3 primarily, in addition to VP5 and VP1, were major factors of BTV8H attenuation in this mouse model. We next aimed to identify the minimal number of BTV8L segments that would restore full virulence to a reassortant based upon the BTV8H backbone (Fig. 3B). None of the single or double reassortants was virulent in IFNAR−/− mice. Furthermore, BTV8H+S1/2/6/10L also possessed an attenuated phenotype. These data suggested that other viral proteins besides VP1, VP2, VP5, and NS3 play a role in the virulence of BTV8L. To achieve 100% mouse mortality at an inoculation dose of either 300 or 3,000 PFU, it was required that at least five proteins of BTV8H be replaced by the BTV8L equivalents, including either VP4 or VP7 in conjunction with the aforementioned VP1, VP2, VP5, and NS3 (Fig. 3B).
FIG 3.
Virulence of BTV8L/BTV8H reassortants in IFNAR−/− mice. Multiple reassortant viruses were rescued using reverse genetics as described in Materials and Methods. Mortality of IFNAR−/− mice (n = 5 per group) inoculated intraperitoneally with 300 or 3,000 PFU of individual reassortants within the BTV8L or the BTV8H backbone is shown. Note that the combination of segments that resulted in rescue of fully attenuated reassortants is shown in blue boxes. The combination of at least five segments of BTV8L was required to recapitulate the fully virulent phenotype (shown in red boxes).
Replication kinetics of BTV8L monoreassortants in CPT-Tert cells.
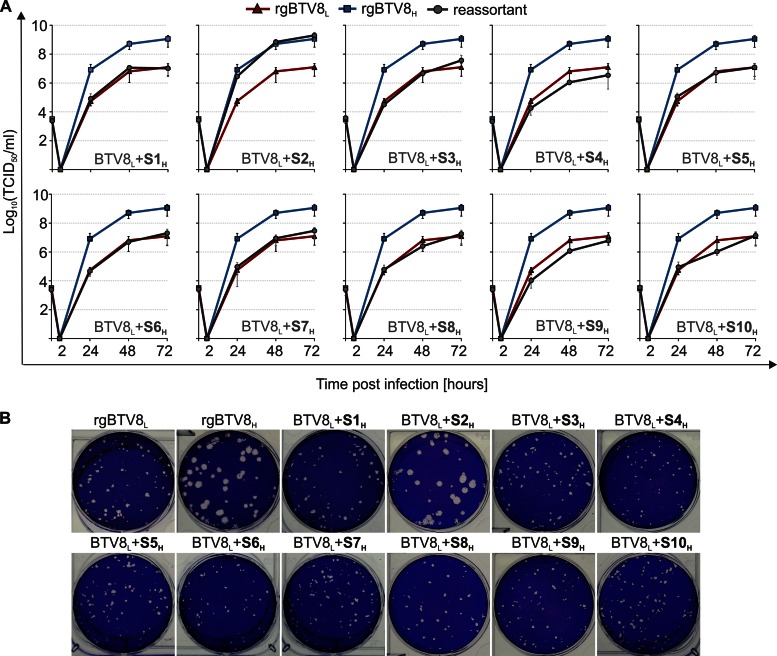
As shown above, the replacement of specific genome segments of BTV8L with the homologous segments from BTV8H resulted in attenuation in vivo. To establish whether the replication kinetics of these reassortants were compromised in vitro, we assessed the full set of 10 monoreassortants containing the BTV8L backbone in CPT-Tert cells (Fig. 4A). Overall, 9 of the 10 monoreassortants displayed similar replication kinetics to those of the parental rgBTV8L. Remarkably, BTV8L+S2H showed replication kinetics essentially identical to those of rgBTV8H and reached over 100-fold higher titers than rgBTV8L at 72 h p.i. Both rgBTV8H and BTV8L+S2H produced larger plaques than those induced by rgBTV8L and all the other monoreassortants (Fig. 4B). Overall, these data indicated that VP2 was the sole determinant of increased replication efficiency of BTV8H in vitro in CPT-Tert cells.
FIG 4.
In vitro replication properties of BTV8L/BTV8H monoreassortants. (A) Growth curves of parental rgBTV8L, rgBTV8H, and monoreassortants containing the BTV8L backbone (gray circle) in CPT-Tert cells. Monolayers were infected with the indicated viruses at an MOI of 0.01, and supernatants were collected at 2, 24, 48, and 72 h p.i. Viral titers were determined by endpoint dilutions. All reassortants, with exception of BTV8L+S2H, showed replication kinetics similar to those of the parental rgBTV8L. (B) Plaques produced in CPT-Tert cells by parental rgBTV8L and rgBTV8H and derived monoreassortants at 48 h p.i. Note the increased plaque size in rgBTV8H and BTV8L+S2H.
Cells cultured in vitro tend to display increased expression of glycosaminoglycans (GAGs) at the cell membrane. Interestingly some viruses, like foot and mouth disease virus, (47) show an increased affinity for heparan sulfate after being passaged in vitro. We hypothesized that VP2 might have acquired higher affinity for GAGs during passage in tissue culture, given that VP2 is the main determinant of BTV serotype and mediates viral entry (22, 25, 48). Therefore, we performed viral replication kinetic assays in CHO cells and in a derived cell line (pgsA-745) deficient in xylotransferase and thus lacking heparan sulfate GAGs. rgBTV8L and a BTV8H reassortant with the VP2 of BTV8L (BTV8H+S2L) grew equally well in both cell lines. However, rgBTV8H and BTV8L+S2H reached approximately 10-fold higher titers in CHO cells (P < 0.001) (Fig. 5A) and induced a more pronounced cytopathic effect (Fig. 5B) than rgBTV8L and BTV8H+S2L, respectively. Hence, BTV8H VP2 has a higher affinity for GAGs and facilitates BTV8 replication in vitro but not in vivo.
FIG 5.
Segment 2 of rgBTV8H favors replication in cells expressing glycosaminoglycans. (A) Viral titers produced in CHO and pgsA-745 cells infected with rgBTV8L, rgBTV8H, and reassortant viruses (MOI of 0.01). Supernatants were collected at 72 h p.i. and titrated in BSR cells by limiting dilution analysis. Mean values from three experiments performed in duplicate are shown (error bars correspond to standard deviations). Note that significant differences were observed between titers produced in CHO and pgsA-745 cells by BTV8L+S2H and BTV8H+S2L (***, P < 0.001; two-way analysis of variance followed by a Bonferroni test to identify interactions). (B) Cytopathic effect in CHO and pgsA-745 cells infected with rgBTV8L, rgBTV8H, and reassortant viruses (MOI of 0.01). Cells were stained with crystal violet at 72 h p.i.
Replication of rgBTV8L, rgBTV8H, and BTV8 monoreassortants in IFN-competent OvEC.
BTV8H does not replicate efficiently in IFN-competent primary OvEC. Thus, we further explored this model in order to characterize the interplay between rgBTV8L/rgBTV8H genomic segments and the cell-autonomous innate immune system. First, we compared the replication kinetics of BTV8L and the BTV8H monoreassortants in OvEC (Fig. 6). BTV8L+S2H reached higher titers in culture than the other viruses tested, which suggested that mutations in VP2H conferred an advantage to the replication of this reassortant in vitro, irrespective of the ability of the cell to produce IFN. Several reassortants demonstrated delayed growth compared to that of rgBTV8L. In particular, BTV8L+S4H and BTV8L+S9H yielded substantially lower titers than the parental BTV8L at 48 and 72 h p.i. (Fig. 6). None of the monoreassortants, however, replicated as poorly as rgBTV8H, which indicated that the growth restriction of this virus was the cumulative result of mutations in several of its genome segments.
FIG 6.
Replication kinetics of parental and monoreassortant viruses in primary OvEC. OvEC were infected at an MOI of 0.01 with rgBTV8L, rgBTV8H, and monoreassortants containing the BTV8L backbone (gray circle). Viral titers were determined at the specified time points by limiting dilution assays. Each panel shows growth curves of both parental viruses and a specific monoreassortant (as labeled).
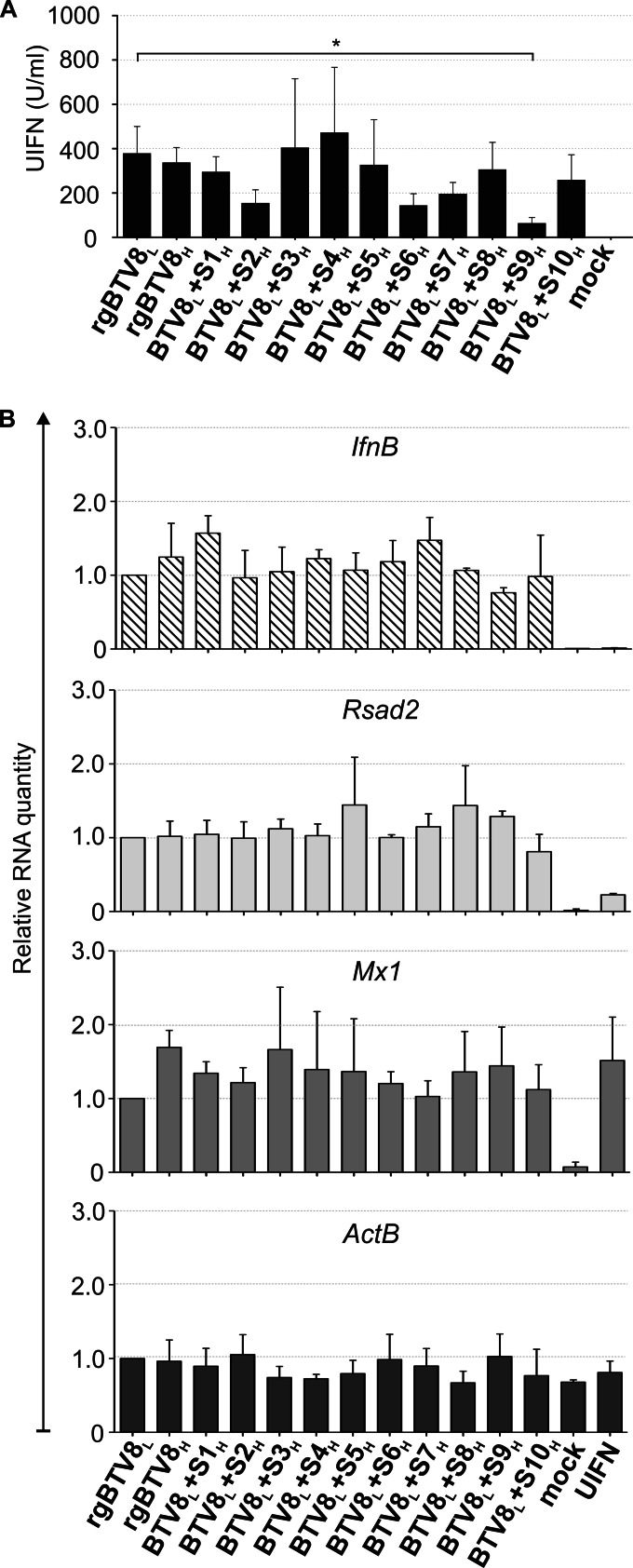
Next, we wanted to establish whether BTV8H was a more potent IFN inducer than the parental BTV8L. To this end, we measured IFN production in the supernatants of OvEC infected with the same set of viruses as above. rgBTV8H and most of the monoreassortants induced similar amounts of IFN. However, statistically significant differences (P < 0.05) were observed between rgBTV8L and BTV8L+S9H (Fig. 7A).
FIG 7.
IFN production and gene expression induced by infection of OvEC by rgBTV8L, rgBTV8H, and BTV8L/BTV8H monoreassortants. OvEC were infected with rgBTV8L, rgBTV8H, and monoreassortants within the BTV8L backbone (MOI of 1). (A) IFN protection assay. Supernatants were collected at 18 h p.i., inactivated by UV treatment, and used in a biological assay to estimate the amount of IFN present, as described in Materials and Methods. The only major differences were observed in cells infected with BTV8L+S9H, where the amount of IFN released was significantly lower than what was found in cells infected with rgBTV8L (P < 0.05; one-way analysis of variance followed by Dunnett's multiple-comparison test to dissect individual interactions). (B) IFN-β, ActB, RSAD2, and Mx1 gene expression. mRNA was measured by qPCR in OvEC at 18 h p.i. with parental and reassortant viruses (MOI of 1) as described in Materials and Methods. Mock-treated and UIFN-treated cells were used as controls. Panels show gene expression relative to rgBTV8L and normalized to GAPDH gene levels.
We also assessed the relative quantities of the IFN-β mRNA, selected IFN-stimulated gene (ISG) mRNAs (Mx1 and RSAD2), and β-actin in OvEC infected with either rgBTV8H, rgBTV8L, or one of the various monoreassortants as above. Mock-infected cells and cells treated with universal IFN (UIFN) were used as controls. We detected no IFN-β mRNA in either of the control samples, while it was readily detectable at similar levels in cells infected with rgBTV8L, rgBTV8H, and the rgBTV8L/rgBTV8H monoreassortants (Fig. 7B). Mx1 and RSAD2 were also upregulated in virus-infected cells, but no significant differences were found between cells infected with the various monoreassortants and the parental viruses (Fig. 7B). β-Actin levels were consistently uniform in all samples analyzed.
Effect of IFN pretreatment on the replication of rgBTV8L, rgBTV8H, and rgBTV8L/rgBTV8H monoreassortants.
The data described above, showed that the reduced replication kinetics of rgBTV8H, BTV8L+S4H, and BTV8L+S9H in OvEC were not due to increased IFN induction in these cells. In light of these data, we wanted to establish whether rgBTV8L, rgBTV8H, and the rgBTV8L/rgBTV8H reassortants were better or worse equipped to overcome restriction in cells already in an antiviral state prior to infection. For these experiments, we used CPT-Tert cells as they do not produce IFN but can respond to IFN treatment (5, 40). We pretreated CPT-Tert with either 1,000 units of UIFN or control medium for 18 h prior to infection with rgBTV8L, rgBTV8H, and monoreassortants with a BTV8L backbone at an MOI of 0.01. At 48 h p.i. supernatants of infected cells were collected and titrated by endpoint dilution analysis. Comparison of viral yields in cells treated and untreated with UIFN showed that the replication of all viruses was inhibited by at least 100-fold by UIFN (P < 0.001) (Fig. 8). The reduction in yield of rgBTV8L in UIFN-treated cells compared to that in untreated cells was approximately (5 × 103)-fold. Strikingly, this ratio was more than a millionfold (1.7 × 106) for rgBTV8H. In UIFN-treated cells, most reassortants reached yields similar to those obtained by rgBTV8L under the same conditions. A notable exception was BTV8L+S4H, which had yields 10-fold lower than those of rgBTV8L (P < 0.05) in the UIFN-treated samples. BTV8L+S2H showed the highest degree of inhibition in treated CPT-Tert cells among all 10 monoreassortants. However, the yield of BTV8L+S2H in treated cells was equivalent to the one obtained by rgBTV8L under the same conditions.
FIG 8.
Replication of rgBTV8L, rgBTV8H, and BTV8L/BTV8H monoreassortants in CPT-Tert cells pretreated with universal IFN (UIFN). Viral titers produced in untreated and IFN-pretreated CPT-Tert cells by parental and reassortant viruses at 48 h p.i. at an MOI of 0.01. Mean values from three experiments performed in duplicate are shown (error bars correspond to standard deviations). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way analysis of variance followed by Dunnett's multiple-comparison test to dissect individual interactions).
DISCUSSION
Several BTV incursions have occurred in Europe in the last 20 years. Some strains, such as the northern European BTV8 strain used in this study, have caused major animal health and economic problems while others did not cause any clinical signs in the field (49, 50).
Although the concept of BTV serotype/strain-related virulence is often quoted in the literature, only a few studies have attempted to understand what the viral molecular determinants that underscore this property are (29, 39, 51). In this study, we aimed to map the molecular determinants of virulence of the northern European strain of BTV8. We found that nonsynonymous mutations in four segments (Seg1, -2, -6, and -10) encoding VP1, VP2, VP5, and NS3 contributed to the attenuation of BTV8 in vivo in IFNAR−/− mice. Reassortants where segments encoding the VP2 or NS3 of the attenuated rgBTV8H were used in the context of the virulent rgBTV8L backbone caused no mortality in mice inoculated at the lower dose (300 PFU) used in this study. The combination of Seg2 with either Seg1, Seg6, or Seg10 of rgBTV8H in the context of the rgBTV8L backbone resulted in a fully attenuated reassortant that was nonpathogenic at either the low or high doses.
The multigenic nature of BTV attenuation has been previously suggested by us as we showed that tissue culture-attenuated strains of different BTV serotypes that show reduced virulence in IFNAR−/− mice tend to accumulate mutations consistently in segments 1, 2, and 8 (39). Interestingly, Seg2 with either Seg1, Seg6, or Seg10 of rgBTV8L did not restore virulence in the rgBTV8H backbone. The fact that a degree of virulence in rgBTV8H was achieved only after combining Seg7 with Seg2, Seg6, and Seg10 of rgBTV8L might suggest that interactions between specific viral proteins (or genomic segments) could play a role in the pathogenicity of BTV. Both VP2 and VP5 have previously been shown to interact with VP7 trimers in BTV virions (52). Additionally, other studies demonstrated the importance of NS3 interactions with outer capsid proteins in virus trafficking and assembly (19, 53). It is therefore possible that while mutations in high-passage-number VP5 or VP7 did not directly affect the functions of these proteins, they did influence virus virulence through their interactions with VP2 and NS3.
Interestingly, two mutations found in VP2 of BTV8H (positions 321 and 328) were located in the same region that was previously identified as being associated with attenuated BTV strains and as a target for neutralizing antibodies (39, 54, 55). This external and highly exposed area of VP2 was also implicated in attachment to a host cell receptor, and the mutations that arise in this region could be due to the changes in affinity for binding to specific ligands (10). Cryo-electron microscopy showed that VP2 possesses a sialic acid binding region located in its hub domain, which is one of two sites suggested to interact with the cell surface receptor (10). An increased affinity for GAGs has often been cited in the context of tissue culture-adapted strains (47, 56–58). Here, we found that BTV8H or reassortants with the VP2 of BTV8H had, indeed, a greater affinity for GAGs and were attenuated in IFNAR−/− mice. Previous studies have shown that viruses acquiring mutations conferring a higher affinity for heparan sulfate proteoglycans are often attenuated in vivo (58–62).
BTV is a potent inducer of type 1 IFN in vivo and in vitro (5, 21, 63–66). IFNAR−/− mice, due to the lack of expression of the type 1 IFN receptor, are a suitable tool to study determinants of viral virulence unrelated to IFN expression (39, 67, 68). However, it is most likely that other viral factors that are used to counteract the IFN system contribute to BTV virulence. Pathogenicity studies of multiple reassortants in sheep are not feasible; hence, we used primary OvEC as an in vitro surrogate model to identify the genome segments that confer higher sensitivity to the cell-autonomous IFN response. BTV8H (and its derivative rescued by reverse genetics) replicated poorly in primary cells. These data showed that the IFN response played a role in the inhibition of BTV8H replication. Both BTV8L and BTV8H induced similar amounts of IFN in infected primary cells. These data suggest that primary OvEC sense and respond to infection with both viruses in a similar manner but that BTV8L was able to counteract this response better than BTV8H.
Most of the BTV8L monoreassortants containing BTV8H genomic segments were able to replicate to similar titers as the parental low-passage-number virus in primary cells. Two notable exceptions included BTV8L+S4H and BTV8L+S9H. However, the VP4 (encoded by Seg4) of BTV8H also influenced the viral phenotype in IFNAR−/− mice, suggesting that mutations in this protein could also have affected its functions unrelated to the type I IFN response. VP4 of BTV acts as a capping enzyme, and therefore the mutations in VP4H and, in particular, the mutation in the amino acid residue at position 332 of the predicted 2′-O-methyltransferase (2′ OMTase) domain could have an adverse effect on the efficiency of viral mRNA capping (69, 70). Moreover, recent studies have shown that viral mRNA that lacks 2′-O-methylation at its 5′ cap structure induces a more potent innate immune response through MDA5 activation or direct interactions with proteins members of the IFIT (interferon-inducible protein with tetratricopeptide repeats) family (71–73). An inefficient capping mechanism would therefore explain the slight, yet consistent, decrease in virus yields reached by BTV8L+S4H in IFN-deficient CPT-Tert cells and the more pronounced growth inhibition in OvEC. This hypothesis was further supported by comparing the growth kinetics of monoreassortants in the BTV8L backbone in CPT-Tert cells either untreated or pretreated with UIFN. Of all reassortants, BTV8L+S4H showed the lowest yields in UIFN-pretreated cells, which suggested that VP4H was one of the proteins contributing to the inability of rgBTV8H to replicate in cells primed with IFN. These data are in concordance with other studies showing that West Nile virus, coronaviruses, and poxviruses mutants with deficient 2′ OMTase activity were not able to escape IFIT-2 induced restriction in cells stably expressing IFIT-2 (72). It is therefore possible that through viral mRNA capping, the VP4 of BTV8 could play a role in evading host restriction factors in order to allow the virus to replicate in host cells in an antiviral state.
BTV8L+S9H also replicated less efficiently than the parental virus in primary endothelial cells. VP6 is encoded by Seg9 of the BTV genome, as is NS4, which is a nonstructural protein shown to counteract the IFN system (5; unpublished data). We found no mutations in the NS4 open reading frame, but we detected a nonsynonymous mutation in VP6 (RNA-dependent ATPase and helicase). We are not aware of viral helicases being involved in interactions with the innate immune system. We noticed a slight reduction in the replication kinetics of BTV8L+S9H (compared to those of BTV8L) in CPT-Tert cells. Therefore, this reassortant might possess a suboptimal replication capacity that is amplified in the IFN-competent OvEC.
It is important to stress that although the BTV monoreassortants displayed an array of intermediate growth patterns in OvEC, none of them replicated as poorly as rgBTV8H. This indicated that mutations in segments other than Seg4 and Seg6 contributed to the restricted replication of the high-passage-number virus. Altogether, these data reinforce the concept that passaging viruses in an environment with no constraints from the IFN system allows greater flexibility of the viral genome, which in turn leads to the emergence of viruses with optimal replication efficiencies. The mutations that arise under such conditions might not necessarily involve major IFN antagonists but can involve proteins that are normally fine-tuned to evoke a minimal immune response while allowing sufficient (yet suboptimal) transmission in the natural host (74). A recent study looking into interserotype determinants of BTV pathogenesis in sheep showed that replacement of BTV1 Seg2, -6, and -10 with homologous BTV8 segments resulted in reassortants that showed a similar phenotype in sheep as BTV8 (51). However, this study was performed using BTV1 and BTV8 serotypes that are both virulent in sheep. Consequently, a clear distinction between levels of pathogenicity of the parental viruses and their reassortants was difficult to assess.
In conclusion, our data show that multiple genome segments determine the virulence of BTV8. Our study reinforces the concept that a constellation of genome segments determines the virulence of viruses with a segmented genome, as suggested in other studies in rotaviruses, influenza viruses, and bunyaviruses (74–82).
VP2 and NS3 were found to be primary determinants of BTV8 pathogenesis although VP1, VP5, VP4, VP6, and VP7 were also found to contribute to virulence. Given the high diversity of BTV, it is possible that different determinants of pathogenicity will be found in other serotypes. More studies aimed to map the molecular determinants of BTV virulence, using different strains/serotypes and experimental systems, will help to build an intellectual framework to characterize thoroughly BTV pathogenesis. The possibility to determine the pathogenicity of viral isolates on the bases of their genome sequences could help in designing control strategies that fit the risk posed by new emerging viruses.
ACKNOWLEDGMENTS
This work was funded by the Wellcome Trust. A.J. was funded by a BBSRC doctoral training grant.
We are grateful to Mariana Varela for useful suggestions.
REFERENCES
- 1.Price DA, Hardy WT. 1954. Isolation of the bluetongue virus from Texas sheep-Culicoides shown to be a vector. J Am Vet Med Assoc 124:255–258. [PubMed] [Google Scholar]
- 2.MacLachlan NJ, Drew CP, Darpel KE, Worwa G. 2009. The pathology and pathogenesis of bluetongue. J Comp Pathol 141:1–16. doi: 10.1016/j.jcpa.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Mellor PS, Baylis M, Mertens PP. 2009. Bluetongue. Academic Press, London, United Kingdom. [Google Scholar]
- 4.Roy P, Noad R. 2006. Bluetongue virus assembly and morphogenesis. Curr Top Microbiol Immunol 309:87–116. doi: 10.1007/3-540-30773-7_4. [DOI] [PubMed] [Google Scholar]
- 5.Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M. 2011. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog 7:e1002477. doi: 10.1371/journal.ppat.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gouet P, Diprose JM, Grimes JM, Malby R, Burroughs JN, Zientara S, Stuart DI, Mertens PP. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481–490. doi: 10.1016/S0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 7.Patel A, Roy P. 2014. The molecular biology of bluetongue virus replication. Virus Res 182:5–20. doi: 10.1016/j.virusres.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimes JM, Jakana J, Ghosh M, Basak AK, Roy P, Chiu W, Stuart DI, Prasad BV. 1997. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5:885–893. doi: 10.1016/S0969-2126(97)00243-8. [DOI] [PubMed] [Google Scholar]
- 9.Roy P. 2008. Bluetongue virus: dissection of the polymerase complex. J Gen Virol 89:1789–1804. doi: 10.1099/vir.0.2008/002089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Boyce M, Bhattacharya B, Schein S, Roy P, Zhou ZH. 2010. Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc Natl Acad Sci U S A 107:6292–6297. doi: 10.1073/pnas.0913403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forzan M, Marsh M, Roy P. 2007. Bluetongue virus entry into cells. J Virol 81:4819–4827. doi: 10.1128/JVI.02284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan SH, Wirblich C, Forzan M, Roy P. 2001. Expression and functional characterization of bluetongue virus VP5 protein: role in cellular permeabilization. J Virol 75:8356–8367. doi: 10.1128/JVI.75.18.8356-8367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owens RJ, Limn C, Roy P. 2004. Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J Virol 78:6649–6656. doi: 10.1128/JVI.78.12.6649-6656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monastyrskaya K, Gould EA, Roy P. 1995. Characterization and modification of the carboxy-terminal sequences of bluetongue virus type 10 NS1 protein in relation to tubule formation and location of an antigenic epitope in the vicinity of the carboxy terminus of the protein. J Virol 69:2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyce M, Celma CC, Roy P. 2012. Bluetongue virus non-structural protein 1 is a positive regulator of viral protein synthesis. Virol J 9:178. doi: 10.1186/1743-422X-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lymperopoulos K, Noad R, Tosi S, Nethisinghe S, Brierley I, Roy P. 2006. Specific binding of Bluetongue virus NS2 to different viral plus-strand RNAs. Virology 353:17–26. doi: 10.1016/j.virol.2006.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kar AK, Bhattacharya B, Roy P. 2007. Bluetongue virus RNA binding protein NS2 is a modulator of viral replication and assembly. BMC Mol Biol 8:4. doi: 10.1186/1471-2199-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butan C, Tucker P. 2010. Insights into the role of the non-structural protein 2 (NS2) in bluetongue virus morphogenesis. Virus Res 151:109–117. doi: 10.1016/j.virusres.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Beaton AR, Rodriguez J, Reddy YK, Roy P. 2002. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc Natl Acad Sci U S A 99:13154–13159. doi: 10.1073/pnas.192432299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celma CC, Roy P. 2009. A viral nonstructural protein regulates bluetongue virus trafficking and release. J Virol 83:6806–6816. doi: 10.1128/JVI.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauveau E, Doceul V, Lara E, Breard E, Sailleau C, Vidalain PO, Meurs EF, Dabo S, Schwartz-Cornil I, Zientara S, Vitour D. 2013. NS3 of bluetongue virus interferes with the induction of type I interferon. J Virol 87:8241–8246. doi: 10.1128/JVI.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huismans H, Erasmus BJ. 1981. Identification of the serotype-specific and group-specific antigens of bluetongue virus. Onderstepoort J Vet Res 48:51–58. [PubMed] [Google Scholar]
- 23.Kahlon J, Sugiyama K, Roy P. 1983. Molecular basis of bluetongue virus neutralization. J Virol 48:627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek-Bankowska K, Belaganahalli MN, Attoui H, Mertens PP. 2011. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS One 6:e26147. doi: 10.1371/journal.pone.0026147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw AE, Ratinier M, Nunes SF, Nomikou K, Caporale M, Golder M, Allan K, Hamers C, Hudelet P, Zientara S, Breard E, Mertens P, Palmarini M. 2013. Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment. J Virol 87:543–557. doi: 10.1128/JVI.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brenner J, Batten C, Yadin H, Bumbarov V, Friedgut O, Rotenberg D, Golender N, Oura CA. 2011. Clinical syndromes associated with the circulation of multiple serotypes of bluetongue virus in dairy cattle in Israel. Vet Rec 169:389. doi: 10.1136/vr.d4629. [DOI] [PubMed] [Google Scholar]
- 27.Spreull J. 1905. Malarial catarrhal fever (bluetongue) of sheep in South Africa. J Comp Pathol Ther 18:321–339. doi: 10.1016/S0368-1742(05)80073-6. [DOI] [Google Scholar]
- 28.Anderson GA, Stott JL, Gershwin LJ, Osburn BI. 1985. Subclinical and clinical bluetongue disease in cattle: clinical, pathological and pathogenic considerations. Prog Clin Biol Res 178:103–107. [PubMed] [Google Scholar]
- 29.Waldvogel AS, Anderson CA, Higgins RJ, Osburn BI. 1987. Neurovirulence of the UC-2 and UC-8 strains of bluetongue virus serotype 11 in newborn mice. Vet Pathol 24:404–410. [DOI] [PubMed] [Google Scholar]
- 30.Parsonson IM. 1990. Pathology and pathogenesis of bluetongue infections. Curr Top Microbiol Immunol 162:119–141. [DOI] [PubMed] [Google Scholar]
- 31.MacLachlan NJ. 1994. The pathogenesis and immunology of bluetongue virus-infection of ruminants. Comp Immunol Microbiol Infect Dis 17:197–206. doi: 10.1016/0147-9571(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 32.Oura CA, Wood JL, Sanders AJ, Bin-Tarif A, Henstock M, Edwards L, Floyd T, Simmons H, Batten CA. 2009. Seroconversion, neutralising antibodies and protection in bluetongue serotype 8 vaccinated sheep. Vaccine 27:7326–7330. doi: 10.1016/j.vaccine.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 33.Caporale M, Di Gialleonorado L, Janowicz A, Wilkie G, Shaw A, Savini G, Van Rijn PA, Mertens P, Di Ventura M, Palmarini M. 2014. Virus and host factors affecting the clinical outcome of bluetongue virus infection. J Virol 88:10399–10411. doi: 10.1128/JVI.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbers A, Backx A, van der Spek A, Ekker M, Leijs P, Steijn K, van Langen H, van Rijn P. 2008. Epidemiology of bluetongue virus serotype 8 outbreaks in the Netherlands in 2006. Tijdschr Diergeneeskd 133:222–229. (In Dutch.). [PubMed] [Google Scholar]
- 35.Elbers AR, Backx A, Mintiens K, Gerbier G, Staubach C, Hendrickx G, van der Spek A. 2008. Field observations during the bluetongue serotype 8 epidemic in 2006. II. Morbidity and mortality rate, case fatality and clinical recovery in sheep and cattle in the Netherlands. Prev Vet Med 87:31–40. doi: 10.1016/j.prevetmed.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Perrin JB, Ducrot C, Vinard JL, Morignat E, Gauffier A, Calavas D, Hendrikx P. 2010. Using the National Cattle Register to estimate the excess mortality during an epidemic: application to an outbreak of bluetongue serotype 8. Epidemics 2:207–214. doi: 10.1016/j.epidem.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Coetzee P, Van Vuuren M, Stokstad M, Myrmel M, Venter EH. 2012. Bluetongue virus genetic and phenotypic diversity: towards identifying the molecular determinants that influence virulence and transmission potential. Vet Microbiol 161:1–12. doi: 10.1016/j.vetmic.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Moulin V, Noordegraaf CV, Makoschey B, van der Sluijs M, Veronesi E, Darpel K, Mertens PP, de Smit H. 2012. Clinical disease in sheep caused by bluetongue virus serotype 8, and prevention by an inactivated vaccine. Vaccine 30:2228–2235. doi: 10.1016/j.vaccine.2011.11.100. [DOI] [PubMed] [Google Scholar]
- 39.Caporale M, Wash R, Pini A, Savini G, Franchi P, Golder M, Patterson-Kane J, Mertens P, Di Gialleonardo L, Armillotta G, Lelli R, Kellam P, Palmarini M. 2011. Determinants of bluetongue virus virulence in murine models of disease. J Virol 85:11479–11489. doi: 10.1128/JVI.05226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnaud F, Black SG, Murphy L, Griffiths DJ, Neil SJ, Spencer TE, Palmarini M. 2010. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J Virol 84:4415–4425. doi: 10.1128/JVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, McGregor E, Piras IM, Shaw A, Lamm C, Janowicz A, Beer M, Glass M, Herder V, Hahn K, Baumgartner W, Kohl A, Palmarini M. 2013. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog 9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyce M, Celma CC, Roy P. 2008. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J Virol 82:8339–8348. doi: 10.1128/JVI.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunes SF, Hamers C, Ratinier M, Shaw A, Brunet S, Hudelet P, Palmarini M. 2014. A synthetic biology approach for a vaccine platform against known and newly emerging serotypes of bluetongue virus. J Virol 88:12222–12232. doi: 10.1128/JVI.02183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, Samuel AR, Mertens PP. 2007. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods 143:132–139. doi: 10.1016/j.jviromet.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet—next generation sequence assembly visualization. Bioinformatics 26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baranowski E, Sevilla N, Verdaguer N, Ruiz-Jarabo CM, Beck E, Domingo E. 1998. Multiple virulence determinants of foot-and-mouth disease virus in cell culture. J Virol 72:6362–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mertens PP, Pedley S, Cowley J, Burroughs JN, Corteyn AH, Jeggo MH, Jennings DM, Gorman BM. 1989. Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virology 170:561–565. doi: 10.1016/0042-6822(89)90447-9. [DOI] [PubMed] [Google Scholar]
- 49.Maan S, Maan NS, van Rijn PA, van Gennip RG, Sanders A, Wright IM, Batten C, Hoffmann B, Eschbaumer M, Oura CA, Potgieter AC, Nomikou K, Mertens PP. 2010. Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains. PLoS One 5:e10323. doi: 10.1371/journal.pone.0010323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson AJ, Mellor PS. 2009. Bluetongue in Europe: past, present and future. Philos Trans R Soc Lond B Biol Sci 364:2669–2681. doi: 10.1098/rstb.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celma CC, Bhattacharya B, Eschbaumer M, Wernike K, Beer M, Roy P. 2014. Pathogenicity study in sheep using reverse-genetics-based reassortant bluetongue viruses. Vet Microbiol 174:139–147. doi: 10.1016/j.vetmic.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nason EL, Rothagel R, Mukherjee SK, Kar AK, Forzan M, Prasad BV, Roy P. 2004. Interactions between the inner and outer capsids of bluetongue virus. J Virol 78:8059–8067. doi: 10.1128/JVI.78.15.8059-8067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharya B, Roy P. 2008. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J Virol 82:10600–10612. doi: 10.1128/JVI.01274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gould AR, Eaton BT. 1990. The amino acid sequence of the outer coat protein VP2 of neutralizing monoclonal antibody-resistant, virulent and attenuated bluetongue viruses. Virus Res 17:161–172. doi: 10.1016/0168-1702(90)90062-G. [DOI] [PubMed] [Google Scholar]
- 55.Gould AR, Hyatt AD, Eaton BT. 1988. Morphogenesis of a bluetongue virus variant with an amino acid alteration at a neutralization site in the outer coat protein, VP2. Virology 165:23–32. doi: 10.1016/0042-6822(88)90654-X. [DOI] [PubMed] [Google Scholar]
- 56.Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72:7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol 75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 60.Byrnes AP, Griffin DE. 2000. Large-plaque mutants of Sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J Virol 74:644–651. doi: 10.1128/JVI.74.2.644-651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee E, Wright PJ, Davidson A, Lobigs M. 2006. Virulence attenuation of Dengue virus due to augmented glycosaminoglycan-binding affinity and restriction in extraneural dissemination. J Gen Virol 87:2791–2801. doi: 10.1099/vir.0.82164-0. [DOI] [PubMed] [Google Scholar]
- 62.Olmsted RA, Baric RS, Sawyer BA, Johnston RE. 1984. Sindbis virus mutants selected for rapid growth in cell culture display attenuated virulence in animals. Science 225:424–427. doi: 10.1126/science.6204381. [DOI] [PubMed] [Google Scholar]
- 63.Huismans H. 1969. Bluetongue virus-induced interferon synthesis. Onderstepoort J Vet Res 36:181–185. [PubMed] [Google Scholar]
- 64.Jameson P, Schoenherr CK, Grossberg SE. 1978. Bluetongue virus, an exceptionally potent interferon inducer in mice. Infect Immun 20:321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacLachlan NJ, Thompson J. 1985. Bluetongue virus-induced interferon in cattle. Am J Vet Res 46:1238–1241. [PubMed] [Google Scholar]
- 66.Ruscanu S, Pascale F, Bourge M, Hemati B, Elhmouzi-Younes J, Urien C, Bonneau M, Takamatsu H, Hope J, Mertens P, Meyer G, Stewart M, Roy P, Meurs EF, Dabo S, Zientara S, Breard E, Sailleau C, Chauveau E, Vitour D, Charley B, Schwartz-Cornil I. 2012. The double-stranded RNA bluetongue virus induces type I interferon in plasmacytoid dendritic cells via a MYD88-dependent TLR7/8-independent signaling pathway. J Virol 86:5817–5828. doi: 10.1128/JVI.06716-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ortego J, de la Poza F, Marin-Lopez A. 2014. Interferon alpha/beta receptor knockout mice as a model to study bluetongue virus infection. Virus Res 182:35–42. doi: 10.1016/j.virusres.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 68.Calvo-Pinilla E, Nieto JM, Ortego J. 2010. Experimental oral infection of bluetongue virus serotype 8 in type I interferon receptor-deficient mice. J Gen Virol 91:2821–2825. doi: 10.1099/vir.0.024117-0. [DOI] [PubMed] [Google Scholar]
- 69.Sutton G, Grimes JM, Stuart DI, Roy P. 2007. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol 14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 70.Ramadevi N, Burroughs NJ, Mertens PP, Jones IM, Roy P. 1998. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc Natl Acad Sci U S A 95:13537–13542. doi: 10.1073/pnas.95.23.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Sastre A. 2011. 2 Methylate or not 2 methylate: viral evasion of the type I interferon response. Nat Immunol 12:114–115. doi: 10.1038/ni0211-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zust R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K. 2013. Rational design of a live attenuated dengue vaccine: 2′-O-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Cidoncha M, Killip MJ, Oliveros JC, Asensio VJ, Fernandez Y, Bengoechea JA, Randall RE, Ortin J. 2014. An unbiased genetic screen reveals the polygenic nature of the influenza virus anti-interferon response. J Virol 88:4632–4646. doi: 10.1128/JVI.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoshino Y, Saif LJ, Kang SY, Sereno MM, Chen WK, Kapikian AZ. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274–280. doi: 10.1006/viro.1995.1255. [DOI] [PubMed] [Google Scholar]
- 76.Safronetz D, Rockx B, Feldmann F, Belisle SE, Palermo RE, Brining D, Gardner D, Proll SC, Marzi A, Tsuda Y, Lacasse RA, Kercher L, York A, Korth MJ, Long D, Rosenke R, Shupert WL, Aranda CA, Mattoon JS, Kobasa D, Kobinger G, Li Y, Taubenberger JK, Richt JA, Parnell M, Ebihara H, Kawaoka Y, Katze MG, Feldmann H. 2011. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J Virol 85:1214–1223. doi: 10.1128/JVI.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clements ML, Subbarao EK, Fries LF, Karron RA, London WT, Murphy BR. 1992. Use of single-gene reassortant viruses to study the role of avian influenza A virus genes in attenuation of wild-type human influenza A virus for squirrel monkeys and adult human volunteers. J Clin Microbiol 30:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Griot C, Gonzalez-Scarano F, Nathanson N. 1993. Molecular determinants of the virulence and infectivity of California serogroup bunyaviruses. Annu Rev Microbiol 47:117–138. doi: 10.1146/annurev.mi.47.100193.001001. [DOI] [PubMed] [Google Scholar]
- 79.Janssen RS, Nathanson N, Endres MJ, Gonzalez-Scarano F. 1986. Virulence of La Crosse virus is under polygenic control. J Virol 59:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rott R. 1980. Genetic determinants for infectivity and pathogenicity of influenza viruses. Philos Trans R Soc Lond B Biol Sci 288:393–399. doi: 10.1098/rstb.1980.0016. [DOI] [PubMed] [Google Scholar]
- 81.Scholtissek C, Vallbracht A, Flehmig B, Rott R. 1979. Correlation of pathogenicity and gene constellation of influenza A viruses. II. Highly neurovirulent recombinants derived from non-neurovirulent or weakly neurovirulent parent virus strains. Virology 95:492–500. [DOI] [PubMed] [Google Scholar]
- 82.Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, Sears SD, Dolin R, Maassab HF, Murphy BR. 1988. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol 62:488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]