ABSTRACT
Although a large number of immune epitopes have been identified in the influenza A virus (IAV) hemagglutinin (HA) protein using various experimental systems, it is unclear which are involved in protective immunity to natural infection in humans. We developed a data mining approach analyzing natural H1N1 human isolates to identify HA protein regions that may be targeted by the human immune system and can predict the evolution of IAV. We identified 16 amino acid sites experiencing diversifying selection during the evolution of prepandemic seasonal H1N1 strains and found that 11 sites were located in experimentally determined B-cell/antibody (Ab) epitopes, including three distinct neutralizing Caton epitopes: Sa, Sb, and Ca2 [A. J. Caton, G. G. Brownlee, J. W. Yewdell, and W. Gerhard, Cell 31:417–427, 1982, http://dx.doi.org/10.1016/0092-8674(82)90135-0]. We predicted that these diversified epitope regions would be the targets of mutation as the 2009 H1N1 pandemic (pH1N1) lineage evolves in response to the development of population-level protective immunity in humans. Using a chi-squared goodness-of-fit test, we identified 10 amino acid sites that significantly differed between the pH1N1 isolates and isolates from the recent 2012-2013 and 2013-2014 influenza seasons. Three of these sites were located in the same diversified B-cell/Ab epitope regions as identified in the analysis of prepandemic sequences, including Sa and Sb. As predicted, hemagglutination inhibition (HI) assays using human sera from subjects vaccinated with the initial pH1N1 isolate demonstrated reduced reactivity against 2013-2014 isolates. Taken together, these results suggest that diversifying selection analysis can identify key immune epitopes responsible for protective immunity to influenza virus in humans and thereby predict virus evolution.
IMPORTANCE The WHO estimates that approximately 5 to 10% of adults and 20 to 30% of children in the world are infected by influenza virus each year. While an adaptive immune response helps eliminate the virus following acute infection, the virus rapidly evolves to evade the established protective memory immune response, thus allowing for the regular seasonal cycles of influenza virus infection. The analytical approach described here, which combines an analysis of diversifying selection with an integration of immune epitope data, has allowed us to identify antigenic regions that contribute to protective immunity and are therefore the key targets of immune evasion by the virus. This information can be used to determine when sequence variations in seasonal influenza virus strains have affected regions responsible for protective immunity in order to decide when new vaccine formulations are warranted.
INTRODUCTION
Influenza A virus (IAV) is a negative-sense single-stranded RNA virus within the Orthomyxoviridae family. The two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), carry the major antigenic determinants of the virus and are the primary targets of the humoral immune response in humans (1). H1N1 and H3N2 are the main influenza A virus subtypes that have been circulating within the human population in the recent past. Since the first documented case of H1N1 in 1918, the virus has had a major global public health impact. According to the WHO, approximately 5 to 10% of adults and 20 to 30% of children are infected by influenza every year. Of those, 3 million to 5 million infected individuals experience severe illness resulting in between 250,000 and 500,000 deaths annually (http://www.who.int/mediacentre/factsheets/fs211/en/).
From year to year, gradual mutations accumulate in the HA gene that produce immunologically distinct virus strains through a process known as antigenic drift (2). These new drift variants allow the virus to escape preexisting immunity and cause individuals who had previously been infected or vaccinated to again become susceptible to infection. The HA protein is structurally plastic and accumulates mutations in antigenic sites recognized by neutralizing antibodies (Abs) to evade the host immune system while still maintaining its function as the primary receptor binding protein (3).
Several groups have used selection pressure analysis to characterize the evolution of H1N1. Studies of pandemic H1N1 isolates in specific geographic regions (United Kingdom, Italy, Thailand, and Japan) used selection pressure analysis to quantify the rates of evolution and adaptation during the pandemic waves and identify the dominant selected residue during each wave (4–6). Other studies used selection pressure analysis to distinguish the pathogenic profiles of viruses by comparing selected sites in the seasonal versus the pandemic H1N1 viruses (7, 8). While these studies characterized the previous evolution of H1N1, they did not attempt to address the future trajectory of H1N1 evolution.
In this report, we describe a data mining approach designed to identify regions of the HA protein that are targets of a protective immune response in humans. Our approach selects relevant regions by filtering for experimentally determined H1 B-cell/Ab epitopes that are located in regions experiencing diversifying selection. This approach is based on the expectation that if a B-cell/Ab epitope is important for protective immunity, then the epitope will be located in a region that tends to experience evolutionary changes in the amino acid sequence in order to escape immune recognition. We refer to these regions as diversified B-cell/Ab epitopes. We show that these diversified epitope regions can predict the future evolution of IAV.
MATERIALS AND METHODS
H1 HA B-cell/Ab epitope data.
The Influenza Research Database (IRD) (http://www.fludb.org/) Sequence Feature Variant Types component was queried for positive H1 HA B-cell/Ab epitopes from all hosts (9, 10). In IRD, positive B-cell epitopes are defined as regions of the antigen that were found to be positive in at least one experiment curated by the Immune Epitope Database (IEDB) in which the region either activated a B cell through the B-cell antigen receptor (BCR) or bound to soluble antibody with an affinity or avidity exceeding a certain threshold (see http://www.fludb.org/brcDocs/documents/Epitope_Filter_Tutorial.pdf). This search returned 87 epitopes (data as of 7 April 2014). We added the 5 Caton antigenic regions (11) that were originally defined by grouping mutants of the influenza virus A/PR/8/34 virus based on their antigenic reactivities to panels of neutralizing monoclonal antibodies (11, 12) but which were absent from IEDB. Epitopes that were found using screening peptides that did not elicit an immune response were removed. Additional discontinuous epitopes were identified in the Immune Epitope Database (http://www.iedb.org) (13) and added to the list. IEDB is a free resource funded by NIAID that focuses on the dissemination of the immune epitope information. In total, 72 experimentally determined B-cell/Ab epitopes were identified that covered 304 of 566 amino acid positions in HA.
Prepandemic H1 HA sequence data preparation.
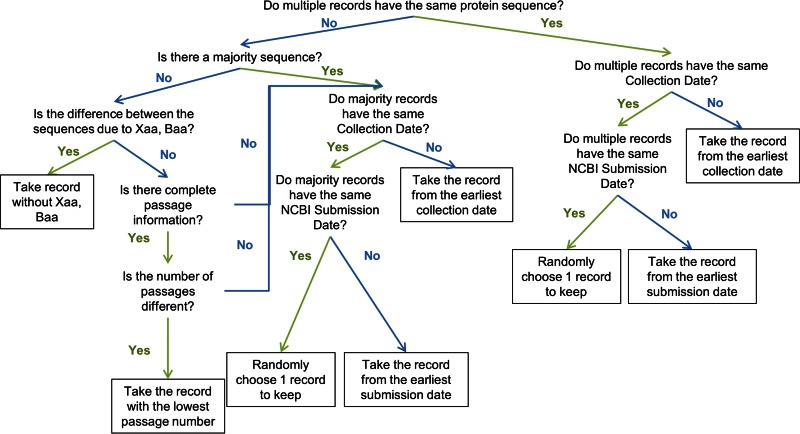
Using the Influenza Research Database (IRD) (http://www.fludb.org), we searched for all human H1N1 HA protein sequences with complete segments that were not pandemic-like, which returned 2,149 sequences (as of 15 April 2014). Sequences with large deletions in comparison with the majority of the other sequences were removed in order to exclude erroneous and incomplete sequences that would disrupt subsequent analyses. The prepandemic H1 sequence collection was further filtered to ensure that there was only one sequence record per strain (see Fig. 1 for the multiple-sequence record curation workflow). A total of 2,003 prepandemic H1 HA protein sequences from unique isolates were used for further analysis.
FIG 1.
Curation workflow to filter sequences from strains with multiple sequence records. The goal of the curation workflow was to select single sequence records that likely represent natural isolates. If a strain has multiple identical sequence records available, the sequence that was collected or submitted to GenBank with the earliest date was selected. If a strain has multiple heterogeneous sequence records, then the earliest sequence record that is identical to the majority sequence was selected. If there is no majority sequence, then the sequence record with the lowest passage number was selected.
The nucleotide sequences associated with these protein sequences were retrieved and aligned by codon using the HCV Sequence Database Codon Alignment tool (http://hcv.lanl.gov/content/sequence/CodonAlign/codonalign.html).
A phylogenetic tree was generated from these prepandemic sequences using the RAXML algorithm with the GTR model of evolution in IRD (http://www.fludb.org/brc/tree.spg?method=ShowCleanInputPage&decorator=influenza). The tree was rooted using the earliest H1N1 strain (A/South Carolina/1/18).
Selection pressure analysis.
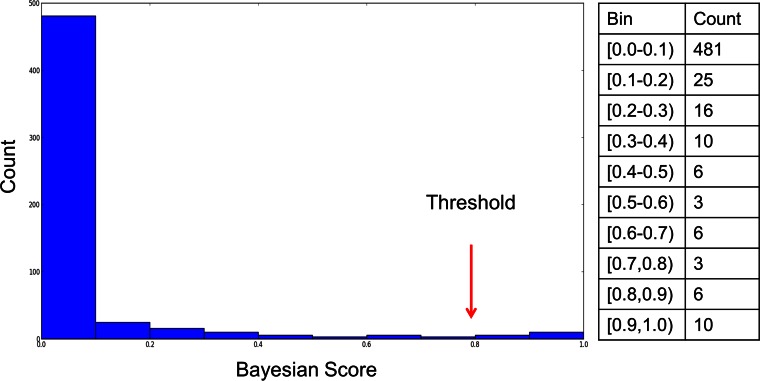
The fast unconstrained Bayesian approximation (FUBAR) method (14), as implemented in the open-source Hypothesis testing using Phylogenies (HyPhy) software (15), was used to identify sites that were experiencing diversifying selection as the prepandemic H1N1 strains evolved in humans, based on the assumption that host protective immunity would be the main driver of diversifying selection of H1N1 influenza viruses as they circulate through the human population. FUBAR takes the prepandemic H1 HA codon aligned nucleotide sequences and the tree topology associated with these sequences as input and uses rates of nonsynonymous (dN) and synonymous (dS) substitutions to test if a site is experiencing diversifying selection. A site is considered to be under diversifying selection if the probability that the observed ratio of nonsynonymous to synonymous mutations being greater than the expected ratio exceeds a threshold of 80%. This threshold was selected based on the distribution of probabilities that a site is experiencing diversifying selection (Fig. 2).
FIG 2.
Threshold to determine which sites are experiencing diversifying selection. For each HA amino acid site, FUBAR returns the probability that the observed ratio of nonsynonymous to synonymous mutations is greater than the expected ratio. A histogram of the number of amino acid positions (count) for a given range of probabilities is plotted. A conservative threshold of 0.8 was chosen within the minimum range observed between the two major modes in order to reduce the number of false-positive calls.
Meta-CATS analysis of pH1N1.
The accuracy of the FUBAR method is dependent on having a large number of diverse sequences to calculate posterior probabilities. Since the pH1N1 lineage has only been evolving in the human population for a short period, calculating diversifying selection for pH1N1 using FUBAR was not feasible. Therefore, in order to identify amino acid positions that are significantly different between the early outbreak and the recent pandemic strains, the metadata-driven Comparative Analysis for Sequences (meta-CATS) tool in IRD was used (16). Meta-CATS performs a chi-squared goodness-of-fit test to identify sites with significant sequence variation between groups of sequences following a multiple-sequence alignment. In this analysis, two groups of pH1N1 HA sequences (early and late) were compared.
Group 1: early 2009 pandemic outbreak sequences.
All pandemic-like human H1N1 HA protein sequences with complete segments were retrieved from the IRD, and the first 45 protein sequences from April 2009 ordered by collection date were selected as representatives of the early outbreak sequences, making sure that each strain provided only a single sequence record (Fig. 1). Forty-five sequences were selected for group 1 in order to maintain a balanced number of sequences when comparing group 1 and group 2 on average.
Group 2: recent (late) pandemic sequences.
All pandemic-like human H1N1 HA protein sequences from complete segments from influenza seasons 2012-2013 and 2013-2014 were retrieved from the IRD. Sequence data were selected from geographic regions that provided at least 10 sequence records in order to produce accurate comparative analysis results, resulting in selections from 6 different states within the United States and from 6 different other countries, as follows: California (USA), 39 protein sequences; Colorado (USA), 19 protein sequences; Florida (USA), 25 protein sequences; Louisiana (USA), 17 protein sequences; New York (USA), 18 protein sequences; Texas (USA), 47 protein sequences; British Columbia (Canada), 31 protein sequences; Quebec (Canada), 37 protein sequences; Czech Republic, 21 protein sequences; Helsinki (Finland), 114 protein sequences; Moscow (Russia), 16 protein sequences; São Paulo (Brazil), 13 protein sequences; and Taiwan, 13 protein sequences.
Multiple meta-CATS analyses were performed where all group 1 sequences consisted of the 45 early 2009 pandemic outbreak sequences and group 2 sequences consisted of the late pandemic sequences from each of the geographic regions separately, giving rise to 13 comparisons with 13 sets of significant sites.
H1 HA numbering.
The amino acid numbering is based on the A/California/04/2009 (H1N1) HA protein sequence (GenBank accession number ACP41105.1). All protein sequences used in this study were aligned against the A/California/04/2009 HA protein sequence in order to calculate the coordinates in reference to this strain. A mapping of selected A/California/04/2009 (H1N1) HA amino acid positions to the H3 coordinate system is provided in Table 1.
TABLE 1.
Amino acid sites with annotations of epitopes, diversifying selection, and postpandemic mutations
| AA site, H1a | AA site, H3b | H1 HA1/HA2 coordinate | H3 HA1/HA2 coordinate | B-cell epitope (IEDB code) | Probability of diversified prepandemic |
P valuec |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| California, USA | New York, USA | Texas, USA | Louisiana, USA | Florida, USA | Colorado, USA | British Columbia, Canada | Czech Republicd | Helsinki, Finlandd | São Paulo, Brazil | Moscow, Russiad | Quebec, Canada | Taiwan | ||||||
| 4 | 4 | Signal peptide | Signal peptide | 0.83 | ||||||||||||||
| 52 | 61 | 35 HA1 | 45 HA1 | 150978 | 0.85 | |||||||||||||
| 111 | 117 | 94 HA1 | 101 HA1 | 172565, 181471 | 0.89 | |||||||||||||
| 114 | 120 | 97 HA1 | 104 HA1 | 172565, 181471 | 1.686E−16 | 2.529E−12 | 4.449E−20 | 4.203E−14 | 4.332E−15 | 1.358E−14 | 2.250E−17 | 4.504E−15 | 6.553E−42 | 1.098E−10 | 7.488E−14 | 1.027E−18 | 1.501E−04 | |
| 142 | 145 | 125 HA1 | 129 HA1 | Sa, Extd Sa, 76963, 164527, 180050, 76962 | 0.85 | |||||||||||||
| 158 | 160 | 141 HA1 | 144 HA1 | 0.91 | ||||||||||||||
| 170 | 172 | 153 HA1 | 156 HA1 | Sb, Extd Sb, 72805, 136074, 179938, 76950 | 0.93 | |||||||||||||
| 172 | 174 | 155 HA1 | 158 HA1 | Sa, Extd Sa, 12284, 72805, 12285, 180309, 164527, 77529, 159269, 194989, 173915, 76950 | 0.93 | |||||||||||||
| 177 | 179 | 160 HA1 | 163 HA1 | Sa, Extd Sa, 12284, 12285, 164527 | 0.99 | |||||||||||||
| 179 | 181 | 162 HA1 | 165 HA1 | Sa, Extd Sa, 12284, 12285, 164527, 180050 | 0.93 | |||||||||||||
| 180 | 182 | 163 HA1 | 166 HA1 | Sa, Extd Sa, 12284, 12285, 133973, 164527, 180050, 190190, 94400 | 1.177E−12 | 2.060E−11 | 4.209E−16 | 4.838E−13 | 1.666E−07 | 1.299E−12 | 6.855E−09 | 1.062E−07 | 1.860E−10 | 1.217E−03 | ||||
| 202 | 204 | 185 HA1 | 188 HA1 | 180309, 159269 | 6.160E−19 | 2.380E−14 | 6.422E−21 | 4.203E−14 | 5.230E−16 | 1.358E−14 | 3.149E−17 | 4.504E−15 | 2.753E−38 | 1.098E−10 | 7.488E−14 | 1.027E−18 | 4.561E−13 | |
| 203 | 205 | 186 HA1 | 189 HA1 | 76960, 180309, 76961, 159269, 179938, 76959, 76962 | 0.96 | |||||||||||||
| 204 | 206 | 187 HA1 | 190 HA1 | 76960, 180309, 76961, 179938 | 1.00 | |||||||||||||
| 220 | 222 | 203 HA1 | 206 HA1 | 127932, 2136 | 6.160E−19 | 2.380E−14 | 6.422E−21 | 4.203E−14 | 5.230E−16 | 1.358E−14 | 2.250E−17 | 4.504E−15 | 2.436E−19 | 4.561E−13 | 7.488E−14 | 1.027E−18 | 4.561E−13 | |
| 239 | 241 | 222 HA1 | 225 HA1 | Ca2, Extd Ca2, 177089, 177084, 177087, 177088, 177121, 159269, 179938, 76965 | 1.00 | |||||||||||||
| 251 | 253 | 234 HA1 | 237 HA1 | 8.998E−03 | 3.146E−02 | 2.380E−05 | 3.230E−05 | 3.617E−13 | 4.100E−33 | 1.030E−11 | 1.331E−04 | 1.501E−04 | ||||||
| 273 | 275 | 256 HA1 | 259 HA1 | 2.957E−11 | 1.410E−09 | 1.049E−15 | 4.838E−13 | 4.662E−07 | 1.299E−12 | 6.855E−09 | 1.062E−07 | 1.860E−10 | ||||||
| 275 | Gap | 258 HA1 | Gap | 0.84 | ||||||||||||||
| 278 | 279 | 261 HA1 | 263 HA1 | 0.95 | ||||||||||||||
| 300 | 301 | 283 HA1 | 285 HA1 | 1.686E−16 | 2.529E−12 | 1.681E−18 | 4.203E−14 | 3.285E−14 | 1.358E−14 | 1.675E−16 | 4.228E−14 | 7.185E−22 | 1.880E−07 | 7.488E−14 | 1.027E−18 | 1.501E−04 | ||
| 391 | 392 | 47 HA2 | 48 HA2 | 180911, 190150 | 6.160E−19 | 2.380E−14 | 6.422E−21 | 4.203E−14 | 5.230E−16 | 1.358E−14 | 2.250E−17 | 4.504E−15 | 1.866E−43 | 4.561E−13 | 9.248E−13 | 1.027E−18 | 4.561E−13 | |
| 468 | 469 | 124 HA2 | 125 HA2 | 0.97 | 6.160E−19 | 2.380E−14 | 6.422E−21 | 4.203E−14 | 6.327E−16 | 1.358E−14 | 2.250E−17 | 4.504E−15 | 6.553E−42 | 1.098E−10 | 9.248E−13 | 1.568E−18 | 4.561E−13 | |
| 516 | 517 | 172 HA2 | 173 HA2 | 181381 | 2.809E−17 | 2.529E−12 | 2.841E−19 | 4.203E−14 | 4.332E−15 | 1.358E−14 | 1.675E−16 | 4.228E−14 | 1.303E−35 | 1.880E−07 | 7.488E−14 | 1.027E−18 | 1.741E−05 | |
| 537 | 538 | 193 HA2 | 194 HA2 | 29690 | 0.85 | |||||||||||||
Amino acid (AA) positions based on the reference sequence A/California/04/2009 (H1N1) (GenBank accession number ACP41105.1).
Amino acid positions based on the reference sequence A/Aichi/2/1968 (H3N2) (GenBank accession number BAF37221.1).
P value based on sequence variation between early versus late pandemic sequences.
Notice that the pairwise comparison between the early strain versus strains from the Czech Republic, Helsinki, and Moscow did not find any significant sequence variation at sites 180 and 273. This is because the majority of strains from these three regions are from the 2012-2013 flu season and the mutations present at sites 180 and 273 became dominant in the 2013-2014 flu season.
Pandemic HA phylogenetic analysis.
A phylogenetic tree using unique human H1N1 pandemic-like HA protein sequences was constructed with the following parameters: algorithm, RaxML (17); bootstrap, none; outgroup, A/New York/3442/2009 (prepandemic strain from 2008-2009 influenza season); and model, GTR (18). The tree was manually ordered by influenza season using the swap branch feature in the IRD Tree Visualization tool; these trees are isomorphic to the original tree and therefore do not affect phylogenetic relatedness quantified in the branch lengths. Ordered trees were highlighted to distinguish amino acid residues at positions identified using the meta-CATS analysis, which identified sites that were significantly different between early 2009 pandemic outbreak isolates and late pandemic isolates.
Natural influenza virus isolation and propagation.
Pandemic H1N1 viruses were isolated from primary swab specimens by passaging twice in Madin-Darby canine kidney (MDCK) cells to avoid selecting for mutations that frequently arise when IAV is isolated in embryonated chicken eggs. Supernatants from passage two (P2) were clarified by centrifugation at 1,800 × g for 10 min at 4°C, aliquoted, and stored as virus stocks at −80°C.
All virus isolates were verified by whole genome sequencing and comparison with reference nucleotide sequences for IAV HA and NA obtained from the NCBI's GenBank. Viral RNAs were amplified from 3 μl of RNA template using a multisegment reverse transcription-PCR (RT-PCR) strategy (19, 20). The amplicons were sequenced using the Ion Torrent PGM (Thermo Fisher Scientific, Waltham, MA).
Recombinant influenza virus rescue.
Recombinant pandemic A/New York/1682/2009 (H1N1), A/Wisconsin/02/2011 (H1N1), A/St. Petersburg/100/2011 (H1N1), and A/California/52/2011 (H1N1) viruses for antigenic testing were generated using gene synthesis (21) and a modified reverse genetics system (19, 20). Briefly, 6:2 reassortant viruses were rescued following transfection with plasmids encoding the 6 internal protein viral RNAs (vRNAs) (PB1, PB2, PA, NP, M, and NS) from A/Puerto Rico/8/1934 (H1N1) and double-stranded linear DNA synthesized to contain the HA and NA genes of the desired pH1N1 viruses. Recombinant viruses are designated with an “r” before the strain name (e.g., rA/New York/1682/2009).
Ethics statement.
The University of Rochester Research Subjects Review Board approved this study protocol, and human experimentation guidelines of the U.S. Department of Health and Human Services and the University of Rochester were followed. Study procedures were in accordance with the ethical standards of the Declaration of Helsinki. All subjects provided written informed consent prior to study participation.
Human antiserum acquisition and HI assay.
Human antisera were generated using a procedure described previously (22). Subjects from two age groups (18 to 32 years and older than 59 years) were enrolled between March and October 2010. Blood samples were obtained before and at days 7, 14, and 28 after administration of an inactivated A/California/07/2009 monovalent vaccine (Novartis, East Hanover, NJ). Younger subjects in the present study all had a prevaccination hemagglutination inhibition (HI) titer of <10, while sera from older subjects ranged from <10 to 80 (Table 2). Subjects with antibody titers exceeding 1,280 HI units on day 14 or 28 to the vaccine strain were selected for use in the HI assays. HI assays were performed as described previously (23) to determine the ability of the selected human antisera to inhibit binding of the IAV isolates to turkey red blood cells (RBCs).
TABLE 2.
Patient specimen metadata
| Participant identifier | Age (yr) | Gendera | Prevaccination titer (day 0)b |
|---|---|---|---|
| 2253 | 61 | F | 80 |
| 1728 | 74 | F | <10 |
| 1551 | 20 | M | <10 |
| 1553 | 67 | F | <10 |
| 1540 | 24 | F | <10 |
| 2364 | 60 | F | 10 |
| 1765 | 26 | M | <10 |
| 1332 | 66 | F | <10 |
| 2419 | 69 | F | 20 |
| 2579 | 78 | F | 40 |
F, female; M, male.
Dilution factor of serum resulting in loss of hemagglutination inhibition before administration of inactivated A/California/07/2009 monovalent vaccine. The postvaccination (day 14 or 28) titer for all participants listed was >1,280.
RESULTS
Predicting targeted B-cell/Ab epitopes.
If a region of a viral protein is a primary target for protective immunity in the infected host, then mutations in that region that result in nonsynonymous amino acid substitutions tend to be positively selected for in order to escape recognition, contributing to a phenomenon termed diversifying selection. This region would show a larger ratio of nonsynonymous (dN) to synonymous (dS) substitutions than would be expected based on random mutation without selection pressure (i.e., neutral evolution). We hypothesized that the identification of sites that are experiencing diversifying selection as the prepandemic H1N1 strains have evolved could be used to identify the immune epitopes that are most relevant for protective immunity in humans, and would also allow us to predict which regions of the HA protein would be most likely to mutate as the 2009 pH1N1 HA proteins evolves during its circulation through the human population.
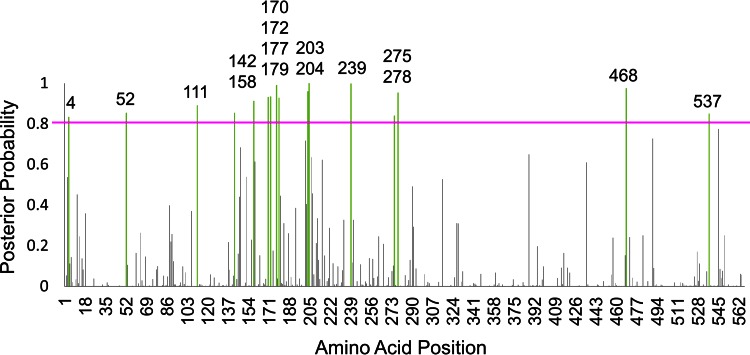
The fast unconstrained Bayesian approximation (FUBAR) method identifies individual sites that experience diversifying selection by estimating and comparing dN and dS rates (14, 15). For each amino acid site, the method calculates and outputs the posterior probability that the observed dN/dS ratio is greater than the expected dN/dS ratio (if the site were experiencing neutral evolution). FUBAR was used to calculate the dN/dS posterior probabilities for each amino acid position in a collection of 2,003 prepandemic H1 HA sequences. An amino acid site is considered to be under diversifying selection if the probability exceeds some threshold. To choose a suitable threshold, the distribution of probabilities that a site experienced diversifying selection was examined (Fig. 2). A conservative threshold of 0.8 was selected to reduce the false-positivity rates based on the assumption that the most strongly selected sites would be the most relevant for our purposes. Sixteen sites were identified as experiencing diversifying selection as the prepandemic H1N1 HA sequences have evolved in humans between the period from 1918 to 1957 and the period from 1977 to 2009: amino acid positions 4, 52, 111, 142, 158, 170, 172, 177, 179, 203, 204, 239, 275, 278, 468, and 537 (Fig. 3).
FIG 3.
Diversifying selection of prepandemic H1N1 strains. The posterior probability that each amino acid site is experiencing diversifying selection is plotted as vertical bars. The pink line denotes a significance threshold at 0.8. The amino acid positions that have a probability of experiencing diversifying selection exceeding the 0.8 posterior probability significance threshold are highlighted in green, with the amino acid position labeled above the bar.
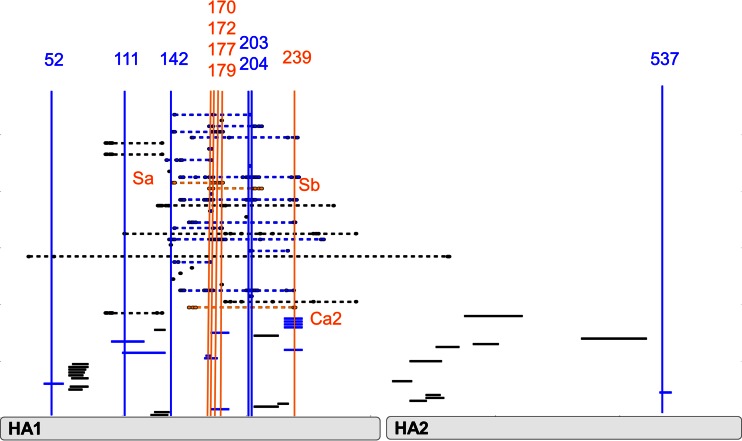
These 16 diversified sites were then compared against a collection of experimentally determined B-cell/Ab epitopes curated from the literature by the Immune Epitope Database (IEDB); 11 of the 16 diversified sites were located within experimentally determined H1 B-cell/Ab epitopes (Fig. 4). These diversified B-cell/Ab epitopes include three previously characterized B-cell/Ab epitopes—Sa, Sb, and Ca2—defined by Caton et al. by grouping mutants of the influenza virus A/PR/8/34 virus based on their antigenic reactivity to panels of neutralizing monoclonal antibodies (11, 12). These epitopes represent distinct neutralizing antigenic regions which have previously been reported to bind to human sera from patients infected with prepandemic H1N1 (24). Thus, this diversifying selection analysis of prepandemic H1N1 HA has identified a subset of experimentally defined H1 HA B-cell/Ab epitopes that appear to be especially important for protective immunity in humans, including the Sa, Sb, and Ca2 Caton epitopes.
FIG 4.
Identification of diversified B-cell/Ab epitopes. Continuous B-cell/Ab epitopes are indicated by solid bars; discontinuous B-cell/Ab epitopes are indicated by solid bars linked by dashed lines. Diversified sites identified from Fig. 3 are depicted as vertical bars. The B-cell/Ab epitopes containing sites experiencing diversifying selection in the evolution of the prepandemic H1N1 strains (diversified epitopes) are highlighted in blue or orange. Caton B-cell/Ab epitopes that contain diversified sites are highlighted in orange (Sa, Sb, and Ca2). B-cell/Ab epitopes that were not diversified are colored black.
Evaluating diversified B-cell/Ab epitope prediction.
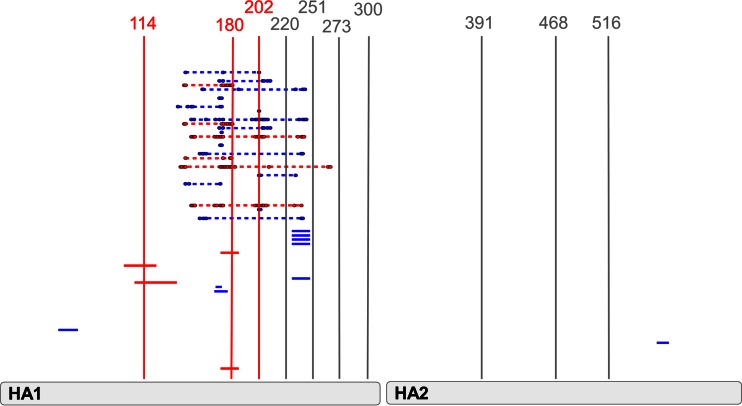
To evaluate the hypothesis that these diversified B-cell/Ab epitope regions would be the targets for ongoing mutation, the evolution of the pH1N1 lineage was assessed based on the idea that once protective immunity has been established in the human population after the initial exposure to early 2009 pandemic outbreak viruses, the virus would mutate in such a way as to disrupt these specific diversified epitope regions through the process of evolutionary genetic drift in order to evade protective immunity. We used the metadata-driven Comparative Analysis Tool for Sequences (meta-CATS) tool (16) to identify which sites were mutated and selected for by comparing sequences from early 2009 pandemic outbreak isolates (early pandemic) to recent pandemic sequences from the 2012-2013 and 2013-2014 influenza seasons (late pandemic) from different geographic regions within (California, New York, Texas, Louisiana, Florida, and Colorado) and outside (British Columbia and Quebec, Canada; Czech Republic; Helsinki, Finland; Moscow, Russia; São Paulo, Brazil; and Taiwan) the United States. The meta-CATS analysis identified 10 sites that were significantly changed in the majority of comparisons between early and late pandemic isolates from each geographic region (Table 3). Of these 10 meta-CATS sites, 3 (positions 114, 180, and 202) were located in the previously identified diversified B-cell/Ab epitope regions (Fig. 5).
TABLE 3.
Amino acid positions significantly different between early and late H1N1 pandemic isolates
Early, amino acids found in the early pandemic isolates (2009 outbreak); Late, amino acids found in the late pandemic isolates (2012-2013 and 2013-2014 flu seasons).
The dominant residue present in the late pandemic sequences is in bold.
—, the pairwise comparison between the early strain versus strains from the Czech Republic, Helsinki, and Moscow did not identify any significant sequence variation at sites 180 and 273. This is likely because the majority of strains from these three regions are from the 2012-2013 flu season and the mutations present at sites 180 and 273 became dominant in the 2013-2014 flu season.
FIG 5.
Diversified B-cell/Ab epitopes containing mutations acquired during pandemic evolution. All continuous and discontinuous diversified epitopes identified from Fig. 4 are shown. The 10 sites identified by meta-CATS analysis that have mutated since the 2009 outbreak are depicted as vertical bars. Meta-CATS sites that are located in the diversified B-cell/Ab epitopes identified in the prepandemic analysis are highlighted in red. Meta-CATS sites that are not located in any diversified B-cell/Ab epitope are highlighted in gray.
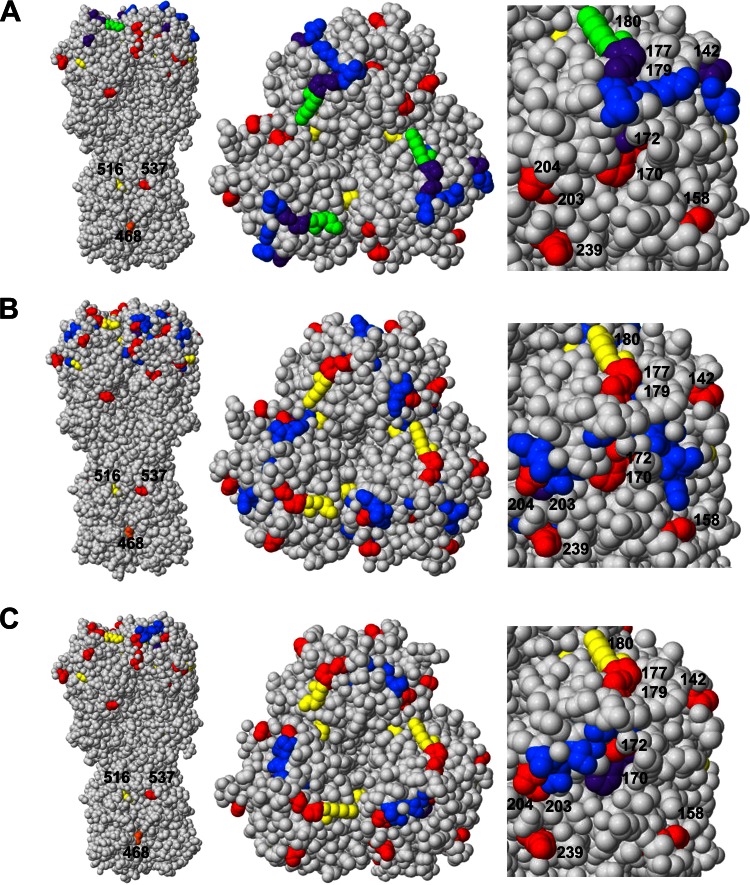
In order to investigate the structural relationships between these amino acid positions, the selected B-cell/Ab epitopes, sites experiencing diversifying selection in prepandemic strains, and sites evolving postpandemic were mapped onto a three-dimensional (3D) protein structure of HA from A/California/04/2009. In the case of both the Sa (Fig. 6A; see also Movie S1 in the supplemental material) and the IEDB:159269 B-cell/Ab epitope (Fig. 6B; see also Movie S2), overlap between the prepandemic diversified sites and postpandemic evolving sites with each of the B-cell/Ab epitopes (highlighted in purple and green) is observed, suggesting that sequence alterations both prepandemic and postpandemic would have major effects on the structure of these B-cell/Ab epitope. In the case of the Sb epitope (Fig. 6C; see also Movie S3), while less direct overlap is observed, the close proximity of these three regions (prepandemic diversified sites, postpandemic mutation sites, and Sb epitope) would also be consistent with a disruption of B-cell receptor/Ab recognition.
FIG 6.
Diversified B-cell/Ab epitopes highlighted on 3D HA trimer structure. Amino acid positions are highlighted on the A/California/04/2009 HA protein structure (PDB ID 3UBQ) based on the A/California/04/2009 full-length HA numbering. Nonoverlapping selected B-cell/Ab epitope positions are highlighted in blue, nonoverlapping positions found to be experiencing diversifying selection in the evolution of the prepandemic H1N1 strains are highlighted in red, nonoverlapping positions that have mutated between the early and late pandemic H1N1 strains are highlighted in yellow, prepandemic diversified sites that are located in the selected B-cell/Ab epitope are highlighted in purple, mutated pandemic sites that are located in the selected B-cell/Ab epitope are highlighted in green, and sites found to be under diversifying selection in the evolution of the prepandemic strains and mutated in the pandemic lineage are highlighted in orange. (A) Caton Sa epitope (see also Movie S1 in the supplemental material). (B) B-cell/Ab epitope IEDB:159269 (see also Movie S2). (C) Caton Sb epitope (see also Movie S3).
Phylogenetic tree analysis of postpandemic evolving sites.
To further explore the postpandemic evolving sites identified by the meta-CATS statistical test, a phylogenetic analysis was performed to determine if the substitutions demonstrated persistence over time, which would be expected if they were indeed being positively selected. A phylogenetic tree was generated using unique pandemic-like human H1N1 HA protein sequences from North America (Fig. 7). (North American isolate sequences were used because information about the respective influenza seasons was consistently available for these isolates. Similar topological characteristics were observed in phylogenetic trees using international isolate sequences [data not shown].) Strains carrying specific amino acid residues at each postpandemic evolving site were highlighted individually. Sites were grouped based on similar patterns of temporal distributions on the tree since sites with similar patterns may occur due to linkage disequilibrium and founder effects. Seven different groups of postpandemic evolving sites were identified: 180 and 273; 114; 300 and 516; 202 and 468; 391; 220; and 251.
FIG 7.
Phylogenetic trees showing the emergence of pandemic mutations. The far left tree is colored and ordered temporally by Northern Hemisphere influenza season and used as a reference tree. The trees to the right of the reference tree are colored by the residues that are present at the site labeled above the specific tree. Strains carrying the amino acid that was dominant in the early 2009 pandemic outbreak sequences (group 1) are colored blue; strains carrying the amino acid that was dominant in the late pandemic sequences (group 2) are colored red. Strains carrying all other residues are colored gray.
Mutations at sites 180 and 273 appeared and dominated in the 2013-14 influenza season (Fig. 7a), suggesting that at least one of the sites has been positively selected. Since the K180Q substitution was found to be significant from the meta-CATS analysis and is located in the diversified Sa epitope region, it is likely that this is the functionally relevant substitution in the group. Similarly, substitutions at sites 202 and 468 first appeared in the 2010-2011 influenza season and then became dominant in subsequent influenza seasons (Fig. 7d), again suggesting that at least one of the sites has been positively selected. Since the S202T substitution was found to be significant from the meta-CATS analysis and is located in the diversified Sb B-cell/Ab epitope region, it is likely that this is the functionally relevant substitution in the group. Substitution at site 114 first appeared in the 2009-2010 influenza season, was predominant in the 2010-2011 season, but then disappeared in the 2011-2012 season, only to reappear and finally be maintained in the 2012-2013 influenza season and onward (Fig. 7b). The strains that contain the 114N substitution in the earlier influenza seasons appear to be the precursors for the strains that appear in the later influenza seasons. Since site 114 is located within both a B-cell/Ab epitope region and the receptor binding site, this phenomenon of stuttering selection may be related to the possibility that the site is somewhat restricted from diversification in order to maintain functionality (25). In this case, a delicate balance between the diversifying effects of protective immunity and the purifying effects of maintaining functionality may be involved.
Substitutions at sites 300 and 516 appear and then disappear several times before being retained together starting in the 2012-2013 season (Fig. 7c), suggesting that individually they may be under weak selection, whereas in combination the selection effect may be stronger. Substitutions at sites 220 and 391 appeared in the 2009-2010 influenza season and have been retained ever since (Fig. 7e and f). Since these mutations appeared early, perhaps even before the establishment of protective immunity in the human population, and were maintained, these changes may have been important for the initial adaptation into humans. Site 220 is located near the receptor binding pocket in the 3D HA trimer structure (PDB code 3UBQ). Site 251 may not actually be undergoing positive selection, since it appeared in the 2012-2013 influenza season and then disappeared in the next influenza season (Fig. 7g).
In summary, three of the postpandemic evolving sites were located in diversified B-cell/Ab epitope regions. While the remaining seven sites that mutated as the pandemic sequences evolved were not located in a targeted B-cell/Ab epitope, they either were linked to these three sites, were located in functional sequence features that may influence influenza viral fitness, or showed weak evidence of selection due to the lack of temporal persistence.
HI analysis with human sera.
In order to test whether the amino acid substitutions that were positively selected were affecting immune system recognition, hemagglutination inhibition (HI) assays were performed using human antiserum from patients who received a pH1N1 vaccine (A/California/07/2009). The HI assay is based on the concept that antibodies that recognize a virus will attach to the virus and prevent the virus from attaching to red blood cells (RBCs) and thereby inhibit the normal hemagglutination observed with influenza virus particles due to the activity of HA on the virion surface. Table 4 shows the maximum dilution of antisera that inhibits agglutination of RBCs by representative virus isolates from different time points since the 2009 outbreak. Each of these viruses contains different amino acid mutations depending on the year of isolation (Table 5). The first column shows the level of ferret antisera raised against the A/California/07/2009 strain that inhibits the agglutination as a positive control. Each of the columns to the right of the first column shows the level of human antisera required to inhibit agglutination. A decreasing trend in reactivity is observed in sera from 8 of the 10 subjects. For example, the maximum dilution of antiserum from patient sample 1553 that still supports agglutination inhibition against the recent A/California/3546/2014 virus (80-fold) is only 13% of the maximum dilution that still supports agglutination inhibition against the A/California/07/2009 virus (640-fold), indicating that this antiserum has lost reactivity against this late pandemic isolate. The two patient samples (1540 and 2419) for which there was not a decreasing trend showed a relatively low dilution of antisera required to inhibit the agglutination of the A/California/07/2009 early pandemic isolate.
TABLE 4.
Loss of serum reactivity in hemagglutination inhibition experiments
| Virus | Measurement result with indicated antisera |
Median % of control | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/CA/07 |
1332 |
1540 |
1551 |
1553 |
1728 |
1765 |
2253 |
2364 |
2419 |
2579 |
|||||||||||||
| 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | 1/dilutiona | % of control | ||
| A/California/07/09_Control | 905 | 100 | 1,810 | 100 | 453 | 100 | 640 | 100 | 640 | 100 | 640 | 100 | 2,560 | 100 | 1,810 | 100 | 453 | 100 | 226 | 100 | 453 | 100 | 100 |
| rA/New York/1682/2009 | 640 | 71 | 1,280 | 71 | 320 | 71 | 905 | 141 | 640 | 100 | 226 | 35 | 1,810 | 71 | 1,280 | 71 | 226 | 50 | 320 | 142 | 226 | 50 | 71 |
| rA/Wisconsin/02/2011 | 226 | 25 | 320 | 18 | 320 | 71 | 160 | 25 | 160 | 25 | 80 | 13 | 453 | 18 | 320 | 18 | 80 | 18 | 226 | 100 | 57 | 13 | 18 |
| rA/St. Petersburg/100/2011 | 57 | 6 | 453 | 25 | 226 | 50 | 160 | 25 | 320 | 50 | 160 | 25 | 1,280 | 50 | 640 | 35 | 226 | 50 | 160 | 71 | 113 | 25 | 35 |
| rA/California/52/2011 | 320 | 35 | 905 | 50 | 226 | 50 | 381 | 60 | 320 | 50 | 113 | 18 | 1,076 | 42 | 640 | 35 | 134 | 30 | 226 | 100 | 113 | 25 | 42 |
| A/California/3546/2014 (16 January 2014) | 160 | 18 | 640 | 35 | 640 | 141 | 190 | 30 | 80 | 13 | 80 | 13 | 640 | 25 | 538 | 30 | 134 | 30 | 640 | 283 | 57 | 13 | 30 |
| A/California/3543/2014 (6 January 2014) | 160 | 18 | 453 | 25 | 640 | 141 | 160 | 25 | 80 | 13 | 80 | 13 | 640 | 25 | 453 | 25 | 113 | 25 | 640 | 283 | 57 | 13 | 25 |
| A/New York/3627/2014 (12 January 2014) | 160 | 18 | 640 | 35 | 640 | 141 | 226 | 35 | 80 | 13 | 80 | 13 | 905 | 35 | 640 | 35 | 113 | 25 | 640 | 283 | 68 | 15 | 35 |
| A/New York/3626/2014 (2 January 2014) | 160 | 18 | 320 | 18 | 640 | 141 | 226 | 35 | 80 | 13 | 80 | 13 | 640 | 25 | 453 | 25 | 113 | 25 | 640 | 283 | 57 | 13 | 25 |
| A/Texas/3668/2014 (3 January 2014) | 160 | 18 | 320 | 18 | 640 | 141 | 226 | 35 | 80 | 13 | 80 | 13 | 640 | 25 | 453 | 25 | 113 | 25 | 640 | 283 | 57 | 13 | 25 |
| A/Texas/3665/2014 (3 January 2014) | 160 | 18 | 380.7 | 21 | 1,280 | 283 | 226 | 35 | 80 | 13 | 80 | 13 | 905 | 35 | 538 | 30 | 113 | 25 | 905 | 400 | 68 | 15 | 25 |
Dilution factor of serum resulting in loss of hemagglutination inhibition (geometric mean of the log dilution factor).
TABLE 5.
Postpandemic amino acid mutations contained within each virus used in the HI assay
| Virus | Meta-CATS mutationa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 114 | 180 | 202 | 220 | 273 | 300 | 391 | 468 | 516 | |
| 2009 outbreak isolates | |||||||||
| A/California/07/2009 | D | K | S | S | A | K | E | S | E |
| A/New York/1682/2009 | . | . | . | . | . | . | . | . | . |
| Intermediate isolates | |||||||||
| A/Wisconsin/02/2011 | . | . | . | T | . | . | . | . | K |
| A/St. Petersburg/100/2011 | . | . | T | T | . | . | K | N | . |
| A/California/52/2011 | . | . | T | T | . | . | K | N | . |
| Recent isolates | |||||||||
| A/California/3546/2014 (16 January 2014) | N | Q | T | T | T | E | K | N | K |
| A/California/3543/2014 (6 January 2014) | N | Q | T | T | T | E | K | N | K |
| A/New York/3626/2014 (2 January 2014) | N | Q | T | T | T | E | K | N | K |
| A/New York/3627/2014 (12 January 2014) | N | Q | T | T | T | E | K | N | K |
| A/Texas/3668/2014 (3 January 2014) | N | Q | T | T | T | E | K | N | K |
| A/Texas/3665/2014 (3 January 2014) | N | Q | T | T | T | E | K | N | K |
A period (.) indicates the same amino acid as in the A/California/07/2009 sequence.
Overall, the amount of human antiserum required to inhibit agglutination of the late pandemic isolates is 3- to 4-fold higher (equivalent to a 25 to 35% dilution factor) than the amount required to inhibit agglutination of the early pandemic isolates, presumably due to antigenic evolution.
DISCUSSION
Using a data mining approach, we identified 16 HA sites experiencing diversifying selection as the prepandemic H1N1 strains have evolved in the human population from 1918 to 1957 and 1977 to 2009: amino acid positions 4, 52, 111, 142, 158, 170, 172, 177, 179, 203, 204, 239, 275, 278, 468, and 537. Based on the combined information from immune epitope and phylogenetic analysis, there appear to be two main drivers of diversifying selection: sequence variation within key antigenic sites that drive escape from immune system pressure (antigenic drift) and sequence variation that improves virus replication and transmission in the human hosts (host adaptation), since some of these diversified sites are found in previously defined antigenic sites and others are found to impact sites predicted to affect HA binding avidity for cell surface glycan-containing receptors (11, 12, 26).
Based on our analysis, three B-cell/Ab epitope regions appear to be key targets of protective immunity in humans against influenza H1N1 viruses; these B-cell/Ab epitope regions experienced diversifying selection as prepandemic H1N1 has evolved, and they were targeted for mutation as the postpandemic H1N1 lineage has evolved. In support of this interpretation, a recent study identified peptides recognized by pH1N1-infected human sera using panning with an H1N1 genome fragment phase display library (GFPDL) that overlaps our targeted sites (27). Although the field struggles to understand the intricacies of how influenza evolves to escape immune recognition, we hypothesize that by combining an analysis of the natural evolution of influenza A H1N1 virus with experimental data regarding B-cell/Ab recognition, the critical regions that are important for protective immunity in humans can be identified. The loss of human serum reactivity in late outbreak strains described here supports this hypothesis. Furthermore, our analysis adds to the knowledge gained from previous studies about the antigenic evolution of H1N1 influenza viruses. Several studies identified candidate substitutions that could be responsible for antigenic changes because they are located within epitope regions. Li et al. found sites experiencing diversifying selection that are located within B-cell and T-cell epitope regions that were perhaps responsible for antigenic changes in the seasonal H1N1 lineage. All the diversified sites identified in the analysis by Li et al. except one were consistent with the diversified sites found in our analysis of the evolution of the prepandemic H1N1 sequences (7). Furuse et al. found the same diversified sites using the seasonal H1N1 sequences located within the Sb and Ca2 Caton epitopes, suggesting that these sites are involved in determining antigenicity (28). Huang et al. identified 41 mutation sites that had a significantly high entropy and likelihood ratio score using seasonal H1N1 sequences, indicating that these mutations were likely to cause antigenic variants for H1N1 viruses. Four of these sites overlapped with the 16 sites that were diversified in our prepandemic analysis. The differences in the sites found by the two studies are likely due to difference in the methods used; Huang et al. used a genetic evolution approach without considering phylogenetic relationships between viruses as our approach does (29). Overall, the sites that we identified undergoing diversifying selection in the prepandemic sequences were similar to those sites that were previously identified as contributing to the antigenic evolution of the H1N1 influenza virus. However, the study reported here is the first to use the results from the diversifying selection analysis of seasonal H1N1 to predict the evolution of the pandemic H1N1 lineage.
The HI results presented here demonstrate a loss of reactivity between antibodies present in the serum of patients exposed to early pandemic antigens and viruses from recent influenza seasons that have acquired the amino acid substitutions identified by meta-CATS analysis. While the HI assay measures the interference of HA receptor binding by serum antibody and therefore virus neutralization, it should be emphasized that the mechanisms of protective immunity that are driving diversifying selection of these B-cell/Ab epitope regions may not be limited to effects on virus neutralization. Loss of antisera binding to these B-cell/Ab epitopes could also impact antibody-dependent cellular cytotoxicity (ADCC) of virus-infected cells by circulating NK cells, complement-mediated lysis of infected cells, and BCR-mediated endocytosis, processing, and presentation of viral peptides to helper T cells through major histocompatibility complex (MHC) class II. Thus, while our findings indicate that B-cell/Ab epitope recognition is driving diversifying selection and is therefore likely to be key to protective immunity, they do not directly address whether virion neutralization is the key mechanism.
These findings have important implications for vaccine strain selection. The WHO selects virus vaccine strain candidates using HI assay results and antigenic cartography analysis (30), which generates a 2D map of antigenic similarity between viruses from the HI results, to determine if circulating strains have become antigenically distinct from previous vaccine strains. However, these assays are based on the use of polyclonal ferret antisera that likely include reactivities that may contribute to protective immunity in humans and reactivities that may not, and thus do not discriminate between B-cell/Ab epitopes found under experimental conditions versus those that elicit a protective response to a natural infection. The approach described here identified the B-cell/Ab epitopes that are likely to be the most relevant for protective immunity, and therefore, their impact could be weighted more heavily in comparison with to the other epitope regions. This information would be useful in augmenting current strategies for vaccine strain selection. Our findings suggest that the emergence and persistence of strains carrying substitutions in the targeted B-cell/Ab epitopes (position 114 in the 2010-2011 season, position 202 in the 2011-2012 season, and position 180 in the 2013-2014 season) would possibly warrant consideration for changes in the recommended H1N1 strains used for vaccine formulations.
Although our study was focused on the relationship between evolving amino acid positions and B-cell/Ab epitopes due to their importance for protective immunity and vaccine design, our analysis also identified evolving sites that appear to be responding to selective pressure and yet are distinct from B-cell/Ab epitopes. One possibility is that these sites might represent those T-cell epitopes that are important for protective immunity in humans. Site 278 is a prepandemic diversified site that is not located in any B-cell/Ab epitope; however, it is located in an experimentally determined T-cell epitope (IEDB ID 144732). Furthermore, the NetMHCIIpan method found that site 278 would be predicted to occur within a high-affinity T-cell epitope using representative prepandemic and pandemic sequences as input (31). Likewise, site 468 experienced diversifying selection in the prepandemic, was found to be significantly mutation during postpandemic evolution, but was not located in any previously defined B-cell/Ab epitope, suggesting that it is experiencing some other kind of diversifying pressure. The possibility that diversifying analysis could be used to identify relevant T-cell epitopes for protective immunity requires further exploration.
Supplementary Material
ACKNOWLEDGMENTS
We thank the University of Rochester Respiratory Pathogen Research Center and the Air Force Research Laboratory, 711th Human Performance Wing, U.S. Air Force School of Aerospace Medicine, for providing human serum and virus isolate samples.
This work was supported by NIH/NIAID grants HHSN272201200005C and HHSN272201400028C. The human pH1N1 vaccine trials were supported by NYICE grant HHSN266200700008C.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03636-14.
REFERENCES
- 1.Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. 2007. Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. 2013. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A 110:20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehender G, Pariani E, Piralla A, Lai A, Gabanelli E, Ranghiero A, Ebranati E, Amendola A, Campanini G, Rovida F, Ciccozzi M, Galli M, Baldanti F, Zanetti AR. 2012. Reconstruction of the evolutionary dynamics of the A(H1N1)pdm09 influenza virus in Italy during the pandemic and post-pandemic phases. PLoS One 7:e47517. doi: 10.1371/journal.pone.0047517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makkoch J, Suwannakarn K, Payungporn S, Prachayangprecha S, Cheiocharnsin T, Linsuwanon P, Theamboonlers A, Poovorawan Y. 2012. Whole genome characterization, phylogenetic and genome signature analysis of human pandemic H1N1 virus in Thailand, 2009–2012. PLoS One 7:e51275. doi: 10.1371/journal.pone.0051275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galiano M, Agapow PM, Thompson C, Platt S, Underwood A, Ellis J, Myers R, Green J, Zambon M. 2011. Evolutionary pathways of the pandemic influenza A (H1N1) 2009 in the UK. PLoS One 6:e23779. doi: 10.1371/journal.pone.0023779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Shi W, Qiao H, Ho SY, Luo A, Zhang Y, Zhu C. 2011. Positive selection on hemagglutinin and neuraminidase genes of H1N1 influenza viruses. Virol J 8:183. doi: 10.1186/1743-422X-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Ma J, Wang Q. 2009. Evolutionary trends of A(H1N1) influenza virus hemagglutinin since 1918. PLoS One 4:e7789. doi: 10.1371/journal.pone.0007789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. 2012. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir Viruses 6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noronha JM, Liu M, Squires RB, Pickett BE, Hale BG, Air GM, Galloway SE, Takimoto T, Schmolke M, Hunt V, Klem E, Garcia-Sastre A, McGee M, Scheuermann RH. 2012. Influenza virus sequence feature variant type analysis: evidence of a role for NS1 in influenza virus host range restriction. J Virol 86:5857–5866. doi: 10.1128/JVI.06901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 12.Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, Russ G, Gibbs JS, Bennink JR, Yewdell JW. 2013. Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe 13:314–323. doi: 10.1016/j.chom.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. 2010. The immune epitope database 2.0. Nucleic Acids Res 38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 16.Pickett BE, Liu M, Sadat EL, Squires RB, Noronha JM, He S, Jen W, Zaremba S, Gu Z, Zhou L, Larsen CN, Bosch I, Gehrke L, McGee M, Klem EB, Scheuermann RH. 2013. Metadata-driven comparative analysis tool for sequences (meta-CATS): an automated process for identifying significant sequence variations that correlate with virus attributes. Virology 447:45–51. doi: 10.1016/j.virol.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 18.Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J Mol Evol 20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- 19.Zhou B, Wentworth DE. 2012. Influenza A virus molecular virology techniques. Methods Mol Biol 865:175–192. doi: 10.1007/978-1-61779-621-0_11. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, Dattilo BM, Denisova EA, De Souza I, Eickmann M, Dugan VG, Ferrari A, Gomila RC, Han L, Judge C, Mane S, Matrosovich M, Merryman C, Palladino G, Palmer GA, Spencer T, Strecker T, Trusheim H, Uhlendorff J, Wen Y, Yee AC, Zaveri J, Zhou B, Becker S, Donabedian A, Mason PW, Glass JI, Rappuoli R, Venter JC. 2013. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med 5:185ra168. doi: 10.1126/scitranslmed.3006368. [DOI] [PubMed] [Google Scholar]
- 22.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. 2013. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J Infect Dis 207:297–305. doi: 10.1093/infdis/jis684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Network WGIS (ed). 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. WHO Press, Geneva, Switzerland: http://www.who.int/influenza/gisrs_laboratory/manual_diagnosis_surveillance_influenza/en/. [Google Scholar]
- 24.DuBois RM, Aguilar-Yañez JM, Mendoza-Ochoa GI, Oropeza-Almazán Y, Schultz-Cherry S, Alvarez MM, White SW, Russell CJ. 2011. The receptor-binding domain of influenza virus hemagglutinin produced in Escherichia coli folds into its native, immunogenic structure. J Virol 85:865–872. doi: 10.1128/JVI.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mir-Shekari SY, Ashford DA, Harvey DJ, Dwek RA, Schulze IT. 1997. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J Biol Chem 272:4027–4036. [DOI] [PubMed] [Google Scholar]
- 26.Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, Yewdell JW. 2009. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma N, Dimitrova M, Carter DM, Crevar CJ, Ross TM, Golding H, Khurana S. 2012. Influenza virus H1N1pdm09 infections in the young and old: evidence of greater antibody diversity and affinity for the hemagglutinin globular head domain (HA1 domain) in the elderly than in young adults and children. J Virol 86:5515–5522. doi: 10.1128/JVI.07085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuse Y, Shimabukuro K, Odagiri T, Sawayama R, Okada T, Khandaker I, Suzuki A, Oshitani H. 2010. Comparison of selection pressures on the HA gene of pandemic (2009) and seasonal human and swine influenza A H1 subtype viruses. Virology 405:314–321. doi: 10.1016/j.virol.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 29.Huang JW, Lin WF, Yang JM. 2012. Antigenic sites of H1N1 influenza virus hemagglutinin revealed by natural isolates and inhibition assays. Vaccine 30:6327–6337. doi: 10.1016/j.vaccine.2012.07.079. [DOI] [PubMed] [Google Scholar]
- 30.Fouchier RA, Smith DJ. 2010. Use of antigenic cartography in vaccine seed strain selection. Avian Dis 54:220–223. doi: 10.1637/8740-032509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 31.Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. 2013. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics 65:711–724. doi: 10.1007/s00251-013-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.