Abstract
As the fields of tissue engineering and regenerative medicine mature toward clinical applications, the need for online monitoring both for quantitative and qualitative use becomes essential. Resazurin-based metabolic assays are frequently applied for determining cytotoxicity and have shown great potential for monitoring 3D bioreactor-facilitated cell culture. However, no quantitative correlation between the metabolic conversion rate of resazurin and cell number has been defined yet. In this work, we determined conversion rates of Presto Blue™, a resazurin-based metabolic assay, for human periosteal cells during 2D and 3D static and 3D perfusion cultures. Our results showed that for the evaluated culture systems there is a quantitative correlation between the Presto Blue conversion rate and the cell number during the expansion phase with no influence of the perfusion-related parameters, that is, flow rate and shear stress. The correlation between the cell number and Presto Blue conversion subsequently enabled the definition of operating windows for optimal signal readouts. In conclusion, our data showed that the conversion of the resazurin-based Presto Blue metabolic assay can be used as a quantitative readout for online monitoring of cell proliferation in a 3D perfusion bioreactor system, although a system-specific validation is required.
Introduction
The development of cell-based tissue engineering strategies for the repair or replacement of damaged organs and tissues is a rapidly evolving research field.1,2 As these novel cell-based therapies fall under the definition of the advanced therapy medicinal products (ATMPs) of the European Medicines Agency (EMA), the application of the Process Analytical Technology guidelines (PAT) to develop well-characterized products by designing and controlling the manufacturing process through timely measurements of critical quality attributes is essential.3–5 The integration of bioreactor systems in current laboratory-scale processes therefore holds promise for the translation in a clinical and ultimately commercial setting.6,7 Bioreactors have been employed frequently to provide sufficient nutrient and oxygen transport and removal of waste products6,8–12 while allowing for monitoring and control of physicochemical and biological parameters7,13–17 during cell proliferation, differentiation, and the development of cell–carrier constructs. Furthermore, these parameters could be used as nondestructive quality indicators of the cells or the developing construct.
Since in vitro proliferation is an essential process step for the production of cell-based products,9 online measurements of metabolic activity parameters, such as glucose, lactate, and oxygen concentration, could allow for a nondestructive assessment of the quality of the 3D cell culture.14,16–18 Although, for example, correlating a decrease in oxygen concentration to cell proliferation has been shown to be a viable strategy to monitor proliferation for high cell numbers,16 these techniques often fall short when lower cell numbers in the order of magnitude of 104 to 106 cells need to be monitored and quantified.13 Limited sensor sensitivity as well as low oxygen and glucose consumption rates per cell hamper the reliability of such measurements.13 Therefore, alternate methodologies to monitor cell numbers, viability, and proliferations as well as differentiation in bioreactors are essential, especially for the initial crucial expansion steps (postbiopsy expansion) of scarce adult stem cell populations.
Metabolic assays, such as the tetrazolium-based 3-[4-5-dimethylthiazol-2-yl]-2-5-diphenyl bromide tetrazolium bromide (MTT) and the resazurin-based Alamar Blue® (AB) assay, use the redox activity of the cells to qualitatively monitor cell populations based on the generation of an optical or fluorescent detectable component.19–24 MTT and other tetrazolium-based assays are based on the cleavage and reduction of the tetrazolium ring to blue formazan crystals by the mitochondrial dehydrogenases.22,23 These crystals can be solubilized and quantified in the cell lysate as a measure for metabolic cell activity. Despite the frequent use of these assays, the fact that this methodology can only be used as an endpoint analysis is a serious disadvantage.22 The resazurin-based AB and Presto Blue® (PB) assays, on the other hand, use the mitochondrial activity to reduce the nonfluorescent blue resazurin to the fluorescent pink resorufin.22,23,25 This nontoxic water-soluble dye enables continuous cell culture monitoring and has been shown to be a powerful tool to assess cell viability and proliferation in both static and dynamic 3D setups.13,21,26,27 These reports show a clear correlation between the obtained fluorescent signal and the cell number. However, contradictory results showing discrepancies between the cell number and the metabolic conversion of the resazurin when using static culture systems are also present, indicating that the performance of these assays is dependent on the cell type, culture, and measurement setup.22,28
To determine whether the quantitative use of a resazurin-based assay in a bioreactor setup is possible, the influence of the cell culture method and measurement setup on the conversion rate of the resazurin and on the metabolic activity of the cells has to be known. The redox activity of a cultured cell population was already shown to be influenced by long-term proliferation and the induction of differentiation.29,30 The influence of different culture systems and parameters on the metabolic activity of the cells and the conversion rate of the metabolic assays is, however, not known, which hampers the quantitative use of these methods.
In this work, we used the PB metabolic assay to monitor and quantify cell proliferation in a perfusion bioreactor setup for up to 21 days. At first, the influence of different culture setups was investigated by measuring the metabolic activity of cells cultured in 2D and in 3D static and 3D perfusion bioreactor setups, and correlating this to DNA content. Subsequently, a dynamic monitoring method was developed to overcome diffusion limitations that were observed when 3D scaffolds were measured in a static setup affecting the conversion rate. Finally, the influence of perfusion flow rate and shear stress on the conversion of the metabolic assay was determined by using a medium with different viscosities. Based on DNA measurements, conversion rates for the PB assay in the different setups were found to enable the quantitative monitoring of cell proliferation.
Materials and Methods
Human periosteal-derived cells
Human periosteal-derived cells (hPDCs) were isolated from periosteal biopsies obtained from four different donors (aged 11, 13, 14, and 17, with equal distribution of gender) as described previously and pooled for further use.31 This procedure was approved by the ethics committee for Human Medical Research (KU Leuven) and performed with patient informed consent. hPDCs were expanded in Dulbecco's modified Eagle's medium with high-glucose (Life Technologies) containing 10% fetal bovine serum (Gibco), 1% sodium pyruvate (Life Technologies), and 1% antibiotic–antimycotic (100 units/mL penicillin, 100 mg/mL streptomycin, and 0.25 mg/mL amphotericin B; Life Technologies), further mentioned as culture medium. The cells were seeded at 5700 cells/cm2 and passaged at 80–90% confluency. Cell expansion and 2D experiments were performed in standard cell culture conditions (relative humidity: 95%, 5% CO2, 37°C).
Ti6Al4V scaffolds
For the 3D cell culture, additive manufactured Ti6Al4V scaffolds (Ø=6 mm, h=6 mm)32,33 were used as described before.34 The total volume of the scaffolds was 166±3 mm3, the available volume was 130±5 mm,3 and the available surface was 7.5±0.6 mm2 as determined with nanoCT.35 Briefly, scaffolds were ultrasonically cleansed before use in acetone, ethanol, and distilled water for 10 min each, after which they received an alkaline treatment with 5 M sodium hydroxide for 24 h at 60°C. Subsequently, scaffolds were rinsed with distilled water, after which they were sterilized in a steam autoclave. Before seeding, the scaffolds were prewetted by vacuum impregnation with culture medium for 2 h in standard cell culture conditions, after which they were dried in a nonhumidified incubator overnight.36
Viscosity measurements of culture medium
The viscosity of the culture medium was increased by the addition of dextran (average molecular weight (M/W) 150,000; Sigma). The viscosity of solutions containing 0%, 2.5%, 5%, 7.5%, and 10% dextran was determined using an MCR 501 rheometer (Anton Paar). A C-LTD 180/XL measurement chamber was used to enable shear stress measurements at a constant temperature of 37°C. For each measurement, the solution was presheared at a shear rate of 100/s for 30 s, after which a rate sweep from 100/s to 0.1/s shear rate was performed using a log ramp. Data points corresponding with torques lower than 0.001 mNm were discarded due to inaccuracy of the measurements. Since τ=η*γ (with τ, shear stress; η, viscosity; and γ, shear rate), the viscosity of the solution could be determined by correlating the applied shear rate to the measured shear stress.
Two-dimensional AB and PB measurements
For 2D experiments, cells were seeded at the standard density of 5700 cells/cm2 in six-well plates and cultured statically in 2 mL of growth medium. For 6 days, the metabolic activity of the cells was measured daily using both PB and AB for three wells each (both metabolic assays were obtained from Life Technologies). For both reagents, a measurement solution containing 1 unit volume of reagent for 10 unit volumes of culture medium was prepared, resulting in a 9.09% solution (standard concentration measuring solution, unless otherwise mentioned). The medium was removed from the wells and replaced with 2 mL of the PB or AB solution, after which the cells were transferred back to the incubator at normal cell culture conditions. At the different incubation times (15, 30, 45, 60, 90, and 120 min) a 100 μL sample was taken from every well and transferred to a 96-well plate. The fluorescent signal was measured with an automated microplate fluorometer (SerColab Systems) using an excitation wavelength of 544 nm and an emission wavelength of 590 nm, as described earlier.13 The measured fluorescent signal was expressed as arbitrary fluorescent units (FU). For every measurement, the background signal of the measurement solution was determined by incubating an identical volume in the absence of cells in the experimental conditions. This blank signal was subsequently subtracted from the signal measured in the presence of cells to obtain the signal induced by the presence of the cells. After the measurement, the PB or AB solution was removed from the wells and replaced by normal culture medium. For the measurement on day 5, 3 mL of the measuring solution was used to allow for a higher number of sampling points. Additional samples were taken after 3, 4, 5, and 6 h of incubation.
To determine the quantitative correlation between the PB signal and the cell number in a 2D static setting, two different approaches were used. Six-well plates were seeded with 100,000 cells each, and cells were allowed to attach over night, after which the culture medium was removed and 2, 4, or 6 mL of the PB solution was added on triplicate wells. Cells were incubated for 8 h in total and 100 μL samples were taken every hour and transferred to a 96-well plate for measurement. Alternatively, different amounts of cells were seeded in triplicate in six-well plates (25,000, 50,000, 75,000, 100,000, 200,000, and 300,000). Cells were incubated overnight to allow cell attachment, after which the culture medium was removed and 2 mL of PB solution was added. Samples were taken every hour for 8 h and measured as described before.
Three-dimensional static cell culture and PB measurements
For 3D culture experiments, cells were drop-seeded onto the scaffolds at a density of 200,000 cells/60 μL as described in previous studies.13,34,36,37 Forty-five minutes after seeding, 60 μL of culture medium was added and 135 min later the medium volume was topped up to 1 mL. Scaffolds were statically incubated overnight in standard culture conditions (37°C, 5% CO2, 95% relative humidity). Since the seeding process resulted in homogenous and reproducible seeding efficiencies in previous work (∼60%), this was not assessed separately in this work.34,36,37
To determine the quantitative correlation between the PB signal and the cell content in a 3D static culturing system, scaffolds were transferred to 12-well plates containing 3 mL of culture medium after seeding. The medium was refreshed every 2 days and PB measurements were performed on days 1, 6, 10, 15, and 22 by replacing the culture medium with 3 mL of the PB solution and incubating the scaffolds in standard cell culture conditions for 3 h. The PB signal was measured in triplicate as described before. At each PB time point, three scaffolds were sacrificed for DNA measurement as described below.
Three-dimensional perfusion bioreactor culture and PB measurement
For the 3D perfusion bioreactor culture, 3D Ti6Al4V scaffolds were seeded in the bioreactor system using the same methods as described for the static system.34,36,37 Scaffolds were subsequently press-fitted in an in-house developed bioreactor chamber13,34,36,37 ensuring forced perfusion through the scaffold and cultured at a flow rate of 1 mL/min in a nonhumidified incubator (37°C, 5% CO2). The total medium volume in each circuit was 10 mL, of which 7.2 mL was located in the medium reservoir. The medium was refreshed every second day by attaching a falcon tube containing fresh medium to the system. For PB measurements, a falcon tube containing 2.2 mL of a 20.5% PB solution was connected to the circuit resulting in a total volume of 5 mL and a final PB concentration of 9.09% as used for the static measurements. The PB solution was perfused for 1 h at 1 mL/min, after which the resulting PB signal in the solution was measured. To remove remnants of the PB solution in the bioreactor circuits for continued cell culture, two falcon tubes containing 5 mL of culture medium were subsequently connected to the circuits for a perfusion step of 5 min each, after which final medium refreshment was performed as described before. After seeding, the scaffolds were dynamically cultured for 21 days and PB measurements were performed in triplicate on days 2, 7, 12, 17, and 21. After each PB measurement three scaffolds were sacrificed for DNA measurement.
To determine the influence of both culture and the measurement setup (static vs. bioreactor), 3D cell-seeded scaffolds cultured for 14 days in both systems were measured either in the respective culture systems or consecutively both in a static and perfusion bioreactor setup. All conditions were performed and measured in triplicate. In three additional samples cultured in both setups, no PB measurements were performed to verify that the performed handlings had no influence on the cell number. All samples were used for DNA measurement after the experiment.
To determine the influence of different flow rates during 3D perfusion bioreactor culture on the metabolic activity of cells in the 3D scaffolds, and thereby on the conversion rates of the PB, they were cultured at different flow rates (0.5, 1.1, and 2.2 mL/min) for 21 days, after which the metabolic activity of the cells was determined using PB. The flow rates used during the PB measurement were the same as those used during culture for all conditions. The cell content of the scaffolds was also determined based on DNA measurements. Additionally, two conditions, in which the volumetric flow rate was 0.5 mL/min, but the shear stress was equal to, respectively, the 1.1 and 2.2 mL/min condition by increasing the viscosity of the medium as described earlier (Viscosity measurements of culture medium section) using, respectively, 3.37% and 6.36% dextran (average MW 150,000; Sigma), were evaluated.
DNA measurement
The DNA content was determined using a highly quantitative and selective DNA assay (Quant-iT™ dsDNA HS kit; Life Technologies). The scaffolds were rinsed with phosphate-buffered saline and lysed in 350 μL RLT lysis buffer supplemented with 3.5 μL β-mercaptoethanol (Qiagen). The lysed samples were vortexed for 60 s and stored at −80°C before analysis, thawed at room temperature, and spun down for 1 min at 13,000 rpm. Ten microliters of the sample was diluted in 90 μL milliQ water, after which the DNA content was quantified with a Qubit® Fluorometer (Life Technologies) as described by Chen et al.38
Mathematical representation of the PB measurement in the perfusion bioreactor system using ordinary differential equations
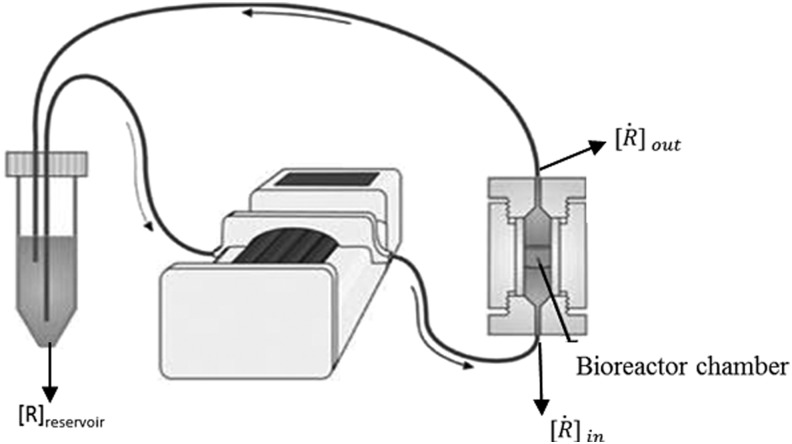
A schematic representation of the bioreactor system is shown in Figure 1. The concentration of the resorufin in the medium reservoir could be determined based on a mass balance over the reservoir taking into account the flow rate dependent on inward and outward fluxes [Eq. (1)]. The outward flux was defined based on the flow rate and the resorufin concentration [Eq. (2)]. The inward flux in the medium reservoir was equal to the outward flux at the scaffold, although a flow rate and tubing volume-dependent time delay had to be implemented to account for the time the measurement suspension required to be perfused from the scaffold back to the medium reservoir [Eq. (3)]. The flux at the outlet of the scaffold was determined by the ingoing flux and the cell-mediated resazurin conversion [Eq. (4)], while the flux at the inlet of the scaffold was defined based on the concentration in the medium reservoir, again implementing the flow rate and tubing volume-dependent time delay to account for the time required to perfuse the solution from the medium reservoir to the scaffold [Eq. (5)].
FIG. 1.
Schematic representation of the perfusion bioreactor system. (Adapted from Zhou et al.13)
At the initial time point the resorufin concentration in the medium reservoir was set at 0, while the maximal value was set at 3500, as these were determined to be the borders of the linear operating window.
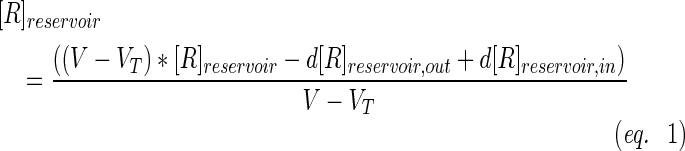 |
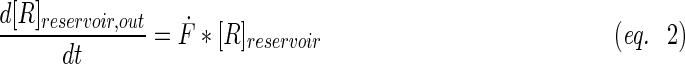 |
 |
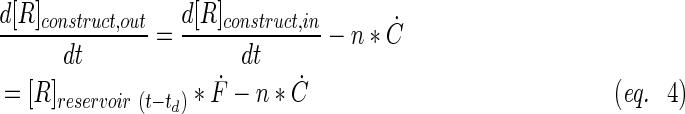 |
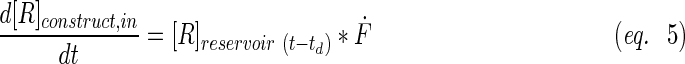 |
With
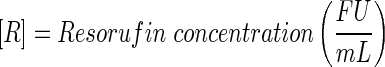 |
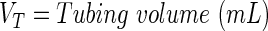 |
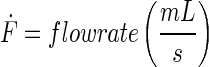 |
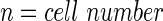 |
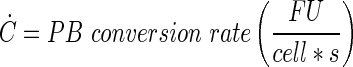 |
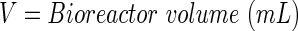 |
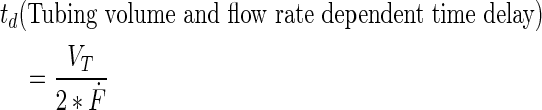 |
Statistical analysis
One-way ANOVA was performed to analyze significant differences between groups. A p-value <0.05 was considered significant.
Results
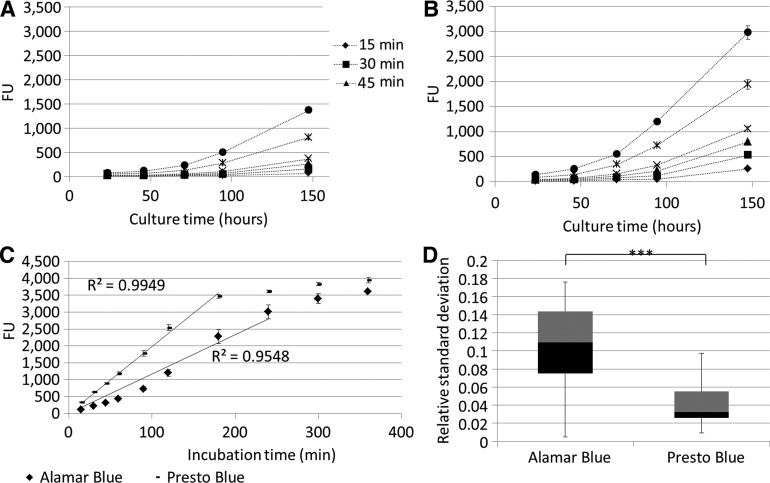
Initially, the PB metabolic assay was compared with the previously validated AB assay13 by monitoring for both assays' cell proliferation in a static 2D setup for different incubation times (Fig. 2A, B). Both metabolic assays showed an increase in metabolic activity in function of culture time, in accordance with the expected exponential phase of cell growth. Although the same incubation times and concentrations were used, the PB signal was significantly higher compared with the AB signal for the same incubation time. This was also visible by the stronger increase in signal shown in Figure 2C, in which the linear operating window of both assays was determined, respectively, up to 3000 FU for the AB assay and 3500 for the PB assay. The relative standard deviation on the measured signal was also significantly lower for the PB measurement as shown in Figure 2D.
FIG. 2.
(A, B) Two-dimensional static expansion of human periosteal-derived cells monitored with Alamar Blue and Presto Blue, respectively, for different incubation times (n=3) (C) Signal saturation determined for Presto Blue and Alamar Blue based on 2D static expansion. The lines indicate the linear operating zone for both assays (R2 values are shown for a fit forced through 0;0) (n=3) (D) Boxplot of relative standard deviation for 2D static Presto Blue and Alamar Blue measurements (n=60). ***p<0.001. Black and grey zones indicate 2nd and 3rd quartile of the boxplot.
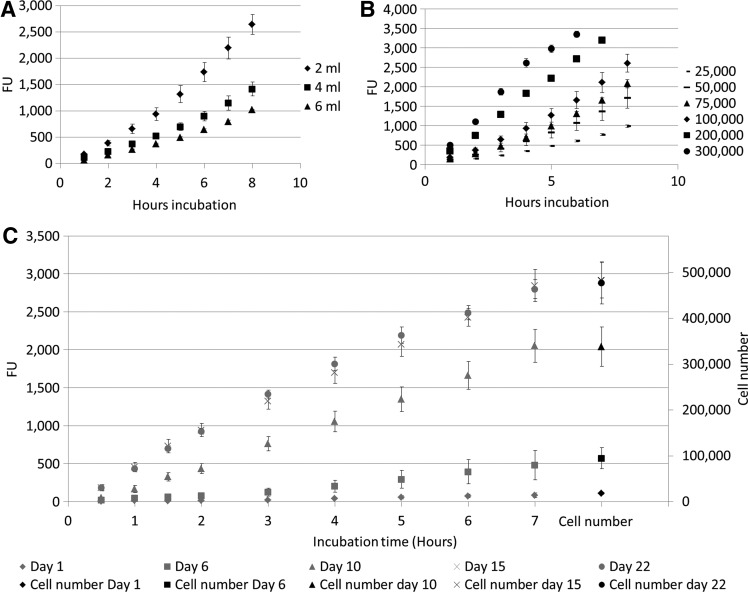
Conversion rates for PB were determined in a static setup for both 2D and 3D cell culture systems. For all conditions, a time-dependent linear increase in the measured signal was observed (Fig. 3). Linear regression was used to determine the slope coefficients, which indicated the increase in FU per hour for all the different conditions (Table 1). These values were corrected for incubation time, cell number, and volume and were then averaged over the different experimental conditions. For the case of the 2D cell culture, the two different setups used to determine the conversion rate resulted in a similar average value of ∼616 FU converted per hour for 100,000. For the measurement in the 3D static culture system, a linear correlation was again observed between the PB obtained signal and the cell number based on DNA measurement as shown in Figure 3C. The resulting PB conversion rate was, however, significantly lower than the one obtained for the 2D static system (Table 1).
FIG. 3.
(A) Fluorescent signal measured for different volumes of Presto Blue solution for 100,000 cells in a 2D static setup (n=3). (B) Fluorescent signal measured for different cell numbers in 2 mL Presto Blue solution in a 2D static setup (n=3). (C) Presto Blue signal in function of incubation time for different time points during 3D static culture and corresponding cell number based on DNA measurements (n=3).
Table 1.
Observed Increase in Presto Blue Signal per Hour for Different Static Culture Conditions Based on Figure 3 and the Representative Presto Blue Conversion Rates
| Increase in fluorescent signal/h | R2value | FU converted by 100,000 cells in 1 h (FU/100,000 cells×h) | ||
|---|---|---|---|---|
| Cell number (2D) | ||||
| 25,000 | 107 | 0.95 | 605±176 | 616±141 |
| 50,000 | 188 | 0.94 | ||
| 75,000 | 225 | 0.93 | ||
| 100,000 | 288 | 0.95 | ||
| 200,000 | 449 | 0.99 | ||
| 300,000 | 592 | 0.98 | ||
| Volume Presto Blue solution (mL) (2D) | 628±38 | |||
| 2 | 295 | 0.95 | ||
| 4 | 157 | 0.95 | ||
| 6 | 111 | 0.95 | ||
| Days of culture (3D) | 237±26 | |||
| 1 | 12.5 | 0.94 | ||
| 6 | 66.8 | 0.93 | ||
| 10 | 275 | 0.98 | ||
| 15 | 415 | 0.99 | ||
| 22 | 421 | 0.99 | ||
FU, fluorescent units.
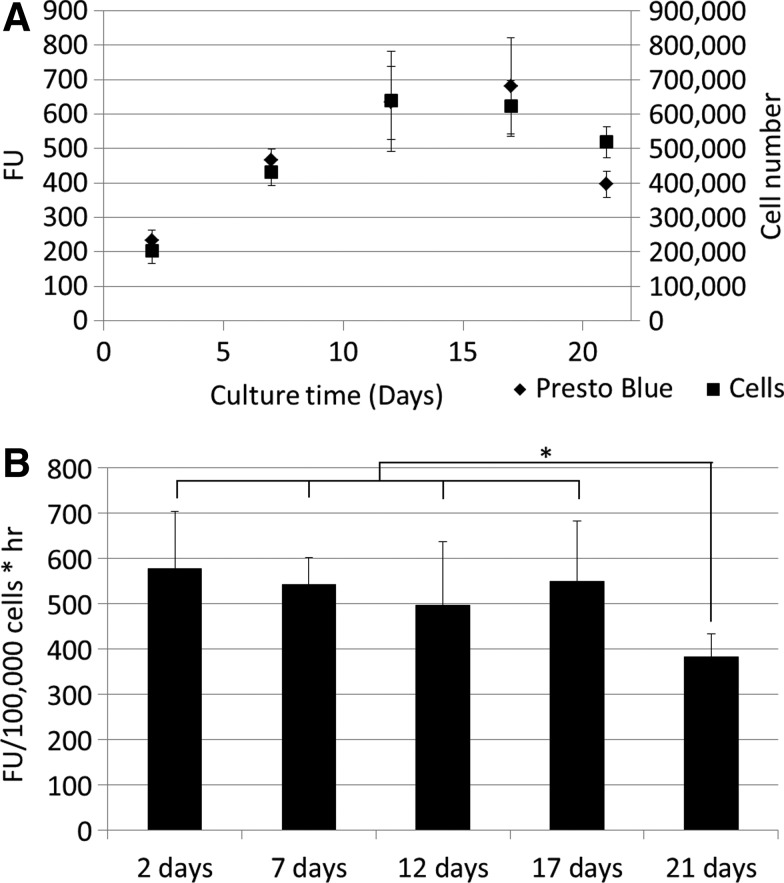
As shown for both the 2D and 3D static culture and measurement setups, the PB signal and DNA content measured for perfusion bioreactor-expanded cells showed no significant differences after fitting the curves as shown in Figure 4A, in which both the increase in FU and the increase in cell number are represented on the Y-axis. Normalizing the signal to the volume and cell number resulted in conversion rates, which were not significantly different from the earlier determined 2D conversion rates during the proliferative phase (Fig. 4B). As also shown in Figure 4A, the metabolic conversion rate of the cells in the scaffold does decrease once confluency is reached as determined with DNA measurements (Fig. 4B).
FIG. 4.
(A) Cell number and Presto Blue signal during 21 days of 3D perfusion bioreactor culture for different time points. (B) Presto Blue conversion rates for different time points during 3D perfusion bioreactor culture. *0.05<p<0.01.
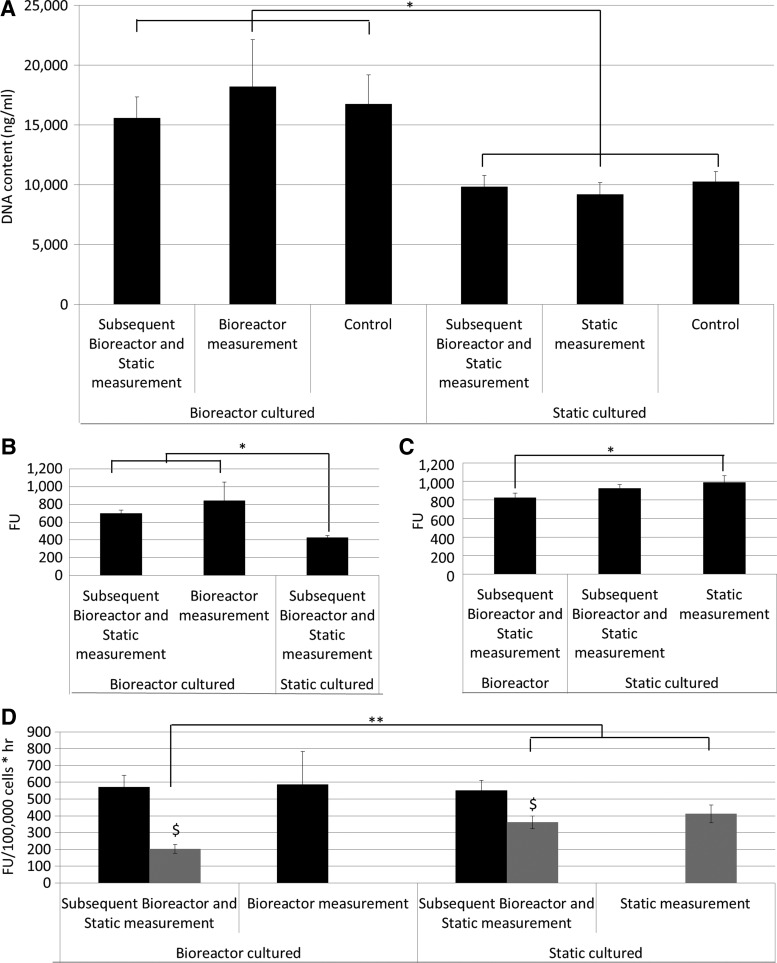
To determine the influence of the culture and measurement setup on the PB signal, bioreactor and static measurements of cells expanded in both 3D bioreactor and static setups were performed. Figure 5A shows a significantly higher DNA content for all 3D bioreactor-expanded cells in comparison with the static 3D expanded cells. No significant difference was observed between the cells expanded under the same conditions, indicating that the handlings performed for the measurements did not influence the cell content. Despite the significant differences observed in DNA content, no differences in PB signal were observed between the 3D static and bioreactor-expanded cells using the static measurement setup (Fig. 5C). However, the measurements performed in the bioreactor system showed a significant difference between the bioreactor and the statically-expanded cells (Fig. 5B). Since different incubation times and PB volumes were used for the bioreactor and static measurements, the obtained signal was corrected for cell number, time, and volume. Figure 5D shows that a similar conversion rate was determined for all bioreactor measured conditions, independently from the culture setup. The conversion rates determined for static measurements were significantly lower in comparison with all bioreactor measured conditions. Additionally, the conversion rates determined for the bioreactor cultured, static measured samples were significantly lower than those for the statically cultured and measured.
FIG. 5.
(A) DNA content (ng/mL) after 3D cell expansion for different culture and measurement setups, as shown on the axis (n=3). (B) Presto Blue signal after either 3D static or perfusion bioreactor cell expansion measured in the perfusion bioreactor system (n=3). (C) Presto Blue signal after either 3D static or perfusion bioreactor cell expansion measured in the static system (n=3). (D) Conversion rates for 3D constructs cultured in a bioreactor or static setup determined using a bioreactor (black) or static (gray) measurement setup (n=3). *0.05<p<0.01, **0.01<p<0.001, $ indicates significant differences between dynamic and static measurement for the same construct, p<0.05.
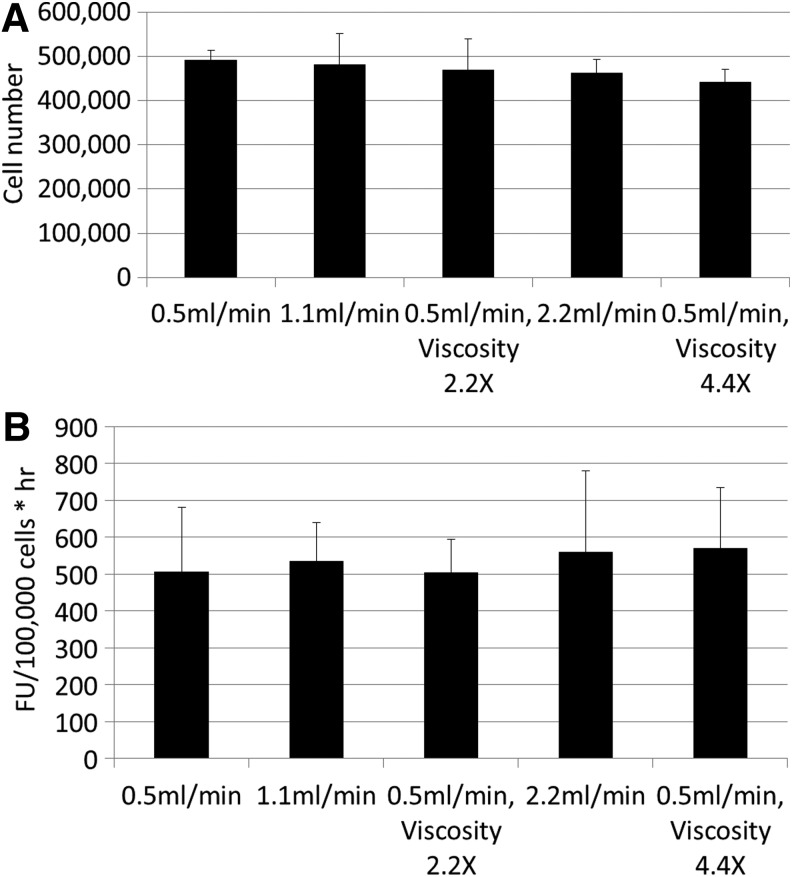
No significant differences in cell proliferation were observed when using different flow rates, nor did the increased viscosity and the correlated shear stress for the low volumetric perfusion influence the final cell number (Fig. 6A). Additionally, the conversion rates determined for the different conditions were not influenced by the perfusion velocity (Fig. 6B) indicating that, next to the proliferation, the PB conversion rates and therefore the metabolic activity of the cell population was not influenced by flow rate within the examined range corresponding with initial shear stress values between 7E-3 and 3.08E-2 Pa.37
FIG. 6.
(A) Cell content for 3D constructs cultured in the perfusion bioreactor based on DNA measurement for different flow rates and medium viscosities (n=3). (B) Presto Blue conversion rate for 3D constructs cultured in the perfusion bioreactor with different flow rates and medium viscosities.
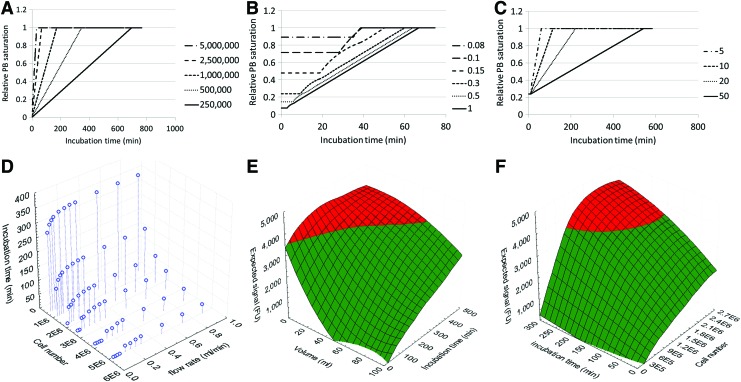
Although flow rate does not influence the conversion rate of the PB assay, it should be high enough to prevent local saturation of the assay at the outlet of the scaffold. To determine at which combinations of flow rate and cell number this could influence the readouts, mass balance equations were used to determine the relative saturation of the linear PB operating window in the medium reservoir and at the scaffold inlet and outlet in functions of flow rate, cell number, measurement volume, and incubation time. The resulting resorufin concentrations at the scaffold outlet obtained from the time-dependent mass balances were subsequently converted to relative PB saturation as shown in Figure 7A–C. Although flow rate did not influence the conversion rate of the assay, flow rate-dependent changes in PB saturation were observed (Fig. 7B). Figure 7D subsequently shows for which combinations of incubation time, cell number, and flow rate saturation at the scaffold outlet is reached, allowing defining an operating window for the measurement.
FIG. 7.
Presto Blue saturation at the bioreactor chamber outlet determined for (A) different cell numbers, (B) different flow rates (mL/min), (C) different measurement volumes (mL), (D) incubation time (min), after which saturation of the Presto Blue assay is reached at the bioreactor chamber outlet for specific combinations of cells and flow rate (mL/min), (E) Operating window for Presto Blue measurement of a bioreactor chamber containing a scaffold with 2,500,000 cells valid for flow rates above 0.4 mL/min, and (F) Operating window for a Presto Blue measurement in a 3D perfusion bioreactor system containing 5 mL of measurement solution valid for flow rates above 0.4 mL/min. The red area shows the measurement conditions, which result in a value outside the linear operating window of the assay. The green area shows the measurement conditions resulting in values within the linear operating window of the assay. Color images available online at www.liebertpub.com/tec
Discussion
A number of reports have shown that metabolic assays such as the AB assay can be used to monitor cell proliferation,13,21 although no quantitative correlation between the conversion rate of the reagent and the cell number has been defined. To enable a quantitative use of this tool, we investigated the influence of three different cell culture methods and correlated measurement setups, including bioreactor culture, on the conversion of the resazurin-based PB metabolic assay.
Initially, a validated metabolic activity assay, the AB assay,13 was compared with the PB assay for conventional 2D cell expansion. As shown in Figure 2C, the maximum signal obtained in the linear operating range was ∼3000 FU for the AB assay and 3500 for the PB assay, indicating that the latter has a significantly larger operating window. Additionally, the time-dependent increase in the signal was significantly higher at each time point for the PB (Fig. 2A–C), demonstrating that this assay has a higher sensitivity power, which was also confirmed by comparing the relative standard deviations obtained for both assays (Fig. 2D).
Table 1 shows that a linear correlation between the PB signal and cell content exists (R2=0.87). Due to the increase in the cell number and the correlated decrease in available surface for proliferation, a decreasing trend in PB conversion in function of cell number was, however, present. Nonetheless, no significant differences were observed between the average conversion rates determined using different volumes and cell numbers and a quantitative correlation between the cell number and PB signal could be established. Although other groups reported discrepancies between resazurin conversion of the AB assay and cell number,22,28 these could probably have been induced by the high cell density in the culture systems used. When performing a similar experiment for static 3D cell culture and monitoring proliferation in function of time, a quantitative relationship between the cell number and PB signal was again observed (Fig. 3C), although a significantly lower conversion rate was obtained (Table 1). For the perfusion bioreactor 3D cell culture, the PB conversion rate was significantly higher than observed for the static 3D system and no difference was found in relation to the 2D setup (Fig. 4B). Since previous studies already showed that cell growth in 3D scaffolds in a static culture setup is predominantly located at the outer edges of the scaffolds due to limitations in nutrient diffusion,34,37,39–41 the difference in the PB conversion rate might be related to limited diffusion.
To confirm that the different conversion rates observed between 3D static and perfused culture systems originated from enhanced mass transport in the perfusion bioreactor system, the conversion rate for cells expanded both in the 3D static or bioreactor system were determined using both static and bioreactor measurement setups. As shown in Figure 5A, the measurement methods and manipulations did not influence the final cell content of the scaffolds. Despite the significant differences in DNA content, the PB signal measured in the static setup was not influenced by the culture conditions (Fig. 5C). Diffusion limitations possibly influenced the accessibility of the cells in the center of the densely populated bioreactor cultured scaffolds.39–41 To confirm this hypothesis, the maximal diffusion depth of the resazurin in the scaffolds was determined using a partial differential equation describing the spatiotemporal evolution of resazurin (cR) as discussed by Demol et al.42 As no data were available on resazurin diffusion in an engineered neo-tissue, the diffusion coefficient (D) of resazurin in aquatic systems was used as an approximation (2.04×10−6 cm2/s).43 The resazurin conversion rate (Q) was based on the results discussed earlier and cells were assumed to be homogenously distributed in the neo-tissue with a density (ccell) based on previous nano-CT volumetric results.34,37
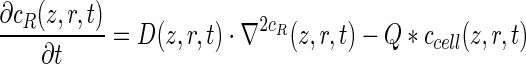 |
The steady-state solution of the system enabled determining a maximal diffusion depth of 700 μm. This results in an active volume of 40% of the total scaffold, implying that only the cells present in this volume can contribute to the resazurin conversion. This corresponds to the discrepancy between the apparent conversion rates determined for bioreactor cultured, static measured scaffolds, and bioreactor cultured and measured scaffolds, in which the resazurin is perfused through the scaffold, for which the static determined conversion rate was only 35% of the dynamic-based value (Fig. 5D). For the bioreactor measured conditions, we did observe an influence of the culture system on PB conversion (Fig. 5B).
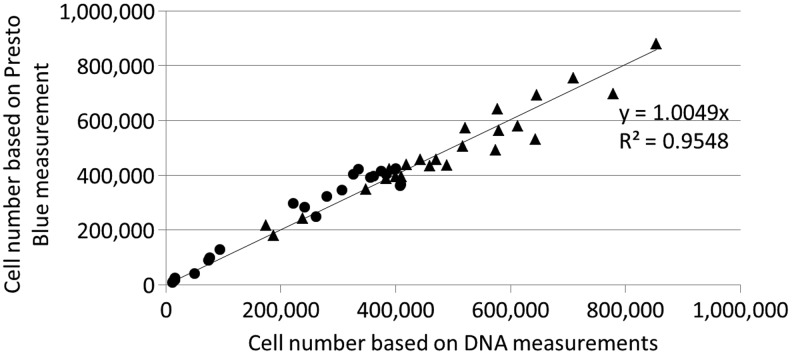
To compare conversion rates between the two measurement setups, the PB signal was corrected for volume, time, and cell number (Fig. 5D), which showed that conversion rates measured with the bioreactor setup were not influenced by the culture system used. As the Péclet number was larger than 1, the convective mass transport in this measurement setup ensured a homogeneous distribution of the PB reagent throughout the scaffold independent from the cell distribution, and therefore a realistic conversion rate could be determined as opposed to apparent conversion rates obtained in the static diffusion-based setups. Since the used culture systems did not influence the conversion rates, the differences observed between the static and bioreactor measurement setups were not caused by changes in cell metabolic activity, but rather by a decreased conversion efficiency caused by diffusion limitations.39–41 Despite the lower conversion rate in the static setup, a correlation between the obtained PB signal and the DNA content still exists as shown in Figure 3C. This indicates that depending on the measurement system different conversion rates should be used and that quantitative use of the methodology requires a system-specific validation of the measurement. This was further validated by generating a correlation plot depicting the cell number based on the PB measurements in function of the cell number based on DNA measurements (Fig. 8). Taking into account the respective conversion rates for the static cultured/measured and dynamic cultured/measured expansion systems an R2 value of 0.9548 was obtained confirming the potential quantitative use of the methodology.
FIG. 8.
Correlation between the cell number determined on DNA measurement and cell number based on Presto Blue measurement for all 3D static and dynamic cell expansion performed for Figures 2–6 using the respective conversion rates determined for the two culture systems (▴, 3D dynamic expansion and measurement; ●, 3D static expansion and measurement).
On day 21 of culture, after the proliferative phase as could be observed in Figure 4A, both the increase in cell content and the PB signal and the conversion rate decreased significantly. As suggested for the 2D system (Fig. 3B and Table 1), the decreased conversion rate of the PB assay for high cell densities might be related to the decrease in cell proliferation. Alternatively, previous work showed a nonlinear correlation between the resazurin conversion and the cell number for high-density cell cultures, which was due to diffusion limitations both in 2D and 3D,28 although the fluid flow in the bioreactor system should circumvent these problems. Additionally, the high cell density in these constructs has been suggested to result in the further reduction of the fluorescent resorufin in the nonfluorescent colorless hydroresorufin,25 thereby resulting in a decreased amount of the fluorescent resorufin. However, since multiple groups showed a linear behavior of the assay within the defined operating conditions,13,21,22 the decrease in cell proliferation remains the most probable explanation for the observed decreased conversion rate of the PB.
To determine the applicability of the proposed methodology for a broader range of operating conditions, we subsequently also determined the influence of the use of different flow rates and shear stresses (using dextran to increase fluid viscosity) on the metabolic conversion rates of the assay. In a recent publication, we showed that the use of a range of flow rates (corresponding to initial shear stress values between 5.59E-4 and 5.59E-2 Pa) did not induce differentiation of the expanding hPDC cell population.37 Furthermore, a functional bone marrow-derived mesenchymal cell population was also expanded in an undifferentiated state under flow conditions in a perfusion bioreactor.44 In correspondence with these findings, we observed that the metabolic activity of the hPDCs is constant and independent from dextran concentration (affecting shear stress) and volumetric flow rate. Taking into account the determined conversion rates, expected number of cells, and the culture and measurement setup, these results now allow to define an operating window for each specific setup, which will allow a rational selection of operating conditions.36,45 Figure 7D shows for which combination of cell number, flow rate, and incubation time saturation of the assay is reached at the scaffold outlet, thus indicating the minimal flow rate and maximal incubation time required for monitoring an expansion system in function of the expected maximal cell number. It should be noted that despite the fixed assay volume and therefore the fixed amount of resazurin that can be converted, the maximal incubation time for a certain cell number varies in function of flow rate.
Next to the saturation of the assay due to high cell numbers and low flow rates, a significant resazurin concentration gradient will be present in the tubing of the system for high cell numbers and low flow rates. For higher flow rates, this concentration gradient will be negligible resulting in a constant, cell-dependent maximal incubation time. For these cases, the operating window of the assay can be determined based on the total circuit volume, incubation time, and expected cell number as shown in Figure 7E and F, in which the expected PB signal is shown in function of measurement volume and incubation time (Fig. 7E) or incubation time and cell number (Fig. 7F). This simplified representation of the system will be valid for flow rates that do not significantly influence the maximal incubation time as shown in Figure 7D.
Next to using this mathematical representation of the system to define optimal operating conditions, the quantitative correlation between PB conversion and the cell number can also be used to determine the cell number in the scaffold at any given time. Additionally, the theoretical boundaries of the window of operation could be determined for the dynamic measurement method, that is, convective perfusion through the constructs. For example, using a flow rate of 1 mL/min as employed for measurements in this study, cell numbers as low as 50,000 cells could be detected using 5 mL of PB solution, although at least 120 min of incubation time would be required to obtain a quantitative signal. On the other hand, using the same flow rate and 50 mL of PB solution, constructs containing up to 25,000,000 cells could be monitored using short incubation times between 15 and 20 min. A further increase in PB solution volume would even allow the further broadening of the window of operations, thereby enabling the methodology to effectively cover the entire range of cell densities used for tissue engineering applications, for which final cell densities in the engineered constructs were reported between 107 and 108 cells/cm3.9,10,16,31,39
In conclusion, we can state that the PB assay is a promising tool for online monitoring of cell proliferation in a 3D perfusion bioreactor system. Since the conversion of the metabolic assay was shown to be constant in function of time during cell proliferation, and the culture parameters such as flow rate and shear stress did not influence the metabolic activity or the conversion efficiency, a quantitative correlation between the cell number and PB conversion could be established. This allowed determining the assay concentration at each location in the setup, thereby enabling to define optimal operating conditions as well as calculating the cell number in the scaffold based on the PB concentration.
Acknowledgments
M.S. is supported by a PhD grant of the Agency for Innovation by Science and Technology (IWT/111457). I.P. is funded by the ENDEAVOUR project G.0982.11N of the Research Foundation Flanders (FWO Vlaanderen). This work is part of Prometheus, the Leuven Research and Development Division of Skeletal Tissue Engineering of the KU Leuven: www.kuleuven.be/prometheus.
Disclosure Statement
No competing financial interests exist.
References
- 1.Martin I., Baldomero H., Bocelli-Tyndall C., Passweg J., Saris D., and Tyndall A. The survey on cellular and engineered tissue therapies in Europe in 2010. Tissue Eng Part A 18, 2268, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Martin I., Baldomero H., Bocelli-Tyndall C., Slaper-Cortenbach I., Passweg J., and Tyndall A. The survey on cellular and engineered tissue therapies in europe in 2009. Tissue Eng Part A 17, 2221, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebulla P., Montemurro T., and Giordano R. Regulation of cell-based medicine: the European experience. ISBT Sci Series 5, 249, 2010 [Google Scholar]
- 4.Schneider C.K., Salmikangas P., Jilma B., Flamion B., Todorova L.R., Paphitou A., et al. . Challenges with advanced therapy medicinal products and how to meet them. Nat Rev Drug Discov 9, 195, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Chew W., and Sharratt P. Trends in process analytical technology. Anal Meth 2, 1412, 2010 [Google Scholar]
- 6.Salter E., Goh B., Hung B., Hutton D., Ghone N., and Grayson W.L. Bone tissue engineering bioreactors: a role in the clinic? Tissue Eng Part B Rev 18, 62, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Martin I., Riboldi S.A., and Wendt D. Bioreactor systems in regenerative medicine. In: Shastri V.P., Altankov G., and Lendlein A., eds. Advances in Regenerative Medicine: Role of Nanotechnology, and Engineering Principles. Netherlands: Springer, 2010, pp. 95–113 [Google Scholar]
- 8.Martin I., Wendt D., and Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol 22, 80, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues C.A.V., Fernandes T.G., Diogo M.M., da Silva C.L., and Cabral J.M.S. Stem cell cultivation in bioreactors. Biotechnol Adv 29, 815, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Haycock J.W. 3D cell culture: a review of current approaches and techniques. Methods Mol Biol 695, 1, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Jakob M., Saxer F., Scotti C., Schreiner S., Studer P., Scherberich A., et al. . Perspective on the evolution of cell-based bone tissue engineering strategies. Eur Surg Res 49, 1, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Grayson W.L., Ma T., and Bunnell B. Human mesenchymal stem cells tissue development in 3D PET matrices. Biotechnol Prog 20, 905, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Zhou X., Holsbeeks I., Impens S., Sonnaert M., Bloemen V., Luyten F.P., et al. . Non-invasive real-time monitoring by alamarBlue(R) during in vitro culture of 3D tissue engineered bone constructs. Tissue Eng Part C Methods 19, 720, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das R.H., van Osch G.J., Kreukniet M., Oostra J., Weinans H., and Jahr H. Effects of individual control of pH and hypoxia in chondrocyte culture. J Orthop Res 28, 537, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Rauh J., Milan F., Gunther K.P., and Stiehler M. Bioreactor systems for bone tissue engineering. Tissue Eng Part B Rev 17, 263, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Santoro R., Krause C., Martin I., and Wendt D. On-line monitoring of oxygen as a non-destructive method to quantify cells in engineered 3D tissue constructs. J Tissue Eng Regen Med 6, 696, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Tsao Y.S., Cardoso A.G., Condon R.G.G., Voloch M., Lio P., Lagos J.C., et al. . Monitoring Chinese hamster ovary cell culture by the analysis of glucose and lactate metabolism. J Biotechnol 118, 316, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Frese J., Morgenroth A., Mertens M.E., Koch S., Rongen L., Vogg A.T., et al. . Nondestructive monitoring of tissue-engineered constructs. Biomed Tech (Berl) 59, 165, 2014 [DOI] [PubMed] [Google Scholar]
- 19.Uzunoglu S., Karaca B., Atmaca H., Kisim A., Sezgin C., Karabulut B., et al. . Comparison of XTT and Alamar blue assays in the assessment of the viability of various human cancer cell lines by AT-101 (-/- gossypol). Toxicol Mech Methods 20, 482, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Back S.A., Khan R., Gan X., Rosenberg P.A., and Volpe J.J. A new Alamar Blue viability assay to rapidly quantify oligodendrocyte death. J Neurosci Methods 91, 47, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Mueller D., Tascher G., Damm G., Nussler A.K., Heinzle E., and Noor F. Real-time in situ viability assessment in a 3D bioreactor with liver cells using resazurin assay. Cytotechnology 65, 297, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quent V.M.C., Loessner D., Friis T., Reichert J.C., and Hutmacher D.W. Discrepancies between metabolic activity and DNA content as tool to assess cell proliferation in cancer research. J Cell Mol Med 14, 1003, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nociari M.M., Shalev A., Benias P., and Russo C. A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J Immunol Methods 213, 157, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Rampersad S.N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel) 12, 12347, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien J., Wilson I., Orton T., and Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267, 5421, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Gloeckner H., Jonuleit T., and Lemke H.D. Monitoring of cell viability and cell growth in a hollow-fiber bioreactor by use of the dye Alamar Blue. J Immunol Methods 252, 131, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Cui Z.F., Xu X., Trainor N., Triffitt J.T., Urban J.P., and Tirlapur U.K. Application of multiple parallel perfused microbioreactors and three-dimensional stem cell culture for toxicity testing. Toxicol In Vitro 21, 1318, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Ng K.W., Leong D.T.W., and Hutmacher D.W. The challenge to measure cell proliferation in two and three dimensions. Tissue Eng 11, 182, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Quinn K.P., Bellas E., Fourligas N., Lee K., Kaplan D.L., and Georgakoudi I. Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials 33, 5341, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice W.L., Kaplan D.L., and Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One 5, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyckmans J., and Luyten F.P. Species specificity of ectopic bone formation using periosteum-derived mesenchymal progenitor cells. Tissue Eng 12, 2203, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Van Bael, S., Kerckhofs G., Moesen M., Pyka G., Schrooten J., and Kruth J.P. Micro-CT-based improvement of geometrical and mechanical controllability of selective laser melted Ti6Al4V porous structures. Mater Sci Eng A Struct Mater 528, 7423, 2011 [Google Scholar]
- 33.Pyka G., Burakowski A., Kerckhofs G., Moesen M., Van Bael S., Schrooten J., et al. . Surface modification of Ti6Al4V open porous structures produced by additive manufacturing. Adv Eng Mater 14, 363, 2012 [Google Scholar]
- 34.Papantoniou I., Sonnaert M., Geris L., Luyten F.P., Schrooten J., and Kerckhofs G. Three-dimensional characterization of tissue-engineered constructs by contrast-enhanced nanofocus computed tomography. Tissue Eng Part C Methods 20, 177, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerckhofs G., Pyka G., Moesen M., Van Bael, S., Schrooten J., and Wevers M. High-resolution microfocus X-ray computed tomography for 3D surface roughness measurements of additive manufactured porous materials. Adv Eng Mater 15, 153, 2013 [Google Scholar]
- 36.Papantoniou I., Chai Y.C., Luyten F.P., and Schrooten J. Process quality engineering for bioreactor-driven manufacturing of tissue engineered constructs for bone regeneration. Tissue Eng Part C Methods 19, 596, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Sonnaert M., Papantoniou I., Bloemen V., Kerckhofs G., Luyten F.P., and Schrooten J. Human periosteal-derived cell expansion in a perfusion bioreactor system: proliferation, differentiation and extracellular matrix formation. J Tissue Eng Regen Med 2014 [DOI] [PubMed] [Google Scholar]
- 38.Chen Y.T., Sonnaert M., Roberts S.J., Luyten F.P., and Schrooten J. Validation of a PicoGreen-based DNA quantification integrated in an RNA extraction method for two-dimensional and three-dimensional cell cultures. Tissue Eng Part C Methods 18, 444, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Bancroft G.N., Sikavitsas V.I., van den Dolder J., Sheffield T.L., Ambrose C.G., Jansen J.A., et al. . Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc Natl Acad Sci U S A 99, 12600, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikavitsas V.I., Bancroft G.N., Holtorf H.L., Jansen J.A., and Mikos A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc Natl Acad Sci U S A 100, 14683, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishaug S.L., Crane G.M., Miller M.J., Yasko A.W., Yaszemski M.J., and Mikos A.G. Bone formation by three-dimensional stromal osteoblast culture in biodegradable polymer scaffolds. J Biomed Mater Res 36, 17, 1997 [DOI] [PubMed] [Google Scholar]
- 42.Demol J., Lambrechts D., Geris L., Schrooten J., and Van Oosterwyck H. Towards a quantitative understanding of oxygen tension and cell density evolution in fibrin hydrogels. Biomaterials 32, 107, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Khazalpour S., and Nematollahi D. Electrochemical study of Alamar Blue (resazurin) in aqueous solutions and room-temperature ionic liquid 1-butyl-3-methylimidazolium tetrafluoroborate at a glassy carbon electrode. RSC Adv 4, 8431, 2014 [Google Scholar]
- 44.Papadimitropoulos A., Piccinini E., Brachat S., Braccini A., Wendt D., Barbero A., et al. . Expansion of human mesenchymal stromal cells from fresh bone marrow in a 3D scaffold-based system under direct perfusion. PLoS One 9, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerontas S., Farid S.S., and Hoare M. Windows of operation for bioreactor design for the controlled formation of tissue-engineered arteries. Biotechnol Prog 25, 842, 2009 [DOI] [PubMed] [Google Scholar]