ABSTRACT
Epstein-Barr virus (EBV) is one of the major oncogenic viruses and is found in nearly 10% of gastric carcinomas. EBV is known to encode its own microRNAs (miRNAs); however, their roles have not been fully investigated. The present report is the largest series to comprehensively profile the expression of 44 known EBV miRNAs in tissue samples from patients with EBV-associated gastric carcinoma. Several miRNAs were highly expressed in EBV-associated gastric carcinoma, and in silico analysis revealed that the target genes of these EBV miRNAs had functions associated with cancer-related pathways, especially the regulation of apoptosis. Apoptosis was reduced in EBV-associated gastric carcinoma tissue samples, and gastric carcinoma cell lines infected with EBV exhibited downregulation of the proapoptotic protein Bid (the BH3-interacting domain death agonist), a member of the Bcl-2 family. The luciferase activity of the reporter vector containing the 3′ untranslated region of BID was inhibited by an ebv-miR-BART4-5p mimic in gastric cancer cell lines. Transfection of an ebv-miR-BART4-5p mimic reduced Bid expression in EBV-negative cell lines, leading to reduced apoptosis under serum deprivation. The inhibition of ebv-miR-BART4-5p expression was associated with partial recovery of Bid levels in EBV-positive cell lines. The results demonstrated the antiapoptotic role of EBV miRNA via regulation of Bid expression in EBV-associated gastric carcinoma. These findings provide novel insights in the roles of EBV miRNAs in gastric carcinogenesis, which would be a potential therapeutic target.
IMPORTANCE This report is the largest series to comprehensively profile the expression of 44 known EBV miRNAs in clinical samples from EBV-associated gastric carcinoma patients. Of the EBV miRNAs, ebv-miR-BART4-5p plays an important role in gastric carcinogenesis via regulation of apoptosis.
INTRODUCTION
Epstein-Barr virus (EBV) is one of the major oncogenic viruses inducing various kinds of malignancies in humans. The virus originally was isolated from Burkitt's lymphoma and is associated with other hematopoietic tumors, including Hodgkin's lymphoma, extranodal nasal-type NK/T-cell lymphoma, diffuse large B-cell lymphoma (DLBCL), and lymphoproliferative disorders in immunosuppressed patients, as well as some epithelial malignancies, such as nasopharyngeal carcinoma (NPC) and gastric carcinoma (1–3). Nearly 10% of all gastric carcinomas are associated with EBV infection and form a distinct subtype of disease, exhibiting characteristic clinicopathological features (4).
Several studies have investigated the roles played by EBV in carcinogenesis and have sought to show that EBV gene transcripts, including EBV nuclear antigen 2 (EBNA2) or latent membrane protein 1 (LMP1), disturb signal transduction in host cells, causing immortalization or transformation (5). Although these studies have shed light on the oncogenic properties of EBV, many unsolved problems remain.
Recently, a novel class of small, noncoding RNA molecules, termed microRNAs (miRNAs), has been identified as a group of important posttranscriptional regulators of gene expression. miRNAs play roles in many complex biological processes, including cellular development and differentiation. An increasing number of studies have shown that dysregulation of certain miRNAs induces carcinogenesis in various organs. To date, more than 4,500 human miRNAs (cellular miRNAs) have been archived in miRBase (http://www.mirbase.org/) (6). In a previous study, we showed that two of these cellular miRNAs, hsa-miR-200a and hsa-miR-200b, were downregulated in EBV-associated gastric carcinoma tissues both in vivo and in vitro (7). This caused a decrease in E-cadherin expression via upregulation of the transcription repressors ZEB1 and ZEB2 and triggered the epithelial-to-mesenchymal transition, an important feature of carcinogenesis (8–11).
Not only mammals but also viruses encode miRNAs. EBV was the first virus in which viral miRNAs were found (12, 13). To date, 25 EBV miRNA precursors and 44 mature EBV miRNAs have been registered in miRBase. The EBV-encoded miRNAs fall into two major clusters, the BHRF-1 and BART clusters. The BHRF-1 cluster contains four mature miRNAs expressed only in lytically infected cells or cells with latency type III infections (12, 14). The BART cluster is further subdivided into subclusters 1 and 2, containing 38 mature EBV miRNAs in total, and the miRNAs ebv-miR-BART2-5p and ebv-miR-BART2-3p are located downstream of these two clusters. Several studies have profiled EBV-encoded miRNA expression in EBV-associated malignancies, including NPC and DLBCL, and have revealed that specific viral miRNAs play roles in carcinogenesis (15–18). Three studies used either Northern blotting or quantitative reverse transcription-PCR (RT-PCR) to measure the expression levels of a limited number of EBV-encoded miRNAs in gastric carcinoma tissues (19–21). In addition, two recent reports profiled viral miRNA expression levels in gastric carcinoma cell lines infected with EBV (22, 23).
The present study is the largest series to comprehensively profile the expression of 44 known EBV-encoded miRNAs in clinical samples from gastric carcinoma patients. We used quantitative RT-PCR to investigate viral miRNA expression in such samples, in cell lines originally derived from EBV-associated gastric carcinoma cells, and in cells secondarily infected with EBV by the cell-to-cell contact method (24). We compared our expression profiles with those previously reported for patients with EBV-associated malignancies, and we identified several viral miRNAs that are frequently highly expressed in EBV-associated gastric carcinoma tissues. We sought the cellular targets of these viral miRNAs using an in silico approach. We focused specifically on apoptosis-related proteins and further investigated their roles in vitro.
MATERIALS AND METHODS
Tissue samples.
Viral miRNA expression was examined in tissues from 10 patients with surgically resected EBV-associated gastric carcinoma. An additional 5 NPC cases and 5 EBV-positive DLBCL cases were selected to measure the expression of specific EBV miRNAs. The frequency of apoptosis also was analyzed in 54 surgical specimens (from 19 cases of EBV-positive and 35 cases of EBV-negative gastric carcinoma). Samples were collected from the archives of the Department of Pathology of the University of Tokyo Hospital. The study was approved by the University of Tokyo Ethics Committee. Samples were fixed in formalin and embedded in paraffin. Hematoxylin-and-eosin (H&E)-stained slides were reviewed. The presence of EBV in tumor cells was confirmed by in situ hybridization targeting an EBV-encoded small RNA (EBER-ISH) with an EBER1-RNA oligonucleotide probe, as previously described (25).
KT tumors were established by inoculating cells of surgically resected EBV-associated gastric carcinoma tissues into SCID mice, as previously described (26).
Cell lines and culture conditions.
The gastric carcinoma cell lines used in the present study were MKN1, NUGC3, AGS, and SNU719. The former three lines originally were derived from gastric carcinoma tissues without EBV infection. The MKN1 line was obtained from the Riken BioResource Center Cell Bank (Tsukuba, Japan), the NUGC3 line from the Japanese Cancer Research Resource Bank (Osaka, Japan), and the AGS line from the American Type Culture Collection (Manassas, VA). The SNU719 line was derived from EBV-associated gastric carcinoma tissue and was obtained from the Korean Cell Line Bank (Seoul, South Korea). All cell lines were authenticated using the short tandem repeat-PCR method. Cell lines were cultured in RPMI 1640 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% (vol/vol) fetal calf serum (FCS) (MP Biomedicals, Solon, OH), penicillin (40 U/ml), and streptomycin (50 μg/ml) at 37°C under 5% (vol/vol) CO2.
EBV infection.
MKN1, NUGC3, and AGS cells were infected with recombinant EBV using the cell-to-cell contact method (24). Cells of a Burkitt's lymphoma line, Akata, were modified to produce recombinant EBV in which a neomycin resistance gene was inserted into BXLF1; this virus was used in the present study. EBV-infected cells were obtained after bulk selection with G418 (700 μg/ml; Sigma-Aldrich, St. Louis, MO). The establishment of EBV infection was confirmed by detecting EBER-ISH in cells grown on plastic slides after fixation in 10% (vol/vol) formalin. The expression of EBV latent genes (BARF0, EBER, EBNA1, EBNA2, LMP1, and LMP2A) was evaluated using RT-PCR as previously described (26). EBV-infected cells were maintained in bulk in RPMI 1640 medium supplemented with 10% (vol/vol) FCS and G418 (50 μg/ml).
RNA extraction and quantitative RT-PCR.
Total RNAs, including miRNAs, were extracted from paraffin-embedded tissues of surgically resected samples and KT tumors using RecoverAll total nucleic acid isolation kits for formalin-fixed, paraffin-embedded tissues (Life Technologies, Carlsbad, CA). Total RNA was extracted from cultured cells using mirVana miRNA isolation kits (Life Technologies).
For miRNA analysis, mature miRNAs were reverse transcribed and quantitative PCR was performed using TaqMan MiRNA assays detecting ebv-miR-BART1-5p (1), ebv-miR-BART1-3p, ebv-miR-BART2-5p (2), ebv-miR-BART2-3p, ebv-miR-BART4-5p (4), ebv-miR-BART4-3p (4*), ebv-miR-BART5-5p (5), ebv-miR-BART5-3p (5*), ebv-miR-BART6-5p, ebv-miR-BART6-3p, ebv-miR-BART7-5p (7*), ebv-miR-BART7-3p (7), ebv-miR-BART8-5p (8), ebv-miR-BART8-3p (8*), ebv-miR-BART9-5p (9*), ebv-miR-BART9-3p (9), ebv-miR-BART11-5p, ebv-miR-BART11-3p, ebv-miR-BART12, ebv-miR-BART13-5p (13*), ebv-miR-BART13-3p (13), ebv-miR-BART14-5p (14*), ebv-miR-BART14-3p (14), ebv-miR-BART15, ebv-miR-BART16, ebv-miR-BART17-5p, ebv-miR-BART17-3p, ebv-miR-BART18-5p (18), ebv-miR-BART18-3p, ebv-miR-BART19-5p, ebv-miR-BART19-3p (19), ebv-miR-BART20-5p, ebv-miR-BART20-3p, ebv-miR-BART21-5p, ebv-miR-BART21-3p, ebv-miR-BART22, ebv-miR-BHRF1-1, ebv-miR-BHRF1-2-5p (1-2*), ebv-miR-BHRF1-2-3p (1-2), and ebv-miR-BHRF1-3, as well as custom TaqMan small RNA assays detecting ebv-miR-BART3-5p (3*), ebv-miR-BART3-3p (3), ebv-miR-BART10-5p (10*), and ebv-miR-BART10-3p (10). The figures in parentheses indicate the previous mature miRNA identifier. The assays were performed according to the protocols of the manufacturer (Life Technologies). Viral miRNA expression data first were normalized to those for a reference RNA, RNU6B, and expression levels relative to those of ebv-miR-BART19-5p were calculated next.
To quantify mRNAs encoding apoptosis-related proteins (Bax, Bid, Bim, and Bmf), total RNA was reverse transcribed using the SuperScript II first-strand synthesis system (Life Technologies) and random primers. Quantitative RT-PCR was performed using TaqMan Gene Expression assays (Life Technologies) according to the manufacturer's protocol. Data were normalized to the expression level of mRNA encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Quantitative RT-PCR analyses were performed using the 7300 real-time PCR system (Life Technologies).
In silico prediction of cellular targets of viral miRNAs.
The top 10 viral miRNAs (by prevalence) expressed in EBV-associated gastric carcinoma tissues were selected, and seed sequences were obtained from miRBase. These sequences were input to TargetScanHuman Custom 5.2 (http://www.targetscan.org/vert_50/seedmatch.html) to identify potential target genes. To identify pathways possibly dysregulated by EBV miRNAs, listed genes were uploaded to the Functional Annotation Tool of DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov) (27, 28). An enrichment was considered to be significant when the modified Fisher P value was <0.05. Potential target genes related to apoptosis identified by TargetScanHuman Custom 5.2 also were confirmed using two different programs, TargetRank (http://genes.mit.edu/targetrank) and RepTar (http://reptar.ekmd.huji.ac.il).
TUNEL.
The frequency of apoptosis was evaluated in 54 surgically resected specimens using the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) method according to the manufacturer's protocol (ApopTag peroxidase in situ apoptosis detection kit; EMD Millipore, Billerica, MA). The proportions of apoptotic cells were calculated by counting stained cells among all tumor cells. Two pathologists (A. Shinozaki-Ushiku and T. Hibiya) independently evaluated all specimens.
Western blotting.
Whole-cell extracts were dissolved in lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1.0% [vol/vol] Triton X-100, 1.0% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, and 1 mM phenylmethylsulfonyl fluoride [PMSF], with a protease inhibitor cocktail), and protein samples (10 μg) were loaded onto and separated on 15% (wt/vol) SDS polyacrylamide gels. After transfer to polyvinylidene difluoride (PVDF) membranes, proteins were probed using the following primary antibodies: anti-human Bax antibody (clone D2D3; 1:1,000 dilution; Cell Signaling, Danvers, MA); anti-human Bid (clone ab2388; 1:500; Abcam, Cambridge, United Kingdom); anti-human Bim (clone C34C5; 1:1,000; Cell Signaling); anti-human Bmf (clone G81; 1:1,000; Cell Signaling); anti-cleaved caspase-3 (clone asp175; 9661; 1:1000; Cell Signaling); or anti-actin (clone A5441-2ML; 1:10,000; Sigma-Aldrich). Probing with secondary antibodies followed; these were anti-rabbit-horseradish peroxidase (HRP; 1:10,000; Jackson, West Grove, PA) and anti-mouse-HRP (1:10,000; Jackson). Stained proteins were visualized using the ImmunoStar LD system (Wako, Osaka, Japan) according to the manufacturer's protocol. Signal intensity was quantified using the LAS-3000 system (Fujifilm Corp., Tokyo, Japan) and Multi Gauge, version 3.0 (Fujifilm).
Luciferase reporter assay.
To evaluate the effect of ebv-miR-BATR4-5p on the expression of the putative target gene, BID, a luciferase reporter assay was performed. Six cell lines, MKN1, MKN1-EBV, NUGC3, NUGC3-EBV, AGS, and AGS-EBV, were seeded in 96-well plates (2.0 × 104 cells/well) and were transfected with the pEZX-MT01 vector containing the 3′ untranslated region (UTR) fragment of BID (GeneCopoeia, Rockville, MD) using ViaFect transfection reagent (Promega, Madison, WI). Mutations to putative binding sites of ebv-miR-BART4-5p were introduced using a QuikChange lightning site-directed mutagenesis kit (Agilent, Santa Clara, CA), and this mutant was transfected into cell lines similarly. After 24 h, ebv-miR-BART4-5p mimic or negative control was transfected into MKN1, NUGC3, and AGS cells using Lipofectamine RNAiMax transfection reagent (Life Technologies). Luciferase activity was measured 24 h posttransfection using a Dual-Glo luciferase assay system (Promega) and the EnSpire plate reader (PerkinElmer, Waltham, MA) according to the manufacturer's protocol.
Transfection with inhibitors and mimics of miRNA.
EBV-negative cells (MKN1, NUGC3, and AGS) or EBV-infected cells (MKN1-EBV, NUGC3-EBV, and AGS-EBV) were seeded at 2 × 105 cells per well into 6-well plates and transfected with miRNA inhibitors, miRNA mimics, or negative controls (Life Technologies) using the RNAiMAX transfection agent (Life Technologies). The final concentrations of miRNA inhibitors and mimics were 30 nM and 3 nM, respectively. Total RNA was collected for assay at 24 h and 48 h posttransfection.
Induction of apoptosis by serum deprivation.
EBV-negative cell lines MKN1, NUGC3, and AGS were cultured in serum-free RPMI 1640 for 48 h in tissue culture slides and 6-well plates. After fixation with 4% paraformaldehyde and staining with hematoxylin, apoptotic cells were counted and the proportions of these cells among viable cells were calculated. Proteins also were extracted, and Western blotting using anti-cleaved caspase-3 antibody was performed.
Statistical analysis.
Statistical analysis was performed using Student's t test. A comparative difference was considered significant when the P value was <0.05.
RESULTS
Viral miRNA profiling of EBV-associated gastric carcinoma tissues using quantitative RT-PCR.
The clinicopathological features of EBV-associated gastric carcinoma cases examined are summarized in Table 1. EBV-associated gastric carcinoma tended to arise in the upper to middle region of the stomach in older patients and exhibited a striking male predominance. The 5-year survival rate of our patients was 78%. These findings are consistent with those of previous reports (4). Tumor tissue was extracted from formalin-fixed and paraffin-embedded tissues from each case and subjected to quantitative RT-PCR analysis. Figure 1A shows the relative expression levels of viral miRNAs normalized to that of ebv-miR-BART19-5p in each of 10 EBV-associated gastric carcinoma samples. Of 44 known viral miRNAs, 40 were expressed at different levels in the tested samples. However, no miRNA of the BHRF cluster was detected. These results are consistent with those of previous reports to the effect that BHRF miRNAs were not expressed in either NPC or DLBCL samples (15–18). No viral miRNA was detected in normal gastric tissue samples obtained from the same patients (data not shown). EBER-ISH confirmed that EBV was present only in tumor cells, not in lymphocytes infiltrating the tumor or the background normal mucosa. Therefore, the viral miRNAs are considered to originate from tumor cells, not lymphocytes.
TABLE 1.
Clinicopathological features of EBV-associated gastric carcinoma cases
| Parameter | Valuea |
|---|---|
| Age (yr; means ± SD) | 70.1 ± 9.5 (range, 56–90) |
| Male/female ratio | 9:1 |
| Region (n) | Upper (5), middle (4), lower (1) |
| Size (cm; means ± SD) | 7.8 ± 4.8 (range, 2.1–18.0) |
| Stage (n) | IA (1), IB (4), IIA (1), IIB (2), IV (2) |
| No. with lymph node metastasis (present/absent) | 6/4 |
| No. with distant metastasis (present/absent) | 2/8 |
| Outcome (n) | NED (5), AWD (1), DOD (1), DOO (2), LFU (1) |
| 5-yr survival rate | 78% (7/9) |
NED, alive with no evidence of disease; AWD, alive with disease; DOD, died of the disease; DOO, died of other causes; LFU, lost to follow-up.
FIG 1.
Comprehensive profiling of EBV miRNAs expressed in EBV-associated gastric carcinoma. (A) Quantitative RT-PCR analysis of EBV miRNA expression levels in samples from 10 surgically resected EBV-associated gastric carcinoma patients. Relative expression levels (target EBV miRNA levels/ebv-miR-BART19-5p level) are shown. (B) Comparison of the average miRNA profiles of surgically resected samples, KT tumors inoculated into mice, and SNU719 cells cultured in vitro. Relative expression levels (target EBV miRNA levels/ebv-miR-BART19-5p level) are shown. (C) Quantitative RT-PCR analysis of expression levels of ebv-miR-BART7-3p and ebv-miR-BART22 in 10 gastric carcinomas, 5 nasopharyngeal carcinomas, and 5 EBV-positive diffuse large B-cell lymphomas. Relative expression levels (ebv-miR-BART7-3p level/ebv-miR-BART22 level) are shown. (D) Quantitative RT-PCR analysis of the levels of the top 10 viral miRNAs expressed in EBV-negative and -positive cell lines. Relative expression levels (target EBV miRNA levels/ebv-miR-BART15 level) are shown. GC, gastric carcinoma; NPC, nasopharyngeal carcinoma; DLBCL, diffuse large B-cell lymphoma; n.s., not statistically significant (Student's t test).
Figure 1B compares the average miRNA expression levels in the 10 cases to those in KT tumors (inoculated into and maintained in SCID mice) and SNU719 (a cell line derived from EBV-associated gastric carcinoma). The viral miRNA expression profiles were similar in clinical samples, KT tumors and SNU719 cells, but the expression levels of miRNAs in BART subcluster 2 were relatively low in SNU719 cells compared to those of clinical samples and KT tumors. Of all miRNAs detected, ebv-miR-BART7-3p exhibited the highest level of expression, followed by ebv-miR-BART9-3p, ebv-miR-BART1-3p, ebv-miR-BART5-5p, ebv-miR-BART14-3p, ebv-miR-BART10-3p, ebv-miR-BART4-5p, ebv-miR-BART1-5p, ebv-miR-BART2-5p, and ebv-miR-BART15. ebv-miR-BART2-3p, ebv-miR-BART7-5p, ebv-miR-BART9-5p, ebv-miR-BART10-5p, ebv-miR-BART14-5p, ebv-miR-BART18-3p, ebv-miR-BART20-5p, and ebv-miR-BART20-3p were expressed at extremely low levels. In contrast to the previous reports, which demonstrated high expression of ebv-miR-BART22 in NPC and lymphoma, the expression of ebv-miR-BART22 was low in our gastric carcinoma tissues. Therefore, the expression of ebv-miR-BART7-3p and ebv-miR-BART22 additionally were measured in 5 NPC cases and 5 EBV-positive DLBCL cases. The expression levels of ebv-miR-BART7-3p and were ebv-miR-BART22 first were normalized to those of RNU6B, and the relative expression levels (ebv-miR-BART7-3p/ebv-miR-BART22) are shown in Fig. 1C. The expression of ebv-miR-BART22 was low in NPC and DLBCL, similar to the case for gastric carcinoma.
EBV miRNA expression in cell lines.
We next measured the expression levels of the top 10 viral miRNAs in cell line models of EBV-associated gastric carcinoma (Fig. 1D). The relative expression level of each miRNA first was normalized to that of RNU6B and, second, to that of ebv-miR-BART15. All 10 viral miRNAs were expressed in EBV-infected MKN1, NUGC3, and AGS cells, similar to the results obtained from clinical samples. No original cell line (i.e., not EBV infected) expressed EBV miRNAs. Thus, in vitro infection of gastric carcinoma cell lines with EBV induced viral miRNA expression, and such cells can be used as models of EBV-associated gastric carcinoma.
Prediction of the targets of viral miRNAs in EBV-associated gastric carcinoma.
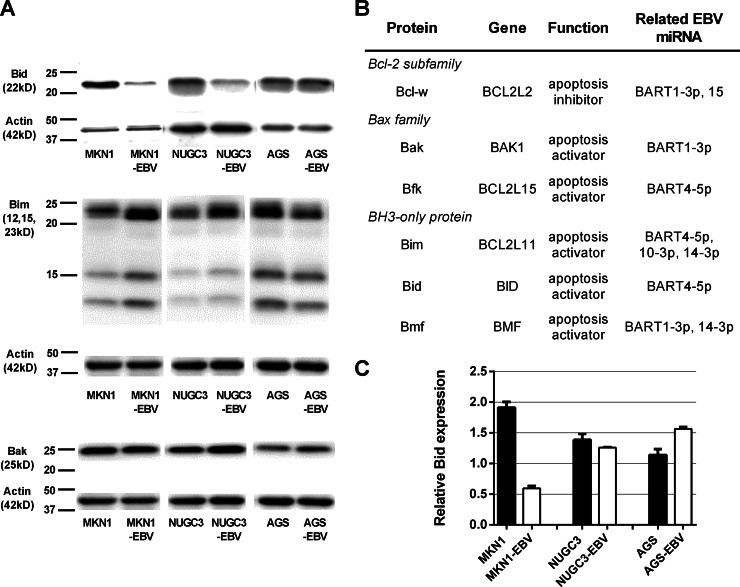
The in silico prediction of target genes was performed using TargetScanHuman Custom 5.2. A total of 2,564 genes were predicted to be targeted by the top 10 viral miRNAs expressed in EBV-associated gastric carcinoma. The list was uploaded to the Functional Annotation Tool of DAVID Bioinformatics Resources 6.7, and analysis showed that 34 pathways (24 KEGG pathways, 8 Reactome pathways, and 2 Biocarta pathways) potentially were targeted (Table 2). Cancer pathways of the KEGG database contained 37 potentially targeted genes, including genes associated with oncogenesis. These were composed of oncogenes (e.g., KRAS), tumor suppressor genes (e.g., APC and PTEN), signal transduction genes (e.g., GRB2, STAT5B, SOS1, PDGFRA, WNT4, and WNT5A), apoptosis-related genes (e.g., BID and CASP3), and others. Cell-to-cell adhesion-associated pathways also were targeted by EBV miRNAs, including focal adhesion pathways (31 genes), tight junction pathways (16 genes), and adherence junction pathways (14 genes). Interestingly, apoptosis-related pathways were listed in all three databases as being targeted by EBV miRNAs. Recently, several studies have investigated the role played by EBV miRNA in inhibition of apoptosis, especially in terms of the activities of the Bcl-2 family proteins, including Bim and PUMA (29, 30). We found that six genes of the Bcl-2 family potentially were targeted by EBV miRNAs, and the findings were confirmed by using different target prediction programs, TargetRank and RepTar. These target genes were BAK1, BCL2L15, BCL2L2, BCL2L11, BID, and BMF, encoding the apoptosis-associated proteins Bak, Bfk, Bcl-w, Bim, Bid, and Bmf, respectively. Bak and Bfk are members of the Bax subfamily, whereas Bim, Bid, and Bmf are BH3-only proteins. These proteins interact with Bcl-2 and induce apoptosis. Bcl-w belongs to the Bcl-2 subfamily and inhibits apoptosis.
TABLE 2.
Functional clustering analysis of EBV miRNA target genes
| Database | Related pathway(s) | Gene count | P value |
|---|---|---|---|
| KEGG | Pathways in cancer | 37 | 0.0009 |
| KEGG | Focal adhesion | 31 | 0.0000 |
| KEGG | Regulation of actin cytoskeleton | 28 | 0.0005 |
| KEGG | Wnt signaling pathway | 25 | 0.0000 |
| KEGG | Ubiquitin mediated proteolysis | 20 | 0.0011 |
| KEGG | Axon guidance | 17 | 0.0080 |
| KEGG | Viral myocarditis | 16 | 0.0000 |
| KEGG | T cell receptor signaling pathway | 16 | 0.0035 |
| KEGG | Tight junction | 16 | 0.0240 |
| KEGG | Neurotrophin signaling pathway | 15 | 0.0270 |
| KEGG | Adherens junction | 14 | 0.0011 |
| KEGG | ECM-receptor interaction | 14 | 0.0025 |
| KEGG | ErbB signaling pathway | 13 | 0.0092 |
| KEGG | Dilated cardiomyopathy | 13 | 0.0140 |
| KEGG | Colorectal cancer | 12 | 0.0180 |
| KEGG | Hypertrophic cardiomyopathy | 12 | 0.0190 |
| KEGG | Fc gamma R-mediated phagocytosis | 12 | 0.0400 |
| KEGG | Pathogenic Escherichia coli infection | 11 | 0.0030 |
| KEGG | p53 signaling pathway | 11 | 0.0110 |
| KEGG | Arrhythmogenic right ventricular cardiomyopathy | 11 | 0.0230 |
| KEGG | Endometrial cancer | 10 | 0.0053 |
| KEGG | Acute myeloid leukemia | 10 | 0.0110 |
| KEGG | Dorsoventral axis formation | 8 | 0.0008 |
| KEGG | SNARE interactions in vesicular transport | 7 | 0.0330 |
| Reactome | REACT_604: hemostasis | 24 | 0.0058 |
| Reactome | REACT_16888: signaling by PDGF | 18 | <0.0001 |
| Reactome | REACT_11061: signaling by NGF | 17 | 0.0380 |
| Reactome | REACT_13552: integrin cell surface interactions | 15 | 0.0001 |
| Reactome | REACT_578: apoptosis | 14 | 0.0280 |
| Reactome | REACT_18266: axon guidance | 13 | <0.0001 |
| Reactome | REACT_11044: signaling by Rho GTPases | 13 | 0.0430 |
| Reactome | REACT_9417: signaling by EGFR | 11 | 0.0002 |
| Biocarta | Apoptotic signaling in response to DNA damage | 7 | 0.0180 |
| Biocarta | D4-GDI signaling pathway | 5 | 0.0340 |
Decreased apoptosis in EBV-associated gastric carcinoma tissue.
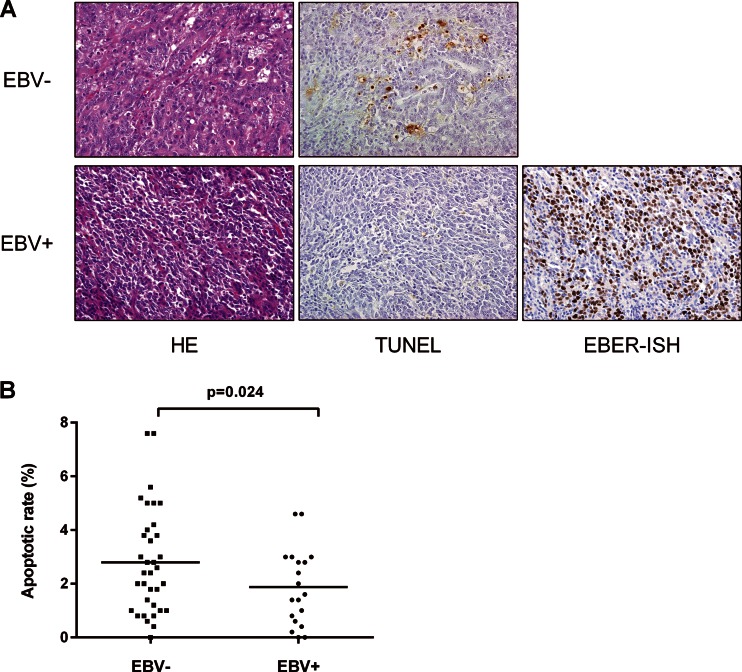
The TUNEL method was used to explore the apoptotic status of 54 surgically resected specimens (Fig. 2). The apoptotic rate was lower in EBV-associated gastric carcinoma tissues (19 cases; average, 1.87%) than in EBV-negative gastric carcinoma tissues (35 cases; average, 3.21%); this difference was statistically significant (P = 0.024 by Student's t test). This result is consistent with that of our previous study showing a decrease in apoptosis in EBV-positive gastric carcinoma cell lines (31).
FIG 2.
Apoptotic status of EBV-positive and -negative gastric carcinoma tissues. (A) Apoptotic cell numbers were quantitated using the TUNEL method and are stained brown. EBER in situ hybridization (EBER-ISH) confirmed that EBV was present in cells of EBV-associated gastric carcinoma. (B) The apoptotic rate was significantly lower in EBV-associated gastric carcinoma tissues than in EBV-negative ones (Student's t test).
Expression of apoptosis-related proteins in vitro.
To explore the antiapoptotic role played by EBV, we used Western blotting to quantify the levels of four proapoptotic proteins, Bak, Bim, Bid, and Bmf, in vitro. Figure 3 shows that the level of Bid expression was significantly less in MKN1-EBV and NUGC3-EBV cells than in control (uninfected) cell lines. AGS-EBV cells exhibited a slight decrease in the level of Bid expression, but this did not attain statistical significance. The expression level of Bim was slightly lower in EBV-AGS than in AGS cells. EBV infection did not affect the expression level of Bak. Expression of Bmf was undetectable in all EBV-positive and -negative cell lines (data not shown). Quantitative RT-PCR analysis showed that Bid mRNA expression was reduced in MKN1-EBV cells, but not in NUGC3-EBV or AGS-EBV cells, compared to the levels of control uninfected cell lines. The results obtained from target prediction programs showed that Bid was a potential target of ebv-miR-BART4-5p; thus, we selected it as a potential regulator of apoptosis in EBV-associated gastric carcinoma and further investigated its role in vitro.
FIG 3.
Expression of apoptosis-associated proteins in EBV-positive and -negative cell lines. (A) Western blotting for Bid, Bim, and Bak. (B) The list of apoptosis-associated proteins targeted by EBV miRNAs expressed at high levels in EBV-associated gastric carcinoma tissue. (C) Quantitative RT-PCR-based analysis of Bid expression levels. Expression of Bid mRNA first was normalized to that of GAPDH and next compared to that of SNU719.
Regulation of Bid protein expression by ebv-miR-BART4-5p.
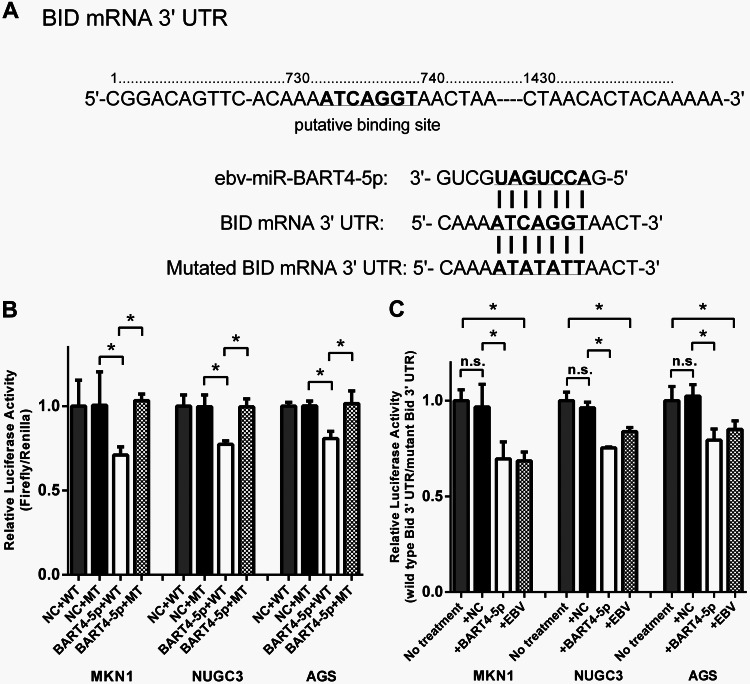
To test the effect of ebv-miR-BART4-5p on the expression of Bid, a luciferase reporter assay was performed in gastric carcinoma cell lines. The luciferase activity was measured by introducing the reporter vector containing the wild-type 3′ UTR of Bid and the vector containing mutations in the putative binding site of ebv-miR-BART4-5p (Fig. 4A). Bid expression was reduced in cells transfected with wild-type vector and the ebv-miR-BART4-5p mimic compared to that of the negative control, while no significant reductions were observed when they were transfected with mutant vector (Fig. 4B). To evaluate the inhibitory effect of endogenously expressed ebv-miR-BART4-5p, EBV-positive and -negative cells also were transfected with wild-type or mutant vectors. Luciferase activities of the wild-type vector were normalized to that of the mutant vector, and the reduction rates of Bid expression were compared (Fig. 4C). EBV-positive cell lines showed reduced Bid expression compared to that of EBV-negative cells. The reduction in rates of Bid expression by the ebv-miR-BART4-5p mimic were almost similar to those observed in EBV-positive cells.
FIG 4.
Effect of ebv-miR-BART4-5p on Bid expression. (A) Sequence of putative binding site of ebv-miR-BART4-5p within 3′ UTR of BID mRNA. Mutations were introduced to the binding site. (B) Relative luciferase activity (Firefly/Renilla) in EBV-negative cells transfected with the wild-type BID 3′ UTR or mutant vectors and ebv-miR-BART4-5p or negative control. (C) Reduction rates of Bid expression in EBV-negative (no treatment) and -positive cells. Luciferase activities of the wild-type reporter are normalized to those of the mutant. NC, negative control; *, P < 0.05 (Student's t test); n.s., not statistically significant.
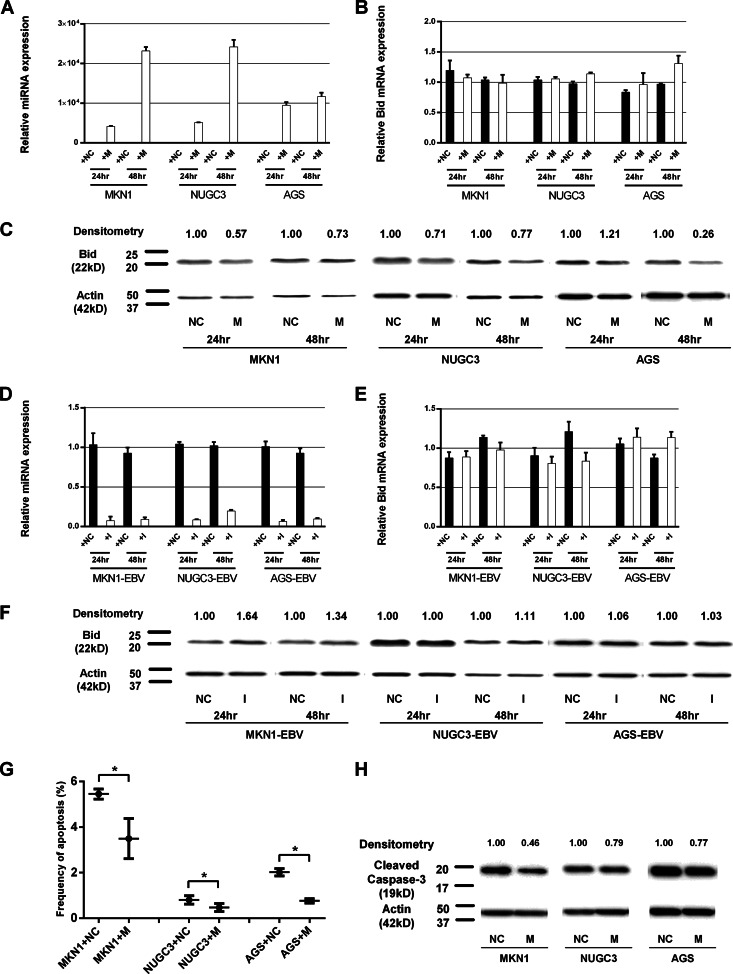
We next performed Western blot analysis to assess the expression of Bid in cell lines transfected with the ebv-miR-BART4-5p mimic (Fig. 5A to C). Transfection of an ebv-miR-BART4-5p mimic into EBV-negative cell lines decreased the level of Bid expression in MKN1 and NUGC3 cells (24 h and 48 h posttransfection) as well as AGS cells (48 h posttransfection).
FIG 5.
Effect of ebv-miR-BART4-5p on regulation of Bid protein expression and apoptosis. The results of transfection of the ebv-miR-BART4-5p mimic (A to C) and ebv-miR-BART4-5p inhibitor (D to F) are shown. (A) Quantitative RT-PCR-based analysis of mature ebv-miR-BART4-5p expression levels in EBV-negative cell lines transfected with ebv-miR-BART4-5p mimic and negative-control miRNA (24 h and 48 h posttransfection). Expression of ebv-miR-BART4-5p first was normalized to that of RNU6B and next compared to that of EBV-positive cell lines. (B) Quantitative RT-PCR-based analysis of Bid expression levels. Expression of Bid mRNA first was normalized to that of GAPDH and next compared to that of EBV-negative cell lines. (C) Western blot analysis of Bid expression levels. Expression levels were densitometrically measured. (D) Quantitative RT-PCR-based analysis of mature ebv-miR-BART4-5p expression levels in EBV-positive cell lines transfected with ebv-miR-BART4-5p inhibitor and negative-control miRNA (24 h and 48 h posttransfection). Expression of ebv-miR-BART4-5p first was normalized to that of RNU6B and next compared to that of EBV-positive cell lines. (E) Quantitative RT-PCR-based analysis of Bid expression levels. Expression of Bid mRNA first was normalized to that of GAPDH and next compared to that of EBV-positive cell lines. (F) Western blot analysis of Bid expression levels. Expression levels were densitometrically measured. (G and H) The antiapoptotic effect of ebv-miR-BART4-5p expression in cells under serum-free culture for 48 h. (G) Frequency of apoptotic cells among viable cells. (H) Western blot analysis of cleaved caspase-3. M, ebv-miR-BART4-5p mimic; NC, negative control; I, ebv-miR-BART4-5p inhibitor; *, P < 0.05 (t test).
In contrast, transfection of an ebv-miR-BART4-5p inhibitor into EBV-positive cells triggered the recovery of Bid expression in MKN1-EBV cells (24 h and 48 h posttransfection) and a slight increase in Bid expression level in NUGC3-EBV cells (48 h posttransfection). The ebv-miR-BART4-5p inhibitor did not affect the Bid level of AGS-EBV cells (Fig. 5D to F). Quantitative RT-PCR analysis showed that the expression level of Bid mRNA was not affected by transfection of the ebv-miR-BART4-5p mimic or the ebv-miR-BART4-5p inhibitor. These results show that ebv-miR-BART4-5p miRNA regulates Bid expression at the posttranscriptional level, although the recovery level of Bid expression by inhibiting ebv-miR-BART4-5p was relatively lower than that expected from the results of its mimic transfection experiment.
Effect of ebv-miR-BART4-5p expression on apoptosis.
To explore the effect of ebv-miR-BART4-5p expression on apoptosis, MKN1, NUGC3, and AGS cells were transfected with ebv-miR-BART4-5p mimic or negative control and cultured for 48 h in serum-deprived RPMI 1640. The frequency of apoptotic cells was significantly higher in MKN1, NUGC3, and AGS cells transfected with the negative control (averages of 5.45%, 0.81%, and 2.02%, respectively) compared to those of cells transfected with the ebv-miR-BART4-5p mimic (averages of 3.49%, 0.47%, and 0.77%, respectively). Similarly, Western blot analysis demonstrated the reduced expression of cleaved caspase-3 levels in all three cell lines transfected with the ebv-miR-BART4-5p mimic, also indicating reduced apoptosis.
DISCUSSION
In the time since Pfeffer et al. first reported that EBV encoded miRNAs, increasing numbers of EBV miRNAs have been sequenced and deposited in databases (12). Several groups have profiled the expression of these viral miRNAs in the tissues or cell lines of various EBV-associated malignancies using different methods, including miRNA microarrays, multiplex RT-PCR, and deep sequencing (15–18, 20, 21, 32–35). The present study is the largest to conduct comprehensive profiling of 44 known EBV miRNAs in EBV-associated gastric carcinoma tissues. The comparison of our data with previous works revealed that some EBV miRNAs (ebv-miR-BART1-3p, ebv-miR-BART1-5p, ebv-miR-BART2-5p, ebv-miR-BART3-3p, ebv-miR-BART4-5p, ebv-miR-BART5-5p, ebv-miR-BART9-3p, ebv-miR-BART10-3p, ebv-miR-BART14-3p, ebv-miR-BART16, ebv-miR-BART19-3p, ebv-miR-BART20-5p, ebv-miR-BART20-3p, and ebv-miR-BART22) commonly were highly expressed in various kinds of tumors. High expression of ebv-miR-BART7-3p was evident in almost all reports, suggesting that ebv-miR-BART7-3p is important in EBV-associated malignancies. The expression levels of ebv-miR-BART1-3p, ebv-miR-BART2-5p, ebv-miR-BART3-3p, ebv-miR-BART4-5p, ebv-miR-BART5-5p, and ebv-miR-BART9-3p were high in epithelial tumors. The expression of EBV miRNA also was associated with latency types; ebv-miR-BART14-3p was expressed at high levels in tumors of latency type I (gastric carcinoma and some forms of lymphoma), whereas ebv-miR-BART6-3p and ebv-miR-BART8-3p were highly expressed in tumors of latency type II (NPC). ebv-miR-BART20-5p and ebv-miR-BART20-3p were highly expressed in cell lines derived from NPC and B-cell lymphomas but not in tissue samples. While expression of ebv-miR-BART22 was low in gastric carcinoma, its expression level was nearly as high as that of ebv-miR-BART7-3p in lymphomas and NPCs in previous studies. However, our additional experiment to compare expression levels of ebv-miR-BART7-3p and ebv-miR-BART22 in NPC and DLBCL showed that relatively low expression of ebv-miR-BART22 was observed in these tumors as well as in gastric carcinoma. The discrepancy may be caused by the different sample types (cell lines or tissues), the differences among individuals, and different detection methods. It also is assumed that the oncogenic roles played by EBV miRNAs in different types of EBV-associated malignancy exhibit both similarities and differences. These findings indicate that caution should be exercised when seeking to apply data from in vitro experiments at a clinical level.
In silico analysis showed that the genes targeted by highly expressed EBV miRNAs were associated with oncogenesis, cell adhesion, signal transduction, and apoptosis, all of which are essential for the development and progression of cancer. As EBV-associated gastric carcinoma exhibited less apoptosis than control EBV-negative gastric carcinoma, we specifically focused on expression of apoptosis-associated proteins and found that Bid was downregulated in EBV-positive cell lines. Bid, also termed the BH3 interacting domain death agonist, belongs to the BH3-only protein of the Bcl-2 family. The protein is a death agonist forming heterodimers with either the agonist Bax or the antagonist Bcl-2, and it mediates mitochondrial damage induced by caspase-8. In silico analysis indicated that Bid was a target of ebv-miR-BART4-5p, and this was confirmed by luciferase assay. Transfection of ebv-miR-BART4-5p mimic caused Bid suppression, leading to reduced apoptosis. These results suggest that reduction of apoptosis in EBV-associated gastric carcinoma is attributable to the expression of ebv-miR-BART4-5p, which in turn controls the level of the proapoptotic protein Bid.
Another finding is that Bid suppression was more significant in EBV-infected cells than in cells transfected with ebv-miR-BART4-5p mimic alone. Furthermore, recovery of Bid expression by ebv-miR-BART4-5p inhibitor was partial in EBV-positive cell lines. From these results, it is assumed that there are some other factors that regulate Bid expression. EBV latent genes may be the candidates, since they are known to play roles in regulating cell proliferation and apoptosis, although our preliminary experiment failed to show direct effects of these genes on Bid expression (data not shown). In addition, considering the fact that transfection of ebv-miR-BART4-5p mimic alone reduced apoptosis, it is speculated that ebv-miR-BART4-5p also targets other apoptosis-related genes as well as Bid. For example, Marquitz et al. demonstrated that the expression of the proapoptotic protein Bim was downregulated by several EBV miRNAs located in ebv-miR-BART cluster 1, to which ebv-miR-BART4-5p belongs (29). Several miRNAs in cluster 1 have been demonstrated to show both antiapoptotic and proapoptotic effects, and it is possible that several EBV miRNAs interact to cooperatively regulate apoptosis via different apoptosis-related proteins (30, 36, 37). Further work is necessary to clarify the complex mechanism by which EBV miRNAs regulate apoptosis-related proteins and the potential relationship with latent genes and tumor subtypes.
Viral miRNAs may be useful targets in cancer detection and therapy. Some EBV miRNAs are secreted and may be detected in plasma of nasopharyngeal carcinoma patients (38–40). Thus, EBV miRNA can be used as a biomarker predicting tumor progression and therapeutic response. As some EBV miRNAs inhibit apoptosis, specific targeting of such miRNAs and their precursors may be therapeutically effective and are associated with fewer side effects than is the use of conventional cytotoxic anti-cancer drugs.
In conclusion, we present the comprehensive profile of the expression of 44 known EBV miRNAs in clinical samples from EBV-associated gastric carcinoma patients. We found that, of several highly expressed EBV miRNAs, ebv-miR-BART4-5p plays a partial role in suppressing proapoptotic protein Bid, leading to reduced apoptosis. Although further investigation is necessary to elucidate other possible factors related to the antiapoptotic effect caused by EBV, our findings provide novel insights in the roles played by EBV miRNAs in gastric carcinogenesis and identify future potential therapeutic targets.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (JSPS) (grant number 25860262) to A.S-U. and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (grant number 25114702) to M.F.
We have no conflicts of interest to report.
We thank Kei Sakuma and Kimiko Takeshita (The University of Tokyo) for their technical assistance.
REFERENCES
- 1.Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 2.Fukayama M, Ibuka T, Hayashi Y, Ooba T, Koike M, Mizutani S. 1993. Epstein-Barr virus in pyothorax-associated pleural lymphoma. Am J Pathol 143:1044–1049. [PMC free article] [PubMed] [Google Scholar]
- 3.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 4.Fukayama M, Hino R, Uozaki H. 2008. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci 99:1726–1733. doi: 10.1111/j.1349-7006.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young LS, Murray PG. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, Uozaki H, Takada K, Fukayama M. 2010. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res 70:4719–4727. doi: 10.1158/0008-5472.CAN-09-4620. [DOI] [PubMed] [Google Scholar]
- 8.Korpal M, Lee ES, Hu G, Kang Y. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 10.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 11.Park SM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Barth S, Meister G, Grasser FA. 2011. EBV-encoded miRNAs. Biochim Biophys Acta 1809:631–640. doi: 10.1016/j.bbagrm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ Jr. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res 68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. 2009. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol 83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, Tinguely M, Faggioni A, Trivedi P, Meister G, Renner C, Grasser FA. 2011. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res 39:1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong AM, Kong KL, Tsang JW, Kwong DL, Guan XY. 2012. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 18.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DN, Chae HS, Oh ST, Kang JH, Park CH, Park WS, Takada K, Lee JM, Lee WK, Lee SK. 2007. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Virol 81:1033–1036. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lung RW, Tong JH, To KF. 2013. Emerging roles of small Epstein-Barr virus derived non-coding RNAs in epithelial malignancy. Int J Mol Sci 14:17378–17409. doi: 10.3390/ijms140917378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, Shapiro M, Thorley-Lawson DA. 2011. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog 7:e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DN, Seo MK, Choi H, Kim SY, Shin HJ, Yoon AR, Tao Q, Rha SY, Lee SK. 2013. Characterization of naturally Epstein-Barr virus-infected gastric carcinoma cell line YCCEL1. J Gen Virol 94:497–506. doi: 10.1099/vir.0.045237-0. [DOI] [PubMed] [Google Scholar]
- 23.Marquitz AR, Mathur A, Shair KH, Raab-Traub N. 2012. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc Natl Acad Sci U S A 109:9593–9598. doi: 10.1073/pnas.1202910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai S, Nishikawa J, Takada K. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol 72:4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukayama M, Hayashi Y, Iwasaki Y, Chong J, Ooba T, Takizawa T, Koike M, Mizutani S, Miyaki M, Hirai K. 1994. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab Investig 71:73–81. [PubMed] [Google Scholar]
- 26.Iwasaki Y, Chong JM, Hayashi Y, Ikeno R, Arai K, Kitamura M, Koike M, Hirai K, Fukayama M. 1998. Establishment and characterization of a human Epstein-Barr virus-associated gastric carcinoma in SCID mice. J Virol 72:8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang Da W, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. 2011. The Epstein-Barr virus BART microRNAs target the pro-apoptotic protein Bim. Virology 412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hino R, Uozaki H, Inoue Y, Shintani Y, Ushiku T, Sakatani T, Takada K, Fukayama M. 2008. Survival advantage of EBV-associated gastric carcinoma: survivin up-regulation by viral latent membrane protein 2A. Cancer Res 68:1427–1435. doi: 10.1158/0008-5472.CAN-07-3027. [DOI] [PubMed] [Google Scholar]
- 32.Nourse JP, Crooks P, Keane C, Nguyen-Van D, Mujaj S, Ross N, Jones K, Vari F, Han E, Trappe R, Fink S, Gandhi MK. 2012. Expression profiling of Epstein-Barr virus-encoded microRNAs from paraffin-embedded formalin-fixed primary Epstein-Barr virus-positive B-cell lymphoma samples. J Virol Methods 184:46–54. doi: 10.1016/j.jviromet.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. 2012. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J 31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol 83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motsch N, Alles J, Imig J, Zhu J, Barth S, Reineke T, Tinguely M, Cogliatti S, Dueck A, Meister G, Renner C, Grasser FA. 2012. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS One 7:e42193. doi: 10.1371/journal.pone.0042193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vereide DT, Seto E, Chiu YF, Hayes M, Tagawa T, Grundhoff A, Hammerschmidt W, Sugden B. 2014. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene 33:1258–1264. doi: 10.1038/onc.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi H, Lee H, Kim SR, Gho YS, Lee SK. 2013. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol 87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan JY, Gao W, Ho WK, Wei WI, Wong TS. 2012. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res 32:3201–3210. [PubMed] [Google Scholar]
- 39.Gourzones C, Gelin A, Bombik I, Klibi J, Verillaud B, Guigay J, Lang P, Temam S, Schneider V, Amiel C, Baconnais S, Jimenez AS, Busson P. 2010. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol J 7:271. doi: 10.1186/1743-422X-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gourzones C, Ferrand FR, Amiel C, Verillaud B, Barat A, Guerin M, Gattolliat CH, Gelin A, Klibi J, Chaaben AB, Schneider V, Guemira F, Guigay J, Lang P, Jimenez-Pailhes AS, Busson P. 2013. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients–evidence of nonexosomal transport. Virol J 10:119. doi: 10.1186/1743-422X-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]