ABSTRACT
Sterile alpha motif domain and HD domain-containing protein 1 (SAMHD1) restricts human immunodeficiency virus type 1 (HIV-1) replication in myeloid and resting T cells. Lentiviruses such as HIV-2 and some simian immunodeficiency viruses (SIVs) counteract the restriction by encoding Vpx or Vpr, accessory proteins that are packaged in virions and which, upon entry of the virus into the cytoplasm, induce the proteasomal degradation of SAMHD1. As a tool to study these mechanisms, we generated HeLa cell lines that express a fusion protein termed NLS.GFP.SAM595 in which the Vpx binding domain of SAMHD1 is fused to the carboxy terminus of green fluorescent protein (GFP) and a nuclear localization signal is fused to the amino terminus of GFP. Upon incubation of Vpx-containing virions with the cells, the NLS.GFP.SAM595 fusion protein was degraded over several hours and the levels remained low over 5 days as the result of continued targeting of the CRL4 E3 ubiquitin ligase. Degradation of the fusion protein required that it contain a nuclear localization sequence. Fusion to the cytoplasmic protein muNS rendered the protein resistant to Vpx-mediated degradation, confirming that SAMHD1 is targeted in the nucleus. Virions treated with protease inhibitors failed to release Vpx, indicating that Gag processing was required for Vpx release from the virion. Mutations in the capsid protein that altered the kinetics of virus uncoating and the Gag binding drug PF74 had no effect on the Vpx-mediated degradation. These results suggest that Vpx is released from virions without a need for uncoating of the capsid, allowing Vpx to transit to the nucleus rapidly upon entry into the cytoplasm.
IMPORTANCE SAMHD1 restricts lentiviral replication in myeloid cells and resting T cells. Its importance is highlighted by the fact that viruses such as HIV-2 encode an accessory protein that is packaged in the virion and is dedicated to inducing SAMHD1 degradation. Vpx needs to act rapidly upon infection to allow reverse transcription to proceed. The limited number of Vpx molecules in a virion also needs to clear the cell of SAMHD1 over a prolonged period of time. Using an engineered HeLa cell line that expresses a green fluorescent protein (GFP)-SAMHD1 fusion protein, we showed that the Vpx-dependent degradation occurs without a need for viral capsid uncoating. In addition, the fusion protein was degraded only when it was localized to the nucleus, confirming that SAMHD1 is targeted in the nucleus and thus explaining why Vpx also localizes to the nucleus.
INTRODUCTION
The replication of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses is limited in mammalian cells by host restriction factors that interfere with specific steps in the virus life cycle. To counteract these factors, lentiviruses have evolved accessory proteins that act primarily by inducing their degradation. Sterile alpha motif domain and HD domain-containing protein 1 (SAMHD1) interferes with lentivirus replication in monocytes, macrophages, dendritic cells (DCs), and resting T cells but has no effect in activated T cells (1–4). SAMHD1 is a dGTP-regulated deoxynucleoside triphosphate (dNTP) triphosphohydrolase (5–7) that depletes the pool of dNTPs, preventing reverse transcription of the viral genomic RNA upon infection (8). In addition, SAMHD1 has been found to have 3′→5′ exonuclease activity on single-stranded DNA and RNA, and these activities may play a role in restriction by degrading the viral genomic RNA or reverse transcripts (9, 10). Polymorphisms in the SAMHD1 gene are associated with Aicardi-Goutières syndrome (AGS), a rare childhood neurologic condition characterized by the constitutive production of type I interferon, a situation that resembles congenital infection (11, 12).
HIV-2 and some simian immunodeficiency viruses (SIVs) counteract SAMHD1 by encoding the accessory protein Vpx or, in the case of SIVmus and SIVdeb, Vpr (13). Vpx and Vpr are packaged into virions. Upon virus entry, Vpx induces the proteasomal degradation of target cell SAMHD1 by forming a complex with the CRL4DCAF1 E3 ubiquitin ligase (14). The degradation begins rapidly upon infection, being detected as early as 2 h postinfection (15). In the CRL4DCAF1 E3 ubiquitin ligase complex, the carboxy-terminal domain of Vpx is bound to DCAF1 (16, 17) and the amino-terminal domain binds to the carboxy terminus of SAMHD1 (14). This complex polyubiquitinates SAMHD1, targeting it for degradation by the proteasome (14, 18). The activity of the CRL4DCAF1 E3 ubiquitin ligase is regulated by the conjugation of Nedd8 to Cul4A. Inhibition of neddylation using the drug MLN4924 prevents SAMHD1 degradation (19). DCAF1 also functions in HECT-family EDD/UBR5 E3 ubiquitin ligases (reviewed in reference 20), and HIV-1 Vpr interacts with the EDD-DDB1-DCAF1 E3 ligase complex to inhibit telomerase activity (21).
HIV-1 is susceptible to SAMHD1 restriction, and yet its Vpr does not induce SAMHD1 degradation and it lacks a Vpx. As a result, the virus is limited in its ability to infect myeloid cells such as DCs and monocyte-derived macrophages (MDMs). Vpx is packaged into virus particles by a 10-amino-acid packaging motif at amino acids 17 to 26 of SIVmac Gag p6 (22–24). HIV-1 p6 lacks the Vpx packaging motif and, as a result, does not package Vpx if provided in trans. Placement of the motif into p6 of HIV-1 results in a virus that packages Vpx (25). Vpx-containing HIV-1 infects myeloid cells with a 10- to 100-fold increase in titer.
The carboxy-terminal region of SAMHD1 is sufficient for interaction with Vpx (26), and the kinetics of binding of the CRL4DCAF1-Vpx complex to C-terminal SAMHD1 are the same as to full-length SAMHD1 (27). Furthermore, fusing the carboxy terminus of SAMHD1 to thioredoxin (14) or GFP (26) makes those proteins susceptible to ubiquitination and degradation mediated by CRL4DCAF1-Vpx.
SAMHD1 is a nuclear protein that contains a canonical nuclear localization signal (NLS) near its amino terminus (11, 18, 28–30). Disruption of the NLS does not prevent antiviral activity (28, 29) but renders the protein resistant to Vpx-mediated degradation (18, 28–31). We and others reported that nuclear localization of SAMHD1 is required for Vpx-mediated degradation and that SAMHD1 polyubiquitination and degradation occurred in the nucleus, most likely through the utilization of nuclear proteasomes (18, 28). However, evidence has been reported suggesting that Vpx can target SAMHD1 for proteasomal degradation in the cytoplasm. A cytoplasmic HIV-2 Vpx induced the degradation of an NLS-deleted SAMHD1 (29). In addition, leptomycin B, a drug that blocks the nucleocytoplasmic shuttling of nuclear proteins, was found to block the Vpx-induced degradation of SAMHD1 (32), further suggesting cytoplasmic degradation of the protein.
It is presumed that, following fusion of the virus at the plasma membrane and entry into the cytoplasm, Vpx is released to associate with CRL4 and induce SAMHD1 degradation. However, little is known about when and how this occurs. In this report, we describe the generation of a HeLa cell line that expresses a fusion protein in which an NLS-tagged GFP is fused to the Vpx-binding domain of SAMHD1. The cell line, termed HeLa NLS.GFP.SAM595, serves as a rapid and accurate model to study Vpx-induced SAMHD1 degradation. Using this cell line, we determined that only nuclear localized fusion proteins were subject to Vpx-mediated degradation. Degradation of the fusion protein was rapidly induced upon infection with Vpx-containing virus and was maintained by continued proteasome-dependent degradation for at least 5 days. Vpx-mediated degradation appeared to be independent of uncoating but was dependent on HIV-1 maturation. Small interfering RNA (siRNA) knockdown experiments to determine the role of the CRL4 complex in SAMHD1 degradation showed that DCAF1, but not DDB1, was required, suggesting that Vpx may be able to use another DCAF1-containing E3 ubiquitin. Together, these findings suggest that Vpx is located outside the viral core and, upon cytoplasmic entry, without a need for uncoating, travels to the nucleus, where it serves to induce the degradation of SAMHD1 over an extended time period.
MATERIALS AND METHODS
Cell culture.
293T and HeLa cell lines were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Plasmids.
NLS and/or SAMHD1 Vpx-binding-site-tagged GFP (NLS.GFP.SAM595, GFP.SAM595, and NLS.GFP) constructs and NLS.GFP.muNS.SAM595 and GFP.muNS.SAM595 constructs were generated by overlapping PCR. The amplicons were cloned into pLenti (Addgene) at the BamHI and XhoI sites. pNLS.GFP.SAM595, pNLS.GFP, and pGFP.SAM595 proteins were generated using pLenti.huSAMHD1 (described in reference 8) and pLenti.EGFP as the templates. pNLS.GFP.muNS.SAM595 and pGFP.muNS.SAM595 were generated using pLenti.huSAMHD1 and pcDNA3.1-NLS.GFP.muNS (29). All plasmids were confirmed by nucleotide sequence.
pLenti-mCherry was constructed by cleaving mCherry from HIV-cytomegalovirus (CMV)-mCherry plasmid with XbaI and SalI and ligating it into pLenti-Neo at the XbaI and SalI sites. The myc-tagged Vpx (SIVmac239) plasmid and Gag/Pol p6 chimeric plasmid (pMDL-chp6) have been described previously (25, 33). Unstable K170A and hyperstable CA point mutations E128A/R132A (34) and Gag cleavage mutants CA/p2 and CA/NC have been described previously (35). The Gag mutations were introduced into pMDL-chp6.
Virus and VLP preparation.
HIV-1 reporter viruses containing or lacking Vpx were produced by cotransfecting 293T cells with pLenti-mCherry, pMDL-chp6, pcVSV-G, pRSV-Rev, and pc.myc-Vpx or pcDNA6 by calcium phosphate coprecipitation. pcVSV-G expresses a CMV-driven vesicular stomatitis virus (VSV) envelope glycoprotein (G), and pRSV-Rev expresses a Rous sarcoma virus (RSV) long terminal repeat (LTR)-driven HIV-1 Rev. Virus-like particles (VLPs) containing or lacking Vpx were produced by transfection of 293T cells with pMDL-chp6, pVSV-G, pRSV-Rev, and myc-His-tagged Vpx or pcDNA6. At 48 h posttransfection, supernatants were harvested, filtered, and concentrated 10-fold by ultracentrifugation over a 20% sucrose cushion at 30,000 rpm for 90 min. To produce virions blocked for proteolytic maturation, 3 μM nelfinavir was added to the 293T cells upon transfection. SIV VLPs containing Vpx or Vpr from different HIV-2 or SIV isolates were produced by cotransfecting 293T cells with pSIV3+ Δvpx Δvpr and their respective FLAG-tagged Vpx or Vpr expression vectors for each of the accessory proteins as we have previously described (28).
Lentiviral expression vectors for NLS.GFP.SAM595, GFP.SAM595, NLS.GFP, NLS.GFP.muNS.SAM595, and GFP.muNS.SAM595 were produced by cotransfecting 293T cells with the pLenti expression vector, pRSV-Rev, pMDL Gag-Pol, and pVSV-G. HeLa cell lines that stably expressed NLS and/or Vpx-binding site-tagged GFP fusion protein with or without muNS were generated by lentiviral vector transduction followed by selection in 1 μg/ml puromycin at a limiting dilution. GFP fluorescent cell clones were expanded.
GFP degradation assay.
GFP-expressing HeLa cells (2 × 104) were plated in clear-bottomed 96-well plates. The next day, the cells were infected with HIV-1 mCherry reporter virus or with 10 ng or 50 ng of p24 HIV-1 VLPs per well. The following day, GFP fluorescence was measured on an Envision (Perkin-Elmer) microplate fluorometer. The fluorescence of control HeLa cells was subtracted. At 3 days later, the cells were fixed in 1% paraformaldehyde (PFA) and analyzed by flow cytometry to quantify GFP- and mCherry-positive cells.
For time course experiments, virus was added to the cells at 4°C and the cells were then centrifuged at 16°C for 2 h at 2,500 rpm. Unbound virus was then removed by two washes with 4°C medium, after which 37°C medium was added. The GFP fluorescence was measured over time. At 24 h after infection, either 10 μM MG132 or 1 μM MLN4924 was added or the cells were transfected with nontemplate control (NTC) siRNA (25 nM) or against DCAF1. For time course experiments over 24 h, 10 μM PF74, 20 mM cyclosporine (CsA), 25 μM zidovudine (AZT), or 5 μM nevirapine was added before infection where indicated.
siRNA transfection.
HeLa single-cell clones expressing NLS.GFP.SAM595 (2 × 105) were plated in 6-well plates. The next day, the cells were transfected with 6 μM siRNA (Dharmacon) using Lipofectamine RNAiMAX (Life Technologies). Two days later, the cells were plated in 96-well plates for the subsequent GFP fluorescence assay. For time course experiments, 2 × 104 NLS.GFP.SAM595 HeLa cells were plated in 96-well plates and synchronously infected. The next day, the cells were transfected with 50 nM siRNA. The extent of knockdown was tested by immunoblot analysis. The siRNA target sequences were as follows: for DCAF1, 5′-CGGAGUUGGAGGAGGACGAUUUU-3′ (61); for DDB1, 5′-CCUGUUGAUUGCCAAAAACUU-3′ (61); and for EDD, 5′-AGACAAAUCUCGGACUUGUU-3′. ON-TARGETplus nontargeting siRNA no. 2 (Dharmacon) was used as a control siRNA NTC (target sequence, UGGUUUACAUGUUGUGUGA).
Immunoblot analysis.
Virions were pelleted from the supernatant of transfected 293T cells by ultracentrifugation over a 20% sucrose cushion for 90 min at 30,000 rpm and 4°C and then solubilized in lysis buffer containing 50 mM HEPES, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, and protease inhibitor. The cells were solubilized in lysis buffer, and the protein concentration was quantified by bicinchoninic acid (BCA) assay. Virus and cell lysates were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and probed. The antibodies (Abs) used were anti-EDD rabbit antiserum (Abcam), anti-DDB1 monoclonal Ab (MAb) ZD001, anti-DCAF1 rabbit antiVprBP antiserum (Proteintech Group), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) MAb, anti-myc MAb for Vpx (Covance), and anti-p24 AG3.0 MAb. The membranes were washed and then probed with a goat anti-mouse or anti-rabbit (Sigma) horseradish peroxidase (HRP)-conjugated second antibody and visualized using HRP substrate (Pierce) on an Odyssey Fc dual-mode imaging system (Li-Cor).
Confocal microscopy.
HeLa single-cell clones (4 × 105) expressing transduced GFP were plated in 35-mm-diameter glass-bottomed culture dishes coated with poly-d-lysine (MatTek). Nuclei were stained with Hoechst 333429, and the cells were fixed with 4% PFA. The cells were visualized with a Leica SP5 confocal microscope and analyzed using ImageJ software.
RESULTS
Establishment of a cell line for the analysis of Vpx-induced SAMHD1 degradation.
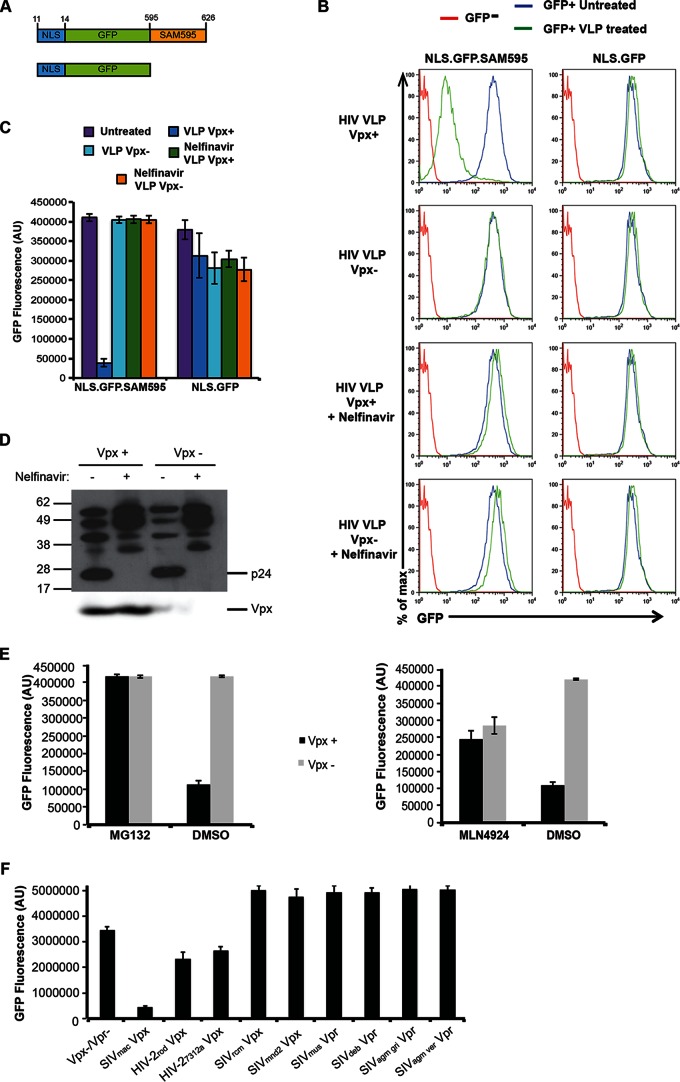
To establish a cell-based assay for Vpx-induced degradation of SAMHD1, we generated a HeLa cell line that expressed GFP fused to the Vpx binding domain of SAMHD1. This part of SAMHD1 is bound by the Vpx of viruses such as SIVmac and HIV-2 and was shown to be sufficient to subject a heterologous protein to Vpx-induced degradation (14, 26). To derive the cell line, we constructed a lentiviral vector that expressed a protein in which a consensus NLS-tagged GFP was fused to the Vpx binding domain of SAMHD1. As a control, we generated a cell line that expressed NLS.GFP (Fig. 1A). Single-cell clones were derived, and highly fluorescent clones were chosen for both proteins. To test the sensitivity of the fusion protein-expressing cell lines to Vpx, we generated VLPs that contained or lacked Vpx by cotransfecting 293T cells with pMDL.chp6, an HIV-1 gag/pol packaging vector in which Gag p6 was modified to contain the Vpx packaging motif and a Vpx expression vector or empty vector control (25). We treated the cells with equivalent amounts of VLPs and, after 24 h, analyzed GFP fluorescence in a microplate fluorometer, which allowed rapid analysis of live cells in 96-well microtiter plates, and by flow cytometry. The fluorometer results showed that Vpx-containing VLPs caused a 10-fold decrease in the fluorescence of cells that expressed NLS.GFP.SAM595 whereas the NLS.GFP protein was unaffected by VLPs containing or lacking Vpx (Fig. 1C). Analysis of the cells by flow cytometry showed similar results and demonstrated the relatively uniform response of the individual cells to the VLPs (Fig. 1B).
FIG 1.
Protease inhibitor prevents Vpx-induced degradation. (A) The structure of GFP.SAMHD1 fusion proteins used to generate HeLa stable cell lines is diagrammed. The fusion proteins were expressed in lentiviral vectors in which an amino-terminal NLS (KRPR) is fused to GFP with or without the Vpx-binding domain of SAMHD1 (NLS.GFP.SAM595 or NLS.GFP, respectively). Numbers in the diagram indicate the corresponding SAMHD1 amino acids. (B) NLS.GFP.SAM595 and NLS.GFP cell lines were treated with Vpx-containing (Vpx+) or control (Vpx−) VLPs and, after 24 h, analyzed by flow cytometry. As shown in the bottom two rows, the VLPs were produced in cells treated with 3 μM nelfinavir protease inhibitor. (C) At 24 h postinfection, GFP fluorescence of the cells treated as described for panel B was measured on a microplate fluorometer. The bars represent the means of the results determined with quadruplicate wells, and the error bars represent the standard deviations. AU, arbitrary units. (D) VLPs produced in the presence or absence of 3 μM nelfinavir were analyzed on an immunoblot probed with anti-CA antibody (top panel) or with anti-myc MAb to detect Vpx (bottom panel). (E) GFP reporter cells were incubated with 10 μM MG132 or 1 μM MLN4924 for 12 h or 2 h, respectively, and then infected with virus containing Vpx (Vpx+) or lacking Vpx (Vpx−), and the GFP fluorescence was measured 24 h postinfection. (F) VLPs were prepared containing HIV-2 and SIV Vpx or Vpr and used to infect NLS.GFP.SAM595 cells. GFP fluorescence was measured 24 h postinfection. The data points represent the means of the results determined with quadruplicate wells, and the error bars represent the standard deviations.
To test whether the degradation of the NLS.GFP.SAM595 reporter protein utilized the same pathway as the full-length SAMHD1, we tested its response to MG132, a proteasome inhibitor, and MLN4924, a neddylation inhibitor, both of which are known to inhibit Vpx function (1, 2, 19). We found that MG132 prevented degradation of the fusion protein and that MLN4924 partially blocked the degradation (Fig. 1E), responses that mimic those of full-length SAMHD1.
To determine the specificity of Vpx in the assay, we tested the sensitivity of the fusion protein to the Vpx and Vpr proteins of different primate lentiviruses. For this, we generated SIV VLPs that contained the Vpx from SIVmac239, HIV-2rod, HIV-27312a, SIVrcm, and SIVmnd2 and the Vpr from SIVmus, SIVdeb, SIVagm gri, and SIVagm ver. Each of the accessory proteins was packaged in the VLPs (28). The VLPs were then tested on the indicator cells. SIVmac239, HIV-2rod, and HIV-2-7312a Vpx degraded the GFP fusion protein and are known to bind to the C terminus of SAMHD1 (36) (Fig. 1F). SIVrcm, SIVmnd2, SIVagm gri, and SIVagm ver Vpr had no effect, and each of these is known to be inactive against human SAMHD1 (13). Vpr of SIVmus and SIVdeb, which target a more N-terminal site on SAMHD1, did not degrade NLS.GFP.SAM595 (36, 37). Taken together, these data demonstrate the specificity of the assay for the interaction of Vpxmac with the C-terminal Vpx-binding site on SAMHD1.
The release of Vpx from virions requires processing of Gag.
Upon entry of the virus into the cytoplasm, Vpx is thought to be released and then transit to the nucleus. Whether this requires uncoating of the capsid is unknown; neither is it known whether Vpx is located inside or outside the capsid. If Vpx is inside the capsid, as might be predicted given its association with p6 at the carboxy-terminal portion of Gag, uncoating would be required for its release. If outside the capsid, Vpx could be released upon removal of the matrix shell without a need for capsid uncoating. To determine the requirements for the release of Vpx, we treated the virus producer cells with the protease inhibitor nelfinavir. Protease inhibitor treatment causes virions to retain an immature morphology in which the p55Gag polyprotein forms a ring-shaped structure underneath the envelope lipid bilayer and the mature capsid does not form (38–40). Such structures cannot uncoat as they lack a functional capsid and are likely to be stable structures. To test the requirement for Gag processing, we produced Vpx-containing VLPs in cells treated with nelfinavir. Immunoblot analysis showed that the virions contained mainly unprocessed or partially processed p55Gag with no fully processed CA and that they packaged normal amounts of Vpx (Fig. 1D). The VLPs were applied to NLS.GFP.SAM595 cells and were analyzed 24 h later by flow cytometry (Fig. 1B) and microplate fluorometry (Fig. 1C). The results showed that nelfinavir-treated VLPs failed to induce the degradation of NLS.GFP.SAM595, suggesting that Gag processing and formation of a mature virion(s) are required to allow the release of Vpx.
SAMHD1 is degraded in the nucleus.
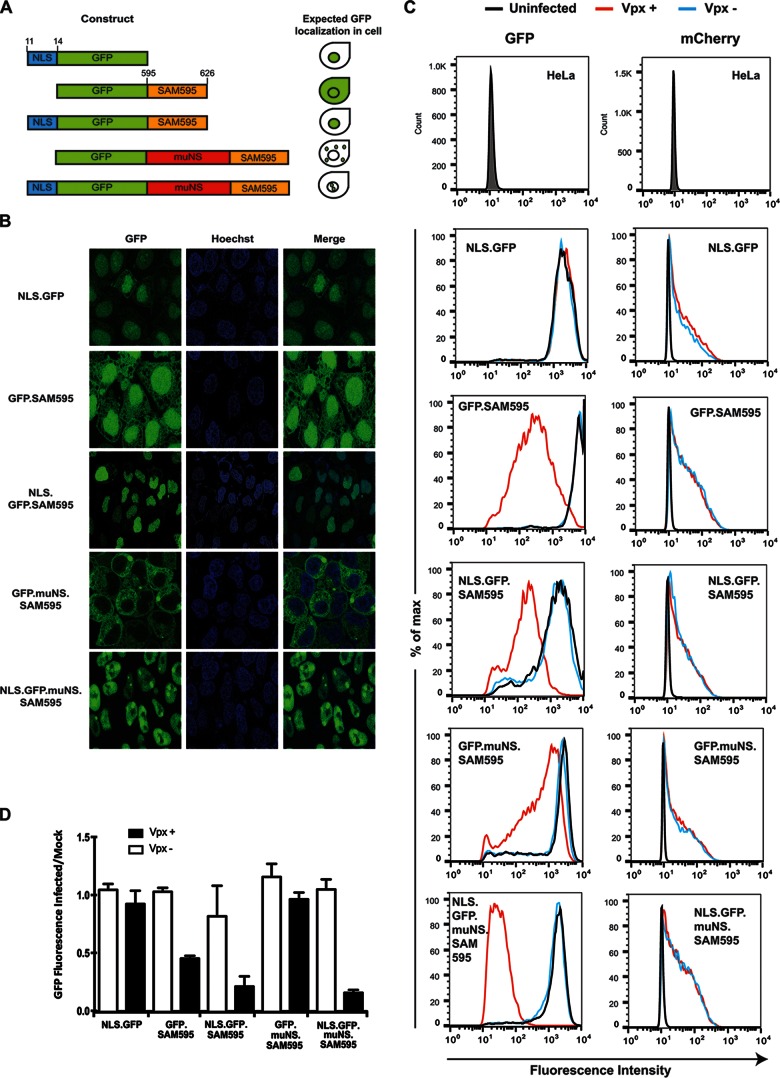
SAMHD1 and Vpx localize to the nucleus, but whether that is the site of SAMHD1 degradation is not clear. We previously reported that deletion of the SAMHD1 NLS rendered the protein resistant to Vpx-induced degradation and that leptomycin B, a drug that prevents the nuclear export of nuclear proteins, did not interfere with Vpx-induced degradation (28), findings that are consistent with those of others (18) and that suggest that SAMHD1 is targeted and degraded in the nucleus. However, it has also been reported that NLS-deleted SAMHD1 is degraded by a cytoplasmic HIV-2 Vpx (29) and that leptomycin B blocks the degradation of SAMHD1, suggesting that the degradation occurs in the cytoplasm as the result of nucleocytoplasmic shuttling (32). To further investigate this issue, we generated cell lines that expressed GFP.SAM595 fusion proteins localized to the nucleus or the cytoplasm. The proteins were constructed with or without an amino-terminal NLS. These included GFP linked to an amino-terminal NLS (NLS.GFP), GFP fused to a Vpx-binding site of SAMHD1 (GFP.SAM595 and NLS.GFP.SAM595), and GFP fused to SAM595 and muNS, a reovirus nonstructural protein that forms inclusion bodies in the cytoplasm (GFP.MuNS.SAM595 and NLS.GFP.muNS.SAM595). Clonal cell lines expressing each of the fusion proteins were established by lentiviral vector transduction. Confocal microscopy showed that as predicted, NLS.GFP localized to the nucleus, GFP.SAM595 was both cytoplasmic and nuclear, NLS.GFP.SAM595 localized to the nucleus, GFP.MuNS.SAM595 was cytoplasmic, and NLS.GFP.muNS.SAM595 localized to the nucleus (Fig. 2A and B).
FIG 2.
Vpx-mediated degradation occurs in the nucleus. (A) The structure of GFP.SAMHD1 fusion proteins used to generate clonal HeLa cell lines is diagrammed. The numbers on the top indicate amino acids corresponding to the NLS and SAMHD1 Vpx-binding sites. The predicted cellular localization of the proteins is diagrammed on the right. (B) The HeLa cell lines were stained with Hoechst 33342 and visualized by confocal microscopy to localize the GFP.SAM595 fusion proteins. (C) HeLa clones expressing the different constructs were infected at an MOI of 0.5 with HIV-1 mCherry reporter virus containing Vpx (Vpx+) or lacking Vpx (Vpx−). GFP and mCherry were measured 3 days postinfection by flow cytometry. The panels on the left show GFP fluorescence. The panels on the right show mCherry fluorescence as a measure of the number of infected cells. Untransduced and uninfected HeLa cells were used as a negative control. max, maximum. (D) The amount of GFP in the infected HeLa cell clones measured 24 h postinfection in a microplate fluorometer and normalized to GFP fluorescence of uninfected cells. The bars represent the means of the results determined with quadruplicate wells, and the error bars represent the standard deviations. Mock, mock infected.
To determine the sensitivity of the fusion proteins to Vpx, we infected the cell lines with mCherry-expressing HIV-1 reporter virus that contained or lacked Vpx and, after 3 days, quantified GFP and mCherry by flow cytometry. mCherry served as an internal control to determine the number of infected cells. While HeLa cells express SAMHD1, no effect of Vpx on infectivity was expected in HeLa cells since SAMHD1 is phosphorylated in cycling cells, which prevents its antiviral function (41, 42). Analysis of GFP showed that the NLS.GFP protein was not affected by Vpx. The intensity of GFP.SAM595 was moderately decreased, while NLS.GFP.SAM595 was efficiently degraded. GFP.muNS.SAM595 was resistant to Vpx, and the addition of an NLS resulted in efficient degradation (Fig. 2C). Analysis of mCherry fluorescence showed that similar numbers of cells were infected and that there was no effect of Vpx on the infectivity of the viruses. Analysis of the GFP fluorescence 24 h after infection in a microplate fluorometer yielded similar results (Fig. 2D). Thus, only those proteins that localized to the nucleus were efficiently degraded, and the extent of nuclear localization was associated with the efficiency of degradation. We conclude that nuclear localization is required for susceptibility of SAMHD1 to Vpx.
The kinetics of Vpx-mediated SAMHD1 degradation.
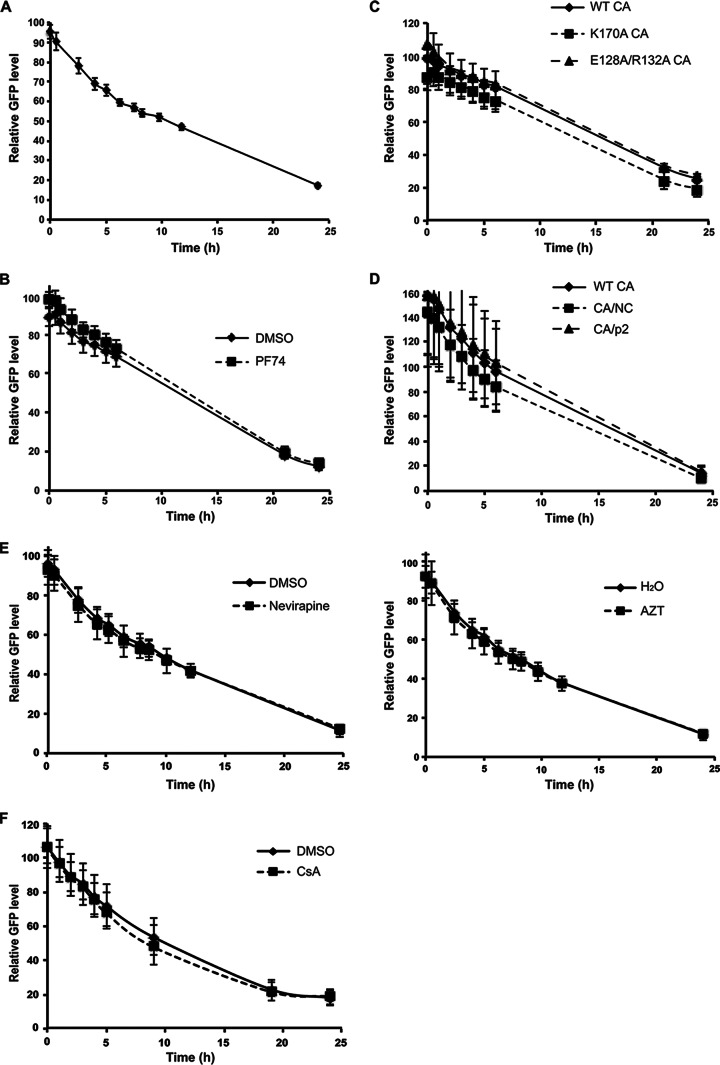
In HIV-infected myeloid cells, SAMHD1 must be degraded before reverse transcription can proceed. This requires that Vpx act as soon as possible upon virus entry into the cytoplasm such that the dNTP pool is increased before the virus becomes inactive. To track the kinetics of Vpx-mediated degradation, we synchronized the infection of the NLS.GFP.SAM595 cells by spin infection with Vpx-containing mCherry reporter HIV-1 at 16°C. Unbound virions were removed, and the cultures were then transferred to 37°C. We measured GFP fluorescence in the synchronously infected cells over a span of 24 h. To determine whether SAMHD1 degradation was associated with uncoating, we tested the effect of PF74, a drug that binds to a groove on the capsid, on the kinetics of SAMHD1 degradation. In the absence of drug, GFP fluorescence began to decrease 30 min postinfection, and the levels continued to drop over 24 h (Fig. 3A). Treatment of the cells with PF74 at 10 μM, a concentration that blocks infection (data not shown), had no effect on the kinetics of SAMHD1 degradation (Fig. 3B). In addition, we tested the effect of capsid point mutations that affect the rate of uncoating by altering capsid stability (34). This included the unstable K170A CA and hyperstable E128A/R132A CA (34). We found that there was no effect of either CA mutation on the kinetics of Vpx-mediated degradation (Fig. 3C). Mutations at the proteolytic processing sites in Gag also affect capsid stability such that mutation at the CA/NC junction blocks uncoating whereas mutation of the CA/p2 junction destabilizes the capsid (35). Analysis of virus containing these two mutations showed that neither affected the kinetics of SAMHD1 degradation (Fig. 3D). A similar analysis using a 5-fold-lower volume of virus showed similar kinetics, indicating the results were not due to the use of an excessive amount of virions (data not shown). In addition, we tested whether the reverse transcriptase (RT) inhibitors nevirapine and AZT would affect SAMHD1 degradation, as it has been suggested that reverse transcription stimulates uncoating (43, 44). Neither had an effect on Vpx-mediated degradation kinetics (Fig. 3E). Cyclophilin A (CypA) binding to CA is required for uncoating, and the treatment of the cells with cyclosporine (CsA) blocks the binding of CypA to CA (45, 46); however, CsA also had no effect on the kinetics of Vpx-mediated degradation (Fig. 3F). Taken together, these findings suggest that Vpx function does not require virus uncoating.
FIG 3.
The kinetics of Vpx-mediated degradation. (A) NLS.GFP.SAM595 cells were synchronously infected with Vpx-containing or Vpx-lacking mCherry reporter HIV-1. GFP was quantified over 24 h. (B) NLS.GFP.SAM595 cells were infected in the presence of 10 μM PF74 or control solvent dimethyl sulfoxide (DMSO). (C) NLS.GFP.SAM595 cells were infected with mCherry viruses containing wild-type CA (WT CA), destabilized CA (K170A CA), or hyperstable CA (E128A/R132A CA). (D) NLS.GFP.SAM595 cells were infected with mCherry reporter HIV-1 containing wild-type CA (WT CA), destabilized gag cleavage mutant (CA/p2), or hyperstable CA gag cleavage mutant (CA/NC). (E) NLS.GFP.SAM595 cells were infected with Vpx-containing or Vpx-lacking mCherry reporter virus in the presence or absence of the RT inhibitor nevirapine (left panel) or AZT (right panel). (F) NLS.GFP.SAM595 cells were infected with Vpx-containing or Vpx-lacking mCherry reporter virus in the presence or absence of the CypA inhibitor cyclosporine (CsA). GFP fluorescence was measured in a microplate fluorometer at multiple time points following infection. Relative levels of GFP fluorescence were determined by comparing GFP fluorescence in cells infected with Vpx-positive (Vpx+) virus to that in cells infected with the corresponding Vpx− virus. The measurements are the means of the results determined with quadruplicate wells, with error bars showing the standard deviations.
Long-lasting depletion of SAMHD1 by Vpx.
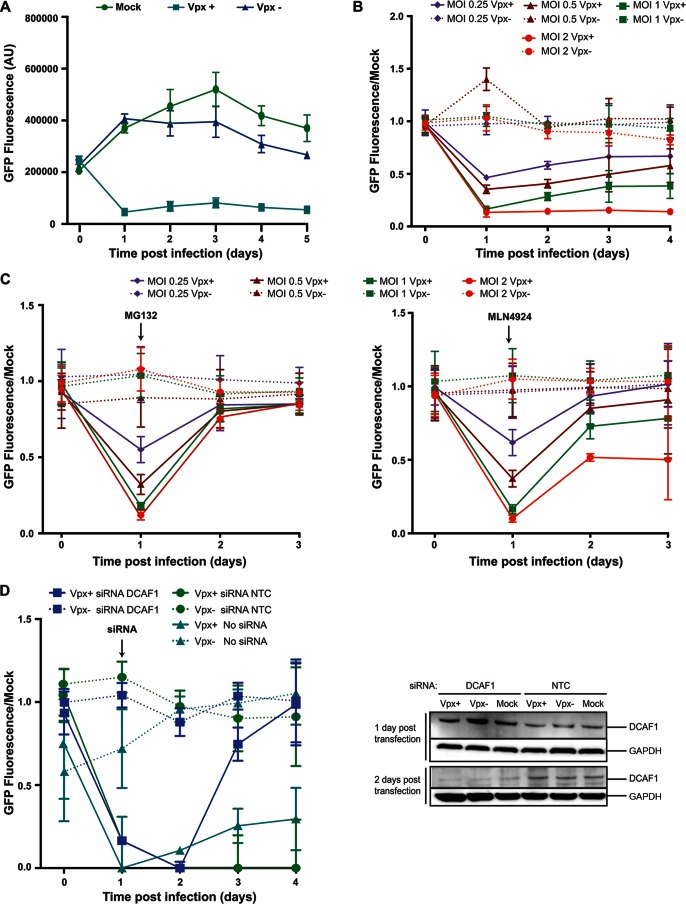
Reverse transcription in macrophages proceeds more slowly than in activated T cells, taking more than 36 h to complete (47). Therefore, increased dNTP levels need to be maintained over that time period in order for infection to occur. The relatively small number of Vpx molecules brought into the cell with the virion must be able to degrade newly made SAMHD1 until the completion of reverse transcription. To determine the length of time that input Vpx molecules continue to suppress SAMHD1 levels, we synchronously infected NLS.GFP.SAM595 cells with Vpx-containing mCherry reporter HIV-1 and measured GFP fluorescence over a span of 5 days. The results showed that NLS.GFP.SAM595 was depleted after 24 h, with the reduced levels being maintained over 5 days, suggesting that Vpx continued targeting newly synthesized NLS.GFP.SAM595 for degradation over several days (Fig. 4A).
FIG 4.
Vpx-mediated degradation is long-lived. (A) NLS.GFP.SAM595 cells were infected with mCherry reporter HIV-1 containing (Vpx+) or lacking (Vpx−) Vpx at an MOI of 1, and GFP fluorescence was measured over 5 days. (B) NLS.GFP.SAM595 cells were infected with Vpx-containing or Vpx-lacking mCherry reporter HIV-1 at the indicated MOI, and GFP fluorescence was measured over 4 days. The data were normalized to the GFP fluorescence of uninfected cells (Mock). (C) NLS.GFP.SAM595 cells were treated with 10 μM MG132 (left) or 1 μM MLN4924 (right) and then infected with Vpx-containing or Vpx-lacking mCherry HIV-1 reporter virus. GFP fluorescence was measured over 4 days. The data were normalized to the fluorescence of uninfected cells (Mock). (D) NLS.GFP.SAM595 cells were infected with Vpx-containing or Vpx-lacking virus and, 24 h later, transfected with 50 nM DCAF1 or nontemplate control (NTC) siRNA. (Left) GFP fluorescence was measured over 4 days and normalized to the GFP fluorescence of uninfected cells (Mock). (Right) DCAF1 was quantified on an immunoblot probed with specific antibody. The blot was probed with anti-GAPDH as a loading control.
We tested lower multiplicities of infection (MOI) to determine whether the sustained degradation was the result of the high MOI used in the initial experiments. At MOI of 2, 1.5, 1, 0.5, and 0.25, Vpx retained its ability to induce the sustained decrease in GFP fluorescence. At an MOI of 0.25, GFP degradation at 24 h was half that seen at the higher MOI, suggesting that the amount of Vpx introduced was limiting. Even at this low MOI, the decrease in GFP fluorescence was maintained over the 4-day time course (Fig. 4B).
The sustained depression in GFP levels suggests that the input Vpx continues to induce SAMHD1 degradation over several days. To determine whether Vpx maintains its ability to induce proteasomal degradation of SAMHD1, we introduced Vpx into the HeLa.NLS.GFP.SAM595 cells and added MG132 or MLN4924 after SAMHD1 was degraded. MG132 caused GFP levels to recover, indicating that proteasomal activity remains high over time (Fig. 4C, left panel). MLN4924 partially restored the GFP, and the effect was more evident at higher MOI, suggesting that some of the E3 ubiquitin ligase complexes are stable and maintain neddylation or that Vpx uses another E3 ubiquitin ligase complex that does not contain cullin (Fig. 4C, right panel). The greater effect at higher MOI could be explained by the fact that if the affinity of Vpx for the other possible complex were lower than its affinity for CRL4DCAF1 at higher MOI, there would be more Vpx available for the other complex to bind.
To determine whether the complexes formed by Vpx binding to the CRL4 E3 ubiquitin ligase complex are stable or continually disassemble and reform, we treated the cells with Vpx-containing mCherry reporter HIV-1. The following day, we knocked down DCAF1 by siRNA transfection and then measured GFP fluorescence. The knockdown of DCAF1 was confirmed by immunoblot analysis (Fig. 4D, right panel). The results showed that the GFP level recovered upon knockdown of DCAF1 (Fig. 4D, left panel). Because DCAF1 knockdown affects only newly produced CRL4 complexes and has no effect on assembled ligase complexes, these results suggest that the Vpx/CRL4 complexes are not stable over time. Rather, they appear to have been continually reforming and assembling with a stable Vpx.
A possible DDB1-independent pathway for Vpx-induced degradation of SAMHD1.
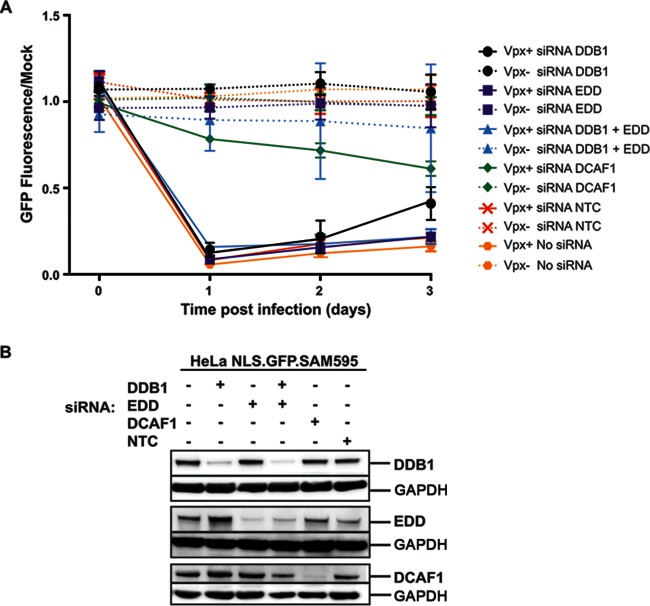
The CRL4DCAF1 complex is composed of DDB1, RBX1, Cul4, and DCAF1, and it is believed that Vpx interacts directly with DCAF1 in the complex (14, 16, 17). While Vpx induces SAMHD1 degradation through CRL4DCAF1, it is possible that it can use other E3 ubiquitin ligases as well. To test the requirement for CRL4, we knocked down the DCAF1 and DDB1 components of CRL4 in NLS.GFP.SAM595 cells by siRNA transfection and tested the ability of Vpx-containing VLPs to induce degradation of the NLS.GFP.SAM595 fusion protein after 24 h. The knockdowns were confirmed by immunoblot analysis (Fig. 5B). We found that DCAF1 knockdown prevented degradation of NLS.GFP.SAM595 but that DDB1 knockdown had no effect on degradation (Fig. 5A). These results suggest that the degradation of SAMHD1 requires DCAF1 but that DDB1 may not be required. DCAF1 associates with the HECT-family EDD/UBR5 E3 ubiquitin ligase complex, which could be an alternative degradation pathway. However, knockdown of both EDD alone and EDD together with DDB1 did not prevent degradation of the fusion protein (Fig. 5A), suggesting that the EDD/UBR5 E3 ubiquitin ligase complex does not serve as a unique alternative degradation pathway.
FIG 5.
DDB1 knockdown does not block Vpx-mediated degradation. (A) NLS.GFP.SAM595 cells were transfected with DDB1 and/or EDD, DCAF1, or nontemplate control (NTC) siRNA. After 3 days, the cells were infected with 10 ng p24 VLPs that contained (Vpx+) or lacked (Vpx−) Vpx. GFP fluorescence was measured over 3 days. Data were normalized to the fluorescence of uninfected cells (Mock). (B) At 3 days posttransfection, DCAF1, DDB1, and EDD were analyzed on an immunoblot probed with specific antibodies and with anti-GAPDH as a loading control.
DISCUSSION
The NLS.GFP.SAM595 fusion protein retains the properties of full-length SAMHD1, making it a useful tool for studying Vpx-induced degradation. Removal of the NLS or fusion to the muNS cytoplasmic protein caused the protein to be resistant to Vpx-induced degradation whereas addition of an NLS to the muNS fusion rendered the protein sensitive to Vpx-induced degradation, further supporting the idea that SAMHD1 is targeted by Vpx and degraded in the nucleus. An analysis of drugs and point mutations in the virion showed that the kinetics with which Vpx induced the degradation of SAMHD1 was independent of capsid uncoating. The effect of Vpx on SAMHD1 was remarkably long-lasting, consistent with findings of Hollenbaugh et al. (48). This effect was caused by the ability of Vpx to continue to stimulate the DCAF1-binding CRL4 E3 ubiquitin ligase over at least 5 days. Degradation of the fusion protein was dependent upon DCAF1 but was not affected by siRNA knockdown of DDB1, suggesting that Vpx can use another E3 ubiquitin ligase or that CRL4 complexes exist that use a different linker protein.
The location of Vpx within virions has not been fully determined. Studies in which HIV-2 or SIV capsids were purified through sucrose density centrifugation differed with respect to whether Vpx is capsid associated (49, 50). We found that Vpx-containing virions composed of hyperstable or unstable capsid mutants induced the degradation of Vpx with normal kinetics. In addition, PF74 or CsA or RT inhibitors such as nevirapine or AZT, compounds that alter the kinetics of uncoating (43–46, 51–53), also had no effect on the kinetics of Vpx-mediated degradation. In contrast, virions produced in the presence of protease inhibitor failed to deliver Vpx-mediated degradation to the fluorescent reporter cells. These findings suggest that the release of Vpx requires that a capsid be formed such that a significant fraction of Vpx is located between the matrix shell and the capsid. This location is unexpected given the interaction of Vpx with p6, which the geometry of the Gag precursor polyprotein would place within the capsid. One possible interpretation of these data is that as the virion matures, Vpx is released from p6, allowing it to move to a location outside the mature capsid. However, further experiments will be needed to test this hypothesis.
Previous studies suggested that SAMHD1 is targeted by Vpx for ubiquitination and degraded in the nucleus (18, 28). Deletion of the NLS in SAMHD1 relocalized the protein to the cytoplasm and caused it to be resistant to Vpx-induced degradation. In addition, leptomycin B, a drug that prevents nuclear/cytoplasmic shuttling, did not interfere with SAMHD1 degradation. The possibility that Vpx could induce degradation of SAMHD1 in the cytoplasm was raised by studies in which cytoplasmic SAMHD1 was degraded by a cytoplasmic HIV-2 Vpx (29) and in which leptomycin B prevented SAMHD1 degradation (32). Using the NLS.GFP.SAM595 cells, we found that fusion of reovirus muNS, a cytoplasmic protein, to GFP.SAM595 blocked the degradation mediated by Vpx. Addition of an NLS localized it to the nucleus and restored its susceptibility to Vpx-induced degradation. In our analysis, the cytoplasmic fusion protein was degraded by Vpx to a small but significant extent, which could account for the divergence from the findings by others. Overexpression of the protein and the ability of DCAF1 to bind to E3 ubiquitin ligases complexes with different affinities (54) might allow for some Vpx-mediated degradation of the cytoplasmic protein. However, this phenomenon is minimal compared to the degradation of the nuclear protein by the E3 ubiquitin ligase complex that binds Vpx-DCAF1 with high affinity.
Following treatment of the NLS.GFP.SAM595 cells with Vpx-containing virions, GFP levels remained low for a prolonged period of time, suggesting that Vpx continues to actively deplete SAMHD1 over a relatively long period of time, consistent with the findings of Hollenbaugh et al. (48). In our analysis, SAMHD1 levels remained low for 5 days, after which the cultures became overgrown, preventing further measurement. The depletion of SAMHD1 was maintained by continued targeting of the CRL4 E3 ubiquitin ligase and proteasomal degradation, as knockdown of DCAF1 or treatment with MG132 during the time course restored SAMHD1 levels. The finding that knockdown of DCAF1 prevents SAMHD1 degradation suggests that the interaction of Vpx with CRL4 is not stable but that the Vpx/CRL4 complexes continually disassemble and reassemble with Vpx. If the initial complexes had been stable, the DCAF1 knockdown would not have interfered with SAMHD1 degradation. Treatment with the neddylation inhibitor MLN4924 partially restored SAMHD1 levels, suggesting that the Nedd8 conjugation is also short-lived, at least in most of the complexes.
Knockdown of DCAF1 prior to infection prevented the Vpx-induced degradation of SAMHD1, consistent with previous findings (1, 2, 37, 55), but knockdown of DDB1 had no effect. It is possible that the siRNA knockdown of DDB1 was not effective; however, the knockdown appeared to be as efficient as the DCAF1 knockdown. The result suggests that Vpx can degrade SAMHD1 using another DCAF1-binding E3 ubiquitin ligase or that the CRL4 complex can use a linker protein other than DDB1. One alternative candidate was the EDD E3 ubiquitin ligase; however, knockdown of EDD had no effect on SAMHD1 degradation.
The events involved in HIV uncoating are not well understood. Uncoating initiates as early as 1 h after viral fusion and may not be completed until the virus docks with the nuclear pore (43, 56–59). Uncoating is facilitated by the process of reverse transcription, in which the growing viral DNA forces the capsid to open (43, 44, 60). The ability of Vpx to act independently of uncoating would provide a means for the rapid degradation of SAMHD1, and the subsequent rise in dNTP levels would facilitate uncoating. Because the reverse transcription complex has a relatively short half-life, a rapid increase in the dNTP level allows the virus to synthesize its DNA genome before becoming inactivated in the cytoplasm.
The ability of Vpx to act without a need for uncoating provides a means for the virus to quickly induce a rise in dNTP levels. If reverse transcription is required to facilitate uncoating, then the release of Vpx prior to uncoating would be a necessity. The ability of Vpx to be released without a need for uncoating suggests that it is localized outside the capsid. Such a localization would require rearrangement of its topology as the virion matures to form the mature capsid. The ability of Vpx to act for a long time ensures that dNTP levels remain high over the several hours required to complete reverse transcription in macrophages.
The NLS.GFP.SAM595 cell line described here expresses a fusion protein that contained only the C-terminal Vpr/Vpx-binding site; however, GFP fusion proteins that contained the N-terminal binding site could be expressed as a means to study the effect of the proteins that recognize this site. The fluorescent cell lines generated here may find useful application in screens for cellular cofactors involved in Vpx-mediated degradation or in screens for small-molecule inhibitors of the ubiquitin proteasome system.
ACKNOWLEDGMENTS
We thank Esperanza Agulló Pascual for assistance with confocal microscopy, Chris Bianco for assistance with pLenti-mCherry cloning, Christopher Aiken for providing HIV plasmids containing point-mutated capsids and PF74, Felipe Diaz-Griffero for providing muNS expression plasmid, and Paul Bieniasz for providing HIV-CMV-mCherry plasmid. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: nelfinavir, zidovudine (AZT), and nevirapine.
This work was supported by grants from the NIH (AI058864 and AI067059).
REFERENCES
- 1.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 6.Powell RD, Holland PJ, Hollis T, Perrino FW. 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem 286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J, Kaur S, DeLucia M, Hao C, Mehrens J, Wang C, Golczak M, Palczewski K, Gronenborn AM, Ahn J, Skowronski J. 2013. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem 288:10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. 2014. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med 20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. 2009. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chahwan C, Chahwan R. 2012. Aicardi-Goutieres syndrome: from patients to genes and beyond. Clin Genet 81:413–420. doi: 10.1111/j.1399-0004.2011.01825.x. [DOI] [PubMed] [Google Scholar]
- 13.Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. 2012. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. 2012. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287:12550–12558. doi: 10.1074/jbc.M112.340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem 287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, Skowronski J. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog 4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergamaschi A, Ayinde D, David A, Le Rouzic E, Morel M, Collin G, Descamps D, Damond F, Brun-Vezinet F, Nisole S, Margottin-Goguet F, Pancino G, Transy C. 2009. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J Virol 83:4854–4860. doi: 10.1128/JVI.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaller T, Pollpeter D, Apolonia L, Goujon C, Malim MH. 2014. Nuclear import of SAMHD1 is mediated by a classical karyopherin alpha/beta1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation. Retrovirology 11:29. doi: 10.1186/1742-4690-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann H, Norton TD, Schultz ML, Polsky SB, Sunseri N, Landau NR. 2013. Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J Virol 87:11741–11750. doi: 10.1128/JVI.02002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa T, Mondal K, Swanson PC. 2013. VprBP (DCAF1): a promiscuous substrate recognition subunit that incorporates into both RING-family CRL4 and HECT-family EDD/UBR5 E3 ubiquitin ligases. BMC Mol Biol 14:22. doi: 10.1186/1471-2199-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Singh S, Jung HY, Yang G, Jun S, Sastry KJ, Park JI. 2013. HIV-1 Vpr protein inhibits telomerase activity via the EDD-DDB1-VPRBP E3 ligase complex. J Biol Chem 288:15474–15480. doi: 10.1074/jbc.M112.416735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Conway JA, Kim J, Kappes JC. 1994. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol 68:6161–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Accola MA, Bukovsky AA, Jones MS, Gottlinger HG. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J Virol 73:9992–9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selig L, Pages JC, Tanchou V, Preveral S, Berlioz-Torrent C, Liu LX, Erdtmann L, Darlix J, Benarous R, Benichou S. 1999. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol 73:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunseri N, O'Brien M, Bhardwaj N, Landau NR. 2011. Human immunodeficiency virus type 1 modified to package simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J Virol 85:6263–6274. doi: 10.1128/JVI.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwefel D, Groom HC, Boucherit VC, Christodoulou E, Walker PA, Stoye JP, Bishop KN, Taylor IA. 2014. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLucia M, Mehrens J, Wu Y, Ahn J. 2013. HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation. J Biol Chem 288:19116–19126. doi: 10.1074/jbc.M113.469007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR. 2012. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J Virol 86:12552–12560. doi: 10.1128/JVI.01657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandariz-Nuñez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. doi: 10.1186/1742-4690-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei W, Guo H, Han X, Liu X, Zhou X, Zhang W, Yu XF. 2012. A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell Microbiol 14:1745–1756. doi: 10.1111/j.1462-5822.2012.01835.x. [DOI] [PubMed] [Google Scholar]
- 31.Guo H, Wei W, Wei Z, Liu X, Evans SL, Yang W, Wang H, Guo Y, Zhao K, Zhou JY, Yu XF. 2013. Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation. PLoS One 8:e66201. doi: 10.1371/journal.pone.0066201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobadilla S, Sunseri N, Landau NR. 2013. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther 20:514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forshey BM, Shi J, Aiken C. 2005. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J Virol 79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. 2013. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog 9:e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W, Guo H, Gao Q, Markham R, Yu XF. 2014. Variation of two primate lineage-specific residues in human SAMHD1 confers resistance to N terminus-targeted SIV Vpx proteins. J Virol 88:583–591. doi: 10.1128/JVI.02866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyma DJ, Jiang J, Shi J, Zhou J, Lineberger JE, Miller MD, Aiken C. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol 78:3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol 72:2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodward CL, Cheng SN, Jensen GJ. 12 November 2014. Electron cryo-tomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. J Virol doi: 10.1128/JVI.02997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 43.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y, Fricke T, Diaz-Griffero F. 2013. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol 87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Iaco A, Luban J. 2014. Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology 11:11. doi: 10.1186/1742-4690-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collin M, Gordon S. 1994. The kinetics of human immunodeficiency virus reverse transcription are slower in primary human macrophages than in a lymphoid cell line. Virology 200:114–120. doi: 10.1006/viro.1994.1169. [DOI] [PubMed] [Google Scholar]
- 48.Hollenbaugh JA, Tao S, Lenzi GM, Ryu S, Kim DH, Diaz-Griffero F, Schinazi RF, Kim B. 2014. dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology 11:63. doi: 10.1186/s12977-014-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kewalramani VN, Emerman M. 1996. Vpx association with mature core structures of HIV-2. Virology 218:159–168. doi: 10.1006/viro.1996.0176. [DOI] [PubMed] [Google Scholar]
- 50.Yu X, Matsuda Z, Yu QC, Lee TH, Essex M. 1993. Vpx of simian immunodeficiency virus is localized primarily outside the virus core in mature virions. J Virol 67:4386–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J Virol 87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fricke T, Brandariz-Nunez A, Wang X, Smith AB III, Diaz-Griffero F. 2013. Human cytosolic extracts stabilize the HIV-1 core. J Virol 87:10587–10597. doi: 10.1128/JVI.01705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol 85:542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharifi HJ, Furuya AK, Jellinger RM, Nekorchuk MD, de Noronha CM. 2014. Cullin4A and cullin4B are interchangeable for HIV Vpr and Vpx action through the CRL4 ubiquitin ligase complex. J Virol 88:6944–6958. doi: 10.1128/JVI.00241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pertel T, Reinhard C, Luban J. 2011. Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology 8:49. doi: 10.1186/1742-4690-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu H, Franks T, Gibson G, Huber K, Rahm N, De Castillia CS, Luban J, Aiken C, Watkins S, Sluis-Cremer N, Ambrose Z. 2013. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 10:70. doi: 10.1186/1742-4690-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hulme AE, Kelley Z, Okocha Z, Hope TJ. 22 October 2014. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hakata Y, Miyazama M, Landau NR. 2014. Interactions with DCAF1 and DDB1 in the CRL4 E3 ubiquitin ligase are required for Vpr-mediated G2 arrest. Virol J 11:108. doi: 10.1186/1743-422X-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]