Abstract
The translation of “next-generation” sequencing directly to the clinic is still being assessed but has the potential for genetic diseases to reduce costs, advance accuracy, and point to unsuspected yet treatable conditions. To study its capability in the clinic, we performed whole-exome sequencing in 118 probands with a diagnosis of a pediatric-onset neurodevelopmental disease in which most known causes had been excluded. Twenty-two genes not previously identified as disease-causing were identified in this study (19% of cohort), further establishing exome sequencing as a useful tool for gene discovery. New genes identified included EXOC8 in Joubert syndrome and GFM2 in a patient with microcephaly, simplified gyral pattern, and insulin-dependent diabetes. Exome sequencing uncovered 10 probands (8% of cohort) with mutations in genes known to cause a disease different from the initial diagnosis. Upon further medical evaluation, these mutations were found to account for each pro-band's disease, leading to a change in diagnosis, some of which led to changes in patient management. Our data provide proof of principle that genomic strategies are useful in clarifying diagnosis in a proportion of patients with neurodevelopmental disorders.
INTRODUCTION
Next-generation sequencing (NGS), in which the whole genome, or a portion thereof, is sequenced, has proven extraordinarily useful for identifying new causes of genetic disease, especially for Mendelian disorders. However, the application of NGS directly in the clinic is not straightforward because of the difficulties in determining which of the thousands of variants of unknown significance (1, 2) are relevant to the individual patient's presenting signs and symptoms. Still, there is great anticipation that NGS, especially whole-exome sequencing, in which the 1% of the genome that codes for proteins is sequenced (3), will improve diagnostic approaches in genetic disease.
Neurodevelopmental disorders affecting 4 to 6% of the general population, most notably children, include intellectual disability, epilepsy, autism, structural brain diseases, and neuromuscular disorders. The U.S. Centers for Disease Control estimates that the annual cost of neuro-developmental disorders accounts for 5 to 10% of total health care expenditure in the United States owing to the lifelong care required for these patients (4). The inaccessibility of neural tissue makes it difficult to arrive at a specific diagnosis for patients with neurodevelopmental disorders, so clinicians are left with categorical diagnoses or long differential diagnoses lists. Low-yield and expensive radiographic, electro-physiological, biochemical, and biopsy evaluations are the only prospect of narrowing these lists, often costing in excess of $10,000 per patient (5). Neurodevelopmental disorders exhibit both clinical and locus heterogeneity; therefore, genetic investigations are often limited to candidate sequencing of a single gene, or a small panel of genes, that requires the clinician to have a clear sense of the likely genetic cause before testing. Although chromosomal and copy number variations account for 10 to 20% of these cases (6, 7), the remaining cases have relatively little chance of achieving a genetic diagnosis. The failure to make a specific diagnosis for neurodevelopmental disorders is a major clinical problem because it limits prognostic information, anticipatory counseling, prevention strategies, quality of life, and initiation of potentially beneficial therapies (8). For these reasons, and the finding that many neurodevelopmental disorders have a genetic basis, the neurodevelopmental disorders clinic represents a fruitful area to explore the use of whole-exome sequencing.
For this project, we identified 118 probands and their families from regions of the world with high rates of consanguinity, which enhances the power to identify recessive genetic mutations using homozygosity mapping (9). About one-sixth of the world's population resides in these areas, making this an important population to study. In such populations displaying recessive disease, heterozygous alleles can usually be excluded as causative (table S1), greatly reducing the number of variants to be considered, and overcoming one of the potential drawbacks of whole-exome sequencing (5). It is estimated that 80% of the variants causing Mendelian disease are located within the exome, making whole-exome sequencing an attractive method to interrogate variants of high effect (10, 11). Furthermore, about 15% of suspected Mendelian disease has a recessive mode of inheritance, and genomic carrier burden for such disease is estimated at 2.8 mutations per genome in outbred populations, making this class of diseases an important part of the neurodevelopmental disorder spectrum (12). Here, we present data on the application of whole-exome sequencing to a large clinical cohort. Our data show that not only is whole-exome sequencing a useful tool for identifying disease-causing genes, but it is also able to correct or modify the diagnosis in ~10% of the families studied (n = 118), thereby providing proof of principle that whole-exome sequencing can be a useful tool for diagnosis in the clinic.
RESULTS
Patient recruitment and diagnostic sequencing
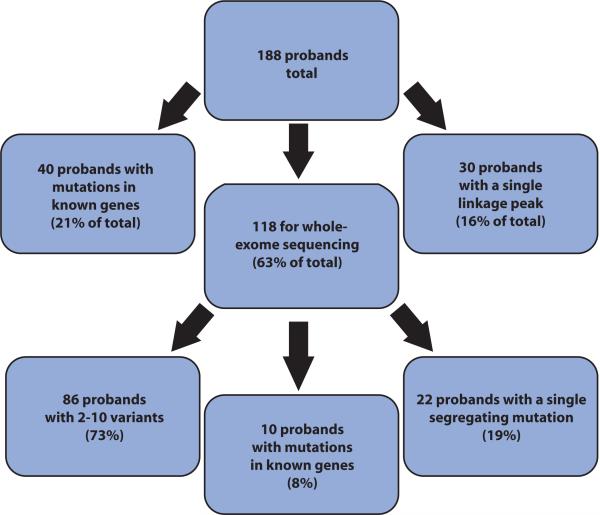
We analyzed a total of 188 families by collecting pedigrees, phenotype information, and blood samples on each genetically informative subject. Initial medical diagnoses were generated by the collective medical team (that is, treating physician, geneticist, and medical specialists) at case conferences consistent with current medical practice (Table 1) and were termed “initial diagnosis” for the purposes of this study. In some instances, the presenting features were too nonspecific to suggest a unique diagnosis, and in such cases, a categorical diagnosis was assigned. All families contained two or more affected individuals born to consanguineous parents. We used a standard protocol to exclude known disease-causing genes either by direct sequencing of all coding exons and splice sites or by excluding known loci with linkage exclusion mapping. Of the 188 probands (Fig. 1), 40 had mutations in one of the genes associated with the initial diagnosis and the mutation segregated with the phenotype in the family according to a recessive model. Such mutations were reported to the referring physician as part of this research protocol, and the families were not further studied. For the remaining families, mutations in known genes were not identified, and these families moved on to the next phase of analysis. In hindsight, whole-exome sequencing analysis of the 40 probands with mutations in known genes might have been more efficient and cost-effective than single gene sequencing methods because most subjects required evaluation at three or more genetic loci (Table 1).
Table 1.
Summary of 10 probands and their families in which whole-exome sequencing corrected diagnosis.
| Family | Initial diagnosis | Clinical profile | Number affected | Number unaffected | Genes excluded | Linkage | Linkage peaks |
|---|---|---|---|---|---|---|---|
| 890 | Pontocerebellar hypoplasia | Cerebellar hypoplasia on MRI, microcephaly, ataxia, mild spasticity, intellectual disability, thoracolumbar scoliosis, high arched palate, triangular face shape, arachnodactyly, nystagmus | 2 | 3 | TSEN2, TSEN34, TSEN54 | Yes | 3 |
| 951 | Intellectual disability | Full-term normal delivery, low birth weight, developmental delay, prominent forehead, wide-set eyes, poor speech, poor hearing | 4 | 3 | VPS13B | Yes | 2 |
| 1002 | Ataxia with vitamin E deficiency | Ataxia, decreased vitamin E levels, decreased HDL, decreased ApoA1 | 3 | 4 | TTPA | Yes | 2 |
| 1004 | Recessive primary microcephaly | Microcephaly at birth, developmental delay, nystagmus onset at 6 months old, hyperreflexia, spasticity, dysarthria, thin corpus callosum, cerebellar vermis hypoplasia | 2 | 1 | MCPH1, CDK5RAP2, MCPH4, ASPM, CENPJ, STIL | Yes | 3 |
| 702 | Microcephaly, intellectual disability | Microcephaly, psychomotor delay, dysarthria, round face, left strabismus, left eye partially closed, wide nose, arachnodactyly, normal karyotype | 3 | 3 | MCPH1, CDK5RAP2, MCPH4, ASPM, CENPJ, STIL | No | — |
| 928 | Infantile neuroaxonal dystrophy | Progressive difficulty walking, hypertonia, spasticity, lower limb dysfunction greater than upper limbs, motor delay, dysarthria, café au lait spots, normal IQ, normal EEG, normal electromyogram (EMG) and nerve conduction | 3 | 1 | PLA2G6 | Yes | 2 |
| 992 | Microcephaly, intellectual disability | Full-term, normal delivery, small for gestational age at 20 months old with microcephaly, psychomotor delay, optic atrophy, normal karyotype | 2 | 5 | MCPH1, CDK5RAP2, MCPH4, ASPM, CENPJ, STIL | Yes | 2 |
| 995 | Hereditary spastic paraplegia | Progressive difficulty walking, spasticity, brisk reflexes, hyperreflexia, tiptoe walking at age 6.5 years old, dysarthria, normal EMG and nerve conduction | 2 | 1 | SPG5A, SPG11, SPG21, SPG20 | Yes | 4 |
| 1409 | Autosomal recessive ataxia | Early-onset ataxia, deep cerebellar white matter changes on MRI, high basal ganglia signal on MRI, staccato speech | 2 | 0 | — | No | — |
| 1436 | Muscle-eye-brain disease | Hypotonia, significantly elevated creatine phosphokinase levels, strabismus, microcephaly, developmental delay, seizures, cerebellar cysts, polymicrogyria, pontocerebellar hypoplasia, arachnodactyly, cleft palate, dysplastic kidneys | 4 | 5 | — | No | — |
Fig. 1.
Summary of probands prioritized for whole-exome sequencing analysis. A total of 188 probands, each from a unique family, were priori-tized for analysis in this study, and of these, 40 (21%) had mutations in known disease-causing genes as determined by candidate gene screening. Thirty demonstrated a single linkage peak (16%) and were thus not candidates for whole-exome sequencing. The remaining 118 were analyzed by whole-exome sequencing, and of these, 10 were found to have mutations in genes known to cause a disease different from their initial diagnosis (8%). In all 10 cases, the whole-exome sequencing information led to a modification of the diagnosis.
Linkage analysis
The remaining 148 probands and their families were subjected to genome-wide parametric linkage analysis using a panel of highly informative single-nucleotide polymorphism (SNP) markers. In 30 families, a single linkage peak was identified, and such families were not considered further because we viewed strategies other than whole-exome sequencing to be a more direct method of mutant gene identification. For these families, in no instance did the identified peak overlap with a genetic locus known to cause the initial diagnosis, suggesting that many of these peaks should reveal previously unidentified causes of disease. In the remaining 118 families, we uncovered between two and eight peaks consistent with linkage, although about 30% of these families were not analyzed with linkage because they came to the study relatively late. These families were instead analyzed using homozygosity mapping from exome data (9).
Exome sequencing and variant discovery
From the 118 families without single linkage peaks, one proband per family was evaluated using whole-exome sequencing, producing an average coverage at >10× read depth for 96% of the exome, which is within the expected coverage and depth for whole-exome sequencing studies (13) and is sufficient to assess most recessive disease variants. On average, a total of 26,393 ± 4971 (SD) variants were identified per proband for evaluation.
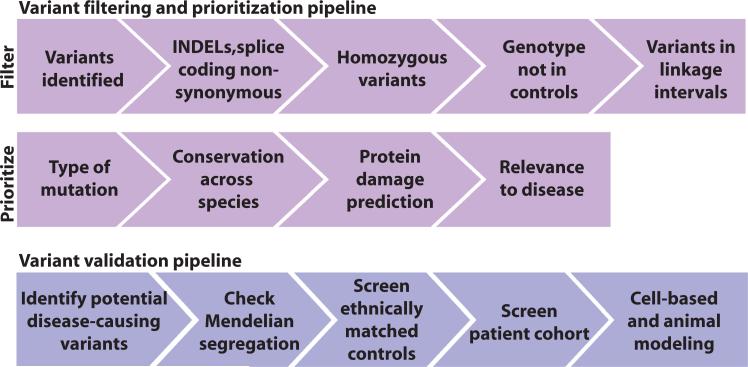
Tabulation of the <10% of the genome that failed adequate recovery from whole-exome sequencing (<10× depth) was generated in case a causative variant could not be identified among those recorded. Variants were then filtered and prioritized according to the presumed recessive disease model to identify variants of high effect size (Fig. 2). On the basis of the HapMap project, the average haplotype block size from an offspring resulting from a first-cousin marriage is >10 centimorgan (cM) (14), so we focused on such blocks of homozygosity identified from either parametric [LOD (logarithm of the odds ratio for linkage) scores] or nonparametric (homozygosity mapping) linkage. The remaining variants were then prioritized according to type of mutation (deletion/insertion > nonsense > missense), amino acid conservation, predicted damage to the protein, and relevance of the candidate gene to the given disease. The final variant list contained a mean of 9 (range, 4 to 21) new, coding, homozygous variants in linkage or homozygous intervals per proband (tables S1 and S2). Variants on the final filtered list were validated by Sanger sequencing, verified as homozygous in affecteds, and tested for segregation in the family to be consistent with the pedigree structure.
Fig. 2.
Variant filter, prioritization, and validation pipelines. All whole-exome sequencing variants were filtered to include coding variants, splice variants or INDELs (insertions-deletions), nonsynonymous amino acid changes, homozygous genotype calls, variants not found in homozygous form in population studies, and those found in linkage intervals. The remaining variants were then prioritized on the basis of type of mutation (INDELs > nonsense > missense), conservation of the amino acid across species, predicted damage to the protein, and relevance to neurodevelopmental disorders. Variant validation pipeline: Variants from the filtering pipeline were then checked for segregation according to a recessive model of disease and their absence in 200 ethnically matched control individuals.
Disease gene identification
In 22 of the 118 probands who were analyzed by whole-exome sequencing, we identified a single variant in a gene not previously implicated in disease, which fell within a region of homozygosity, and suggested a previously unidentified disease gene as the cause of the disorder. Two of these variants in which we have validated segregation are listed in Table 2. Specifically, we identified a mutation in GFM2 in a family with microcephaly, simplified gyral pattern, and insulin-dependent diabetes and a mutation in EXOC8 in a family with Joubert syndrome [Mendelian Inheritance in Man (MIM) number 213300].
Table 2.
Whole-exome sequencing is a useful technique for identifying new disease-causing genes. Summary of two families analyzed in which whole-exome sequencing identified a causative gene not previously associated with disease.
| Family | Diagnosis | Gene | Complementary DNA change | Mutation | Predicted effect | Segregates in family | Supporting evidence |
|---|---|---|---|---|---|---|---|
| 650 | Microcephaly, simplified gyral pattern, and insulin-dependent diabetes | GFM2 | c.T2032A | p.D576E, splice | Altered splicing | Yes | Only segregating variant in linkage interval, not present in 200 healthy controls, fully conserved nucleotide key for splicing, predicted damaging |
| 982 | Joubert syndrome | EXOC8 | c.A794T | p.E265G | Altered pleckstrin homology domain structure | Yes | Only segregating variant in linkage interval, not present in 200 healthy controls, cilia-related gene and Joubert is a known ciliopathy, fully conserved amino acid, and predicted damaging |
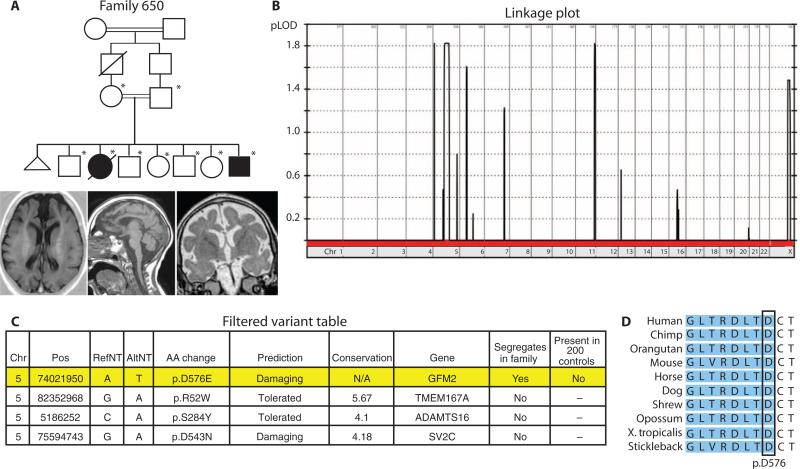
GFM2 (also called EFG2) encodes the mitochondrial elongation factor G2 and is part of the mitochondrial translation complex essential for maintaining energy metabolism. The identified c.T2032A variant in family 650 changes p.D576E, but this variant also occurs in a conserved predicted splice site at the acceptor for exon 17 and is predicted to destroy the splice acceptor function based on NetGene2 and BDGP prediction algorithms (15, 16). The presentation is overlapping with Wolcott-Rallison syndrome (MIM 226980) (17), characterized by early-onset insulin-dependent diabetes and occasional microcephaly. Mutations in EIF2AK3 and IER3IP1, encoding a translational initiation factor kinase and an endoplasmic reticulum stress response factor, respectively, have been linked to Wolcott-Rallison syndrome (18, 19). The p.D576E variant is the single variant found in a homo-zygous interval that segregates in the family, is not present in 200 ethnically matched controls, is predicted to damage the protein, and occurs in an evolutionarily conserved residue (Fig. 3). This mutation in a mitochondrial elongation factor is consistent with the model of Wolcott-Rallison syndrome as a defect in energy and cellular stress homeostasis, leading to altered neurogenesis and apoptosis. These findings suggest that GFM2 is a rational candidate for the disease and further support the use of whole-exome sequencing in identifying previously unidentified disease-causing genes for Mendelian disorders.
Fig. 3.
Genetic data for family 650. (A) Pedigree and MRI scans from the living affected members of family 650. In the pedigree, an asterisk denotes that individual's DNA was used in linkage analysis. The brain MRI (axial, midline sagittal, and coronal views) of one affected member shows a thickened cortex with a paucity of white matter, simplified gyral pattern, and thickened cerebral mantle, compatible with pachygyria. (B) Linkage plot for family 650 shows LOD score plotted against chromosome number and reveals one major linkage block on chromosome 5. (C) Final variant table after whole-exome sequencing analysis reveals four candidate genes, only one of which segregates in the family. Columns in table denote chromo-some (Chr), nucleotide position (Pos; hg19 build), reference nucleotide (RefNT), alternate nucleotide (AltNT), amino acid change (AA change), POLYPHEN damage prediction, conservation as reported by GERP score (GERP >5 is highly conserved), gene name, whether the variant segregates in the family, and whether the variant is present in 200 healthy control individuals. (D) Conservation analysis for the GFM2 variant shows that this amino acid (p.D576) is fully conserved from fish to human. This variant occurs at the exon/intron boundary, and shading denotes the exonic region.
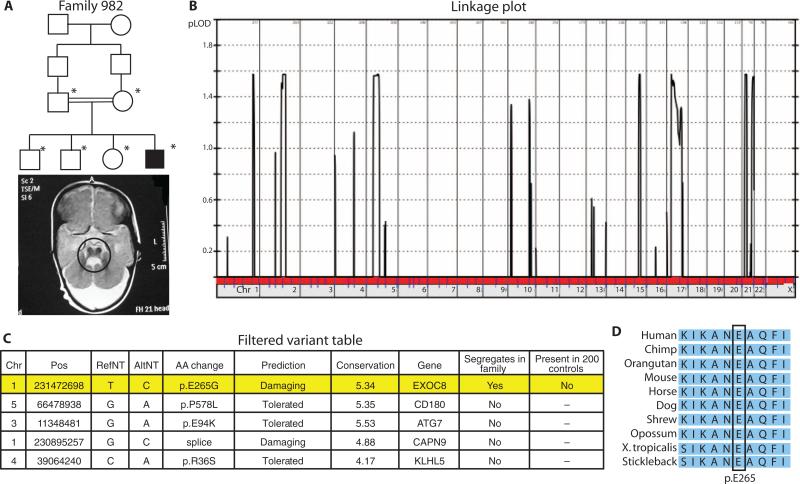
EXOC8 encodes the exocyst 84-kD subunit, one of the critical members of the eight-subunit complex required for targeting secretory vesicles to the plasma membrane during exocytosis (20). The p.E265G variant found in family 982 occurs in the B6 loop of the highly conserved pleckstrin homology (PH) domain, which is involved in binding phosphatidylinositol lipids for vesicular transport. This is the single, segregating variant in the family and is not present in 200 ethnically matched controls. It is predicted to be damaging according to POLYPHEN-2 (15, 16) and occurs in a fully conserved residue. Joubert syndrome is one of the “ciliopathy” diseases, and EXOC8 is part of the ciliary proteome (21). Further, the exocyst complex has been implicated in ciliary function (21). For these reasons, EXOC8 is a rational candidate for this disorder (Fig. 4).
Fig. 4.
Genetic data for family 982. (A) Pedigree and an MRI scan for an affected male member of family 982 showing the hallmark molar tooth malformation characteristic of Joubert syndrome (circled region). (B) Linkage plot of LOD score against chromosome number reveals multiple link age blocks for this family. (C) Final variant table after whole-exome sequencing analysis reveals five potential candidates, only one of which segregated in the family. (D) Conservation analysis for the EXOC8 variant shows that amino acid E265 (p.E265) is fully conserved from fish to human.
In the remaining 86 probands, we found 2 to 10 variants of unknown significance per proband, some of which are good disease-causing candidates. Studies are ongoing in the lab to improve variant annotation and search for probands with similar phenotypes displaying variants in the same gene in an effort to demonstrate causality, similar to published work (22).
Corrected patient diagnoses
In 10 of the 118 probands (Table 3), it was apparent that one of the variants occurred within a gene already listed in Online MIM (OMIM) to cause a neurodevelopmental disease phenotype that at least partially overlapped with the phenotype of the proband, suggesting that it might represent the causative mutation (figs. S1 to S10). In each of these 10 patients, however, the genetic diagnosis suggested from whole-exome sequencing differed from the initial diagnosis, leading us to question the veracity of the initial diagnosis. It was initially surprising to identify mutations in known disease genes, because for each initial diagnosis, we had excluded the genes most frequently mutated. For instance, in a family diagnosed with microcephaly, we excluded the genes for primary microcephaly (MCPH1, CDK5RAP2, MCPH4, ASPM, CENPJ, and STIL); in a family displaying ataxia with vitamin E deficiency, we excluded the causative gene (TTPA); and in a family with intellectual disability, we excluded the most commonly mutated gene for the recessive form of the disease (VSP13B) (Table 1).
Table 3.
Initial diagnosis compared to genetic diagnosis after whole-exome sequencing in 10 probands. Summary of 10 families analyzed in which whole-exome sequencing corrected diagnosis. In each family, an identified mutation in a known disease-causing gene led to a modification of the diagnosis. Only G726E (family 1436) is a previously reported disease mutation. For all mutations leading to a premature stop codon (families 928, 992, 890, 1409, and 951), other stop codons have been reported with the respective disease. For the missense mutations not previously reported (families 1004, 995, and 1002), and the splice mutation (family 702), each was located in an amino acid/base pair that is fully conserved across evolution (Supplementary Materials), located in a protein domain essential for protein function or splicing, is predicted to be damaging, and is not found in 200 ethnically matched controls. All mutations segregated normally with the phenotype in these families. These data, in addition to further scrutiny of the patient's clinical profile, provide evidence that these mutations are the cause of the disorders seen in each family.
| Family | Initial diagnosis | Gene | Complementary DNA change | Mutation | Segregates in family | Corrected diagnosis |
|---|---|---|---|---|---|---|
| 890 | Pontocerebellar hypoplasia | VLDLR | c.1247_53delGTTACAA | p.G1246fsX1305 | Yes | Congenital cerebellar ataxia with intellectual disability |
| 951 | Intellectual disability | MAN2B1 | c.G2088A | p.W695X | Yes | α-Mannosidosis type I |
| 1002 | Ataxia with vitamin E deficiency | SPG11 | c.T5088G | p.A1696G | Yes | Hereditary spastic paraplegia with thin corpus callosum |
| 1004 | Recessive primary microcephaly | GJC2 | c.C94T | p.R35C | Yes | Hypomyelinating leukodystrophy type 2 |
| 702 | Microcephaly, intellectual disability | VPS13B | c.G5220T splice | p.E1765D splice | Yes | Cohen syndrome |
| 928 | Infantile neuroaxonal dystrophy | ALS2 | c.4328_29delCT | p.S1443fsX1461 | Yes | Infantile-onset ascending spastic paralysis |
| 992 | Microcephaly, intellectual disability | VPS13B | c.G664T | p.E222X | Yes | Cohen syndrome |
| 995 | Hereditary spastic paraplegia | ALS2 | c.G529T | p.G177C | Yes | Infantile-onset ascending spastic paralysis |
| 1409 | Autosomal recessive ataxia | SURF1 | c.C817T | p.Q273X | Yes | Leigh syndrome due to mitochondrial complex IV deficiency |
| 1436 | Muscle-eye-brain disease | POMT2 | c.G2378A | p.G726E | Yes | Muscular dystrophy dystroglycanopathy type A2 |
To understand this paradox, we returned to the patient charts to review the presentation and clinical course. In each case, we found that the genetic variant was sufficient to explain the full clinical presentation, suggesting that whole-exome sequencing was able to either modify or correct an initial diagnosis for each of these 10 cases.
Clinical presentations
Family 890: Mutation in VLDLR
This family from Trabzon, Turkey, presented two affecteds at birth with microcephaly, nystagmus, congenital ataxia, mild spasticity, and arachnodactyly. Brain magnetic resonance imaging (MRI) analysis demonstrated severe hypoplasia of the midbrain, consistent with a diagnosis of pontocerebellar hypopla sia (MIM 607596), published as such in 2002 (23). The family was negative for mutations in the three known genes for pontocerebellar hypoplasia—TSEN2, TSEN34, and TSEN54—encoding transfer RNA (tRNA) splicing endonucleases (24), and linkage analysis demonstrated three potential linkage peaks not associated with any known pontocerebellar hypoplasia genes. Whole-exome sequencing identified a homozygous p.G1246fsX1305 alteration, which segregated in the family, leading to a protein frameshift in the VLDLR gene, encoding the very low-density lipoprotein receptor (fig. S1). Reevaluation of the brain MRI was completely consistent with VLDLR-associated congenital cerebellar ataxia with intellectual disability syndrome (MIM 224050), demonstrating the classical very small, smooth cerebellum (25). The team concluded that the initial diagnosis was incorrect because the clinical pheno-type in this family was different from the spectrum previously described for VLDLR-associated disease.
Family 951: Mutation in MAN2B1
This family from Islamabad, Pakistan, presented four affected children with intellectual disability. After a normal pregnancy, labor, and delivery except for low birth weight, there was intellectual disability noted by 2 years of age, as well as mild dysmorphic features including prominent forehead, wide-set eyes, and defects in hearing and speech. Routine metabolic screening and mass spectrometry were noncontributory. The affecteds received an initial diagnosis of recessive intellectual disability and were negative for alterations in the VPSB13B gene, tested because of concordant obesity (26). SNP-based linkage analysis pointed to two potential linkage peaks, neither containing genes for autosomal recessive intellectual disability. Whole-exome sequencing demonstrated a homozygous p.W695* truncating mutation in the MAN2B1 gene that segregated fully in the family (fig. S2). The MAN2B1 gene is mutated in α-mannosidosis (MIM 248500) (27), a metabolic lysosomal storage condition caused by an inability to cleave α-linked mannose residues from the non-reducing end of N-linked glycoproteins. Reevaluation of the phenotype in light of this finding confirmed the typical facial appearance, enlarged liver, and vacuolated lymphocytes typical of type I α-mannosidosis (28). The anticipatory guidance and direction of therapy has been changed to reflect this genetic diagnosis (29). The team concluded that the initial diagnosis did not take into account this disease because of the nonspecific presenting features.
Family 1002: Mutation in SPG11
Family 1002 from Marrakech, Morocco, presented three affected members with progressively unsteady gait from the age of 5 years, interpreted as ataxia. There was areflexia, positive Babinski sign, and loss of proprioception with intact cognition, and a normal brain computed tomography (CT) scan, leading to the initial diagnosis of a progressive ataxia or spasticity. Initial workup included reduced serum levels of ApoA1, high-density lipoprotein (HDL), and vitamin E, consistent with a diagnosis of ataxia with vitamin E deficiency (MIM 277460). The reduced serum levels were within the range of other patients we have evaluated with this condition, although they lacked the common Moroccan 744delA mutation in the TTPA gene (30). However, patients showed nominal improvement in function upon administration of daily exogenous vitamin E, supporting the diagnosis. Full sequence of the TTPA gene was negative for variation, and SNP-based linkage analysis suggested two potential peaks, neither of which contained the TTPA gene or known modulators of vitamin E metabolism.
Whole-exome sequencing analysis identified one splice and two missense variants, two of which were fully conserved across species and one predicted to be damaging. Only a homozygous c.T5088G variant leading to a p.A1696G amino acid transversion in the SPG11 gene segregated according to the predicted mode of inheritance in the seven children in the family, providing compelling evidence that this mutation may cause this neurodevelopmental disorder (fig. S3). The SPG11 gene is a recently reported cause of hereditary spastic paraplegia with thin corpus callosum (MIM 604360) (31). The p.A1696G changes a nonpolar neutral amino acid to a polar negative amino acid and is predicted to be damaging to protein function according to POLYPHEN-2 software (15, 16). The p.A1696 residue is perfectly conserved across evolution and occurs within the leucine-rich repeat 3 domain, supporting its pathogenicity. This variant was not detected in chromosomes from 200 control Moroccan individuals. Subsequent reevaluation of the family led to reinterpretation of the ataxia as spasticity, and brain MRI analysis in two affecteds demonstrated the characteristic thin corpus callosum, consistent with a diagnosis of SPG11-associated disease. Vitamin E therapy has subsequently been halted without clinical consequence. In this situation, the team concluded that the original initial diagnosis was incorrect due to an initial misinterpretation of the clinical signs and false-positive chemistry studies.
Family 1004: Mutation in GJC2
This family from Cairo, Egypt, presented two affecteds with microcephaly and intellectual disability. The initial diagnosis of microcephaly was assigned on the basis of a head circumference of 48 cm at age 8 years (−2.5 SD) in an older male sibling and 45 cm at age 3 years (−2.5 SD) in a younger female sibling. Brain MRI showed thin corpus callosum, mild ventriculomegaly, and cerebellar hypoplasia. The family tested negative for mutations in the known primary microcephaly genes MCPH1, CDK5RAP2, MCPH4, ASPM, CENPJ, and STIL. As the children aged, they displayed signs of nystagmus, hyperreflexia, and spasticity, atypical for primary microcephaly, and the three linkage peaks identified from SNP-based analysis did not suggest any other microcephaly loci. Whole-exome sequencing demonstrated a homozygous c.C94T alteration in the GJC2 gene that segregated fully in the family and is known to cause hypomyelinating leukodystrophy II (MIM 608804) (32). This mutation leads to a p.R35C amino acid transversion in the connexin domain. The p.R35 residue is perfectly conserved across evolution, is predicted to be damaging, and was not found in chromosomes from 200 Egyptian control individuals (fig. S4). Subsequent reevaluation of the family focusing on this variant led us to conclude that the spasticity and nystagmus were progressively worsening, along with the presence of mild peripheral axonal neuropathy. MRI reinterpretation showed a hypomyelinating leukodystrophy consistent with GJC2-associated disease. The team concluded that the initial diagnosis was too broadly categorized due to nonspecific presenting features, which precluded a more accurate diagnosis.
DISCUSSION
The main finding of this work is that whole-exome sequencing is beneficial over individual candidate gene sequencing in identifying mutations in genes not previously suspected in a given patient. This finding provides proof of principle that whole-exome sequencing has the potential to change clinical practice for genetic disease. Specifically, this work demonstrates the use of whole-exome sequencing in the clinic when applied to a group of patients with likely genetic disease for which the cause remained elusive before study. In our study, we found that in 10 cases of 118 probands undergoing whole-exome sequencing, there was a revision of the diagnosis and, in some cases, a change in management. Furthermore, in each of these 10 cases, genetic counseling, prenatal diagnostic options, and carrier testing were altered after diagnosis. We also identified likely causative mutations in two other families with neurodevelopmental disorders, which have the potential to lead to new therapies. Although the ability of NGS to provide an accurate genetic diagnosis has been established for single cases like 3,4-dihydroxyphenylalanine (dopa)–responsive dystonia and Charcot-Marie-Tooth neuropathy (33, 34), this report addresses the benefits of NGS in a large clinical cohort.
In our cohort, we first excluded known genetic causes of disease by sequencing likely mutated genes on the basis of the initial diagnosis. This enriched for patients with new genetic causes of disease and with an incorrect or partially correct diagnosis. From this group, we used whole-exome sequencing to further stratify patients into those with a likely new disease gene (22 of 118, or 19%), those with no obvious single disease gene candidate but rather numerous candidates (86 of 118, or 73%), and those with a mutation in a disease gene known to cause a disorder different from the initial diagnosis (10 of 118, or 8%). These findings should be compared to other diagnostic modalities, such as copy number variant (CNV) or de novo mutation identification in neurodevelopmental disorders, where success rates fall between 10 and 60% in selected populations (35–37). Although it is difficult to compare success rates due to differences in cohort structures, whole-exome sequencing in probands with recessive disease is likely to emerge as an attractive alternative approach to candidate gene sequencing.
Our data show that in a substantial portion of patients in the neuro-developmental disorders clinic, the initial diagnosis might not be accurate or might be too broadly classified. There are several potential reasons why an initial diagnosis might be incorrect or partially correct in the neurodevelopmental disorders spectrum. In our study, we attributed these differences to the following, and it is our experience that these limitations exist in the clinical setting in general: (i) Patient phenotypes differed partially or substantively from the spectrum previously described for a given gene, (ii) medical information or history was incomplete, and (iii) nonspecific clinical features were found in the patients. The field of genetic medicine has literally tens of thousands of unique syndromes. Even an experienced professional might not entertain all possible diagnoses for a given presentation due to the vast number of syndromes to consider. Medical diagnostic software that helps to maintain a broad and systematic differential could help with this issue (38) and would make a powerful partner to help prioritize and filter data. In each case presented here, the medical team returned to the clinical information to determine why the initial diagnosis differed from the genetic diagnosis, and it was found that most differences were due to limitations in the clinical practice of medicine. Whole-exome sequencing was able to overcome many of these limitations. This study demonstrates the clinical use of whole-exome sequencing and points out potential benefits in correcting patient diagnosis.
The current cost for whole-exome sequencing is ~$2000 to $4000 per patient (39) and is expected to drop substantially in the coming years. With similar costs for candidate gene sequencing, whole-exome sequencing should be considered an attractive alternative in families with a presumed genetic cause of disease. Whereas whole-genome sequencing is another technology that is sure to change the face of medicine in the future, whole-exome sequencing has captured the attention of the clinical genetics community because most genetic variants of large effect reside in the exome, because intronic mutations are difficult to interpret and to model, and because genome sequencing is still more expensive than whole-exome sequencing (10, 34). The data presented here suggest that whole-exome sequencing should be considered in a diagnostic context in the appropriate clinical settings.
Whereas whole-exome sequencing was used with some success in this study, it does suffer from limitations—even in the field of Mendelian genetics. Whole-exome sequencing in clinical applications lacks some sensitivity due to its inability to interrogate intronic sequence, the absence of recovery of some exons, and sequencing errors. Even more important is the difficulty in separating causative variants from the vast number of variants of unknown significance identified per patient (10, 13, 15). Furthermore, the recent finding that more than 25% of putative disease-causing variants in available databases are erroneous (12) makes interpretation all that much more difficult. These limitations were partially overcome by restricting analysis to consanguineous families with recessively inherited disease, and allowing exclusion of most variants of unknown significance using criteria specific to this model of disease. In addition, these families allowed for testing of segregation of each variant, thereby providing an additional level of certainty about the causation of each mutation. As human mutation databases become sufficiently populated and carefully curated, the ability to interpret whole-exome sequencing data will greatly improve. The introduction of whole-exome sequencing into routine clinical practice will require careful assessments of these issues. As for the future, the limitations of whole-exome sequencing seem tractable and there are solutions that should allow whole-exome sequencing to ultimately be used routinely in a clinical setting.
MATERIALS AND METHODS
Study participants
The probands for this study were ascertained from the Middle East, North Africa, and Central Asia and were selected based on the criteria of (i) healthy parents with documented consanguinity, (ii) more than one affected child to enrich for recessive disease, and (iii) an initial diagnosis of a neurodevelopmental disorder of unknown genetic etiology. We excluded patients with clear single gene defects in which clinical features are absolutely characteristic, such as for Tay-Sachs disease, Niemann-Pick type C, and spinal muscular atrophy. This study was approved by the Institutional Review Board at the University of California, San Diego, and collaborating institutions; all study participants signed informed consent documents; and the study was performed in accordance with Health Insurance Portability and Accountability Act privacy rules.
Phenotypic assessments
One or more of the authors who are board-certified in pediatrics, neurology, and/or genetics evaluated each patient. Standard evaluation included full general and neurological examination, height, weight, head circumference measurements, intelligence quotient (IQ), brain MRI or CT, and electroencephalogram (EEG) when clinically indicated. Diagnostic criteria were based on those standards in the field, and initial diagnoses were determined by consensus at clinical team meetings where differential diagnoses, genetic counseling, and care plans were discussed.
Linkage analysis
Linkage analysis was performed by genotyping all available and consenting members of the family in the affected and parental generations of the pedigree. DNA was extracted from peripheral blood leukocytes with salt extraction, genotyped with the Illumina Linkage IVb mapping panel (40), and analyzed with easyLINKAGE-Plus (41) software to generate multipoint LOD scores.
Exome sequencing and variant analysis
For each sample, whole-exome sequencing was performed with the Agilent SureSelect Human All Exome 50 Mb Kit, which captures up to 50 Mb of the human exome and includes all exons annotated in the consensus CDS database, as well as 10 bases flanking each targeted exon and small noncoding RNAs. This kit provides >95% coverage at 1×, and >88% at 10× coverage, and paired-end sequencing of 75–base pair sequencing length was done with Illumina GAIIx or HiSeq2000 instruments (42). Depth of sequencing was 30 ± 16 (SD) per exome.
Whole-exome sequencing generated large data sets that required extensive analysis to identify the important genetic variants. The process of isolating potential disease-causing variants involved several major steps: (i) align and ensure quality of DNA sequences for each exome; (ii) identify polymorphisms in the patient's DNA sequence compared to a reference sequence with tested SNP and insertions/ deletions (INDEL) calling software [that is, Genome Analysis Tool Kit (GATK) and in-house generated tools]; (iii) verify the quality, repeatability, and comparability to published results of SNP and INDEL data; (iv) filter out variants that are outside runs of homozygosity, outside of coding/splice regions, and are found in homozygous form in the healthy population; (v) prioritize potential disease-causing variants by type of mutation, conservation of amino acid residue(s) across species, predicted damage to the protein, and relevance to the neuro-developmental disorder; (vi) validate potential disease-causing variants by assuring Mendelian segregation in the family, screening healthy, ethnically matched controls, and identifying mutations in the same gene in other families with the disorder.
Genetic variants were delineated with the GATK software for both SNPs and INDELs (43). Briefly, Illumina “qseq.txt” files from each exome were converted to FASTQ format with Illumina Pipeline Software and then aligned to a reference sequence with Burrows-Wheeler Aligner (BWA) software (SourceForge). Duplicate sequencing reads, which can be produced by polymerase chain reaction amplification, were then removed, and sequence quality scores were recalibrated to correct for sequencing errors and artifacts. Alignments that contain INDELs can lead to mismatches that resemble SNPs; therefore, alignment of sequences with Maq was also necessary to identify and isolate INDEL-containing reads. Next, a Bayesian-based SNP caller and INDEL Genotyper (GATK) were used to filter potential disease-causing variants from the aligned sequences. Each aligned exome was then assessed for sufficient quality with the following parameters: number of SNPs called in each lane (average ~26,000), accuracy, depth of coverage, and error rate per read position. Only exomes of high quality were analyzed further.
Exomes were then filtered to highlight false-positive variants. Likely false positives were flagged with the following criteria: low SNP confidence, frequency of the reference base in the population is overwhelmingly high, low depth of coverage for the SNP, and presence of the SNP in homopolymer runs longer than three bases. Flagged variants were included in the subsequent “variant filtering and prioritization pipeline” but were pursued with caution in the “validation pipeline.” Both pipelines are described below.
Exome call sets underwent the following evaluations: compared number of SNPs to the quantities usually found per cleaned exome data set, determined overlap with the dbSNP database, determined the transition/transversion ratio as a measure of the false-positive rate found in the data set, compared the data to other exomes sequenced in the lab, and compared random variants found in the exome to the Human Genome Browser. These data were used as quality measures to determine the overall integrity of the data set compared to published studies. The end result of this pipeline was a list of quality potential disease-causing variants that were further filtered and prioritized.
The “sequencing analysis pipeline” identified numerous potential disease-causing variants, and on average, an exome from one individual from a first-cousin marriage contains about 26,000 SNPs and 1000 INDELs (from our data sets). Most of the variants identified were known and/or nondeleterious polymorphisms; however, a small number of them represented frameshift, missense, and nonsense mutations in a potential disease-causing gene. Therefore, variants were further filtered with our “potential disease-causing variants filtering and prioritization pipeline.” First, SNPs and INDELs present in stretches of homozygosity, as determined by either linkage analysis or publicly available homozygosity mapping software (Homozygosity Mapper), were isolated (44). Homozygosity mapping in consanguineous families with recessive disease is a proven method for identifying disease-causing mutations given that the DNA sequence flanking the mutation will be preferentially homozygous by descent in children from consanguineous marriages. Next, variants present in coding regions that lead to nonsynonymous amino acid changes, or found in splice sites, were isolated. Only those not found in the homozygous state in dbSNP genotype studies were pursued. The remaining variants were cross compared to the Genome Variation Server (http://gvs.gs.washington.edu/GVS/) and SIFT (http://sift.jcvi.org/) databases to determine single-nucleotide evolutionarily constraint and conservation scores (GERP, PhastCons), protein damage prediction determinations (Polyphen), and relationship of variants to OMIM classifications (45). These measures were used to prioritize variants by high conservation across species, predicted damage to the protein (or causing nonsense-mediated decay), and are not associated with any known, nonrelated diseases. Finally, variants were reviewed for their expression profiles in the brain and their relevance to the neurodevelopmental disorders of interest (Fig. 2). The end result of this pipeline was a list of key potential disease-causing variants that underwent follow-up validation in patients and family members.
Once key potential disease-causing variants were identified, it was then necessary to validate each candidate to determine whether they were the disease-causing mutation in our “potential disease-causing variants validation pipeline.” Specifically, we used direct sequencing to confirm that the polymorphism segregates within the family in a manner that is consistent with recessive inheritance, and we verified that the mutation was not present in a large cohort of healthy, ethnically matched control individuals. Thus, each variant was consistent with variant interpretation category 2 (unreported) and is of the type that is expected to cause the disorder according to the American College of Medical Genetics and Genomics guidelines (1, 2).
Supplementary Material
Acknowledgments
We thank the families for their participation. Funding: Supported by Howard Hughes Medical Institute, NIH [National Institute of Neurological Disorders and Stroke grants R01NS041537, R01NS048453, and R01NS052455; National Human Genome Research Institute grants P01HD070494 (to J.G.G.) and U54HG003067 (to S.B.G. and C.R.); National Institute on Alcohol Abuse and Alcoholism/Center for Inherited Disease Research grant N01-HG-65403] for SNP genotyping, and NSF (grants III-081905 and CCF-1115206 to V.B.).
Footnotes
Author contributions: T.J.D.-S., J.L.S., S.B., and J.G.G. recruited patients and designed and analyzed the experiments. N.U., J.S., A.E.S., J.O., V.B., A.G.F., G.N., and N.A. generated the bioinformatic pipeline for data analysis. M.S.Z., G.H.A.-S., L.A.M., L.S., S.A.-H., N.M., T.B.-O., N.A.A.-S., F.M.S., F.C., and M.A. identified and recruited families for study and ascertained clinical information. K.J.H. and A.C. assembled clinical data in tabular format. K.V.G., C.S., C.R., and S.B.G. generated sequencing results and provided preliminary analysis. T.J.D.-S. and J.G.G. wrote the manuscript.
Citation: T. J. Dixon-Salazar, J. L. Silhavy, N. Udpa, J. Schroth, S. Bielas, A. E. Schaffer, J. Olvera, V. Bafna, M. S. Zaki, G. H. Abdel-Salam, L. A. Mansour, L. Selim, S. Abdel-Hadi, N. Marzouki, T. Ben-Omran, N. A. Al-Saana, F. M. Sonmez, F. Celep, M. Azam, K. J. Hill, A. Collazo, A. G. Fenstermaker, G. Novarino, N. Akizu, K. V. Garimella, C. Sougnez, C. Russ, S. B. Gabriel, J. G. Gleeson, Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 4, 138ra78 (2012).
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Data have been deposited into dbGap (phs000288).
REFERENCES AND NOTES
- 1.Maddalena A, Bale S, Das S, Grody W, Richards S, ACMG Laboratory Quality Assurance Committee Technical standards and guidelines: Molecular genetic testing for ultra-rare disorders. Genet. Med. 2005;7:571–583. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 2.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 3.Biesecker LG. Exome sequencing makes medical genomics a reality. Nat. Genet. 2010;42:13–14. doi: 10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- 4.Center for Disease Control and Prevention (CDC) Economic costs associated with mental retardation, cerebral palsy, hearing loss, and vision impairment—United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 2004;53:57–59. [PubMed] [Google Scholar]
- 5.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: When will it become routine? Sci. Transl. Med. 2011;3:87ps23. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: Promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Stevenson DA, Carey JC. Health-related quality of life measures in genetic disorders: An outcome variable for consideration in clinical trials. Am. J. Med. Genet. C Semin. Med. Genet. 2009;151C:255–260. doi: 10.1002/ajmg.c.30217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, Botstein D. Homozygosity mapping: A way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 10.Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, Lobbens S, Simon A, Bellanné-Chantelot C, Létourneau L, Scharfmann R, Delplanque J, Sladek R, Polak M, Vaxillaire M, Froguel P. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper DN, Krawczak M, Antonorakis SE. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, Kinzler KW, Vogelstein B, editors. McGraw-Hill; New York: 1995. pp. 259–291. [Google Scholar]
- 12.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, Albrechtsen A, Andersen G, Cao H, Korneliussen T, Grarup N, Guo Y, Hellman I, Jin X, Li Q, Liu J, Liu X, Sparsø T, Tang M, Wu H, Wu R, Yu C, Zheng H, Astrup A, Bolund L, Holmkvist J, Jørgensen T, Kristiansen K, Schmitz O, Schwartz TW, Zhang X, Li R, Yang H, Wang J, Hansen T, Pedersen O, Nielsen R, Wang J. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wit MC, de Coo IF, Julier C, Delépine M, Lequin MH, van de Laar I, Sibbles BJ, Bruining GJ, Mancini GM. Microcephaly and simplified gyral pattern of the brain associated with early onset insulin-dependent diabetes mellitus. Neurogenetics. 2006;7:259–263. doi: 10.1007/s10048-006-0061-1. [DOI] [PubMed] [Google Scholar]
- 18.Delépine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 19.Poulton CJ, Schot R, Kia SK, Jones M, Verheijen FW, Venselaar H, de Wit MC, de Graaff E, Bertoli-Avella AM, Mancini GM. Microcephaly with simplified gyration, epilepsy, and infantile diabetes linked to inappropriate apoptosis of neural progenitors. Am. J. Hum. Genet. 2011;89:265–276. doi: 10.1016/j.ajhg.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Guo W. The exocyst complex in exocytosis and cell migration. Protoplasma. 2011 doi: 10.1007/s00709-011-0330-1. 10.1007/s00709-011-0330-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr., Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dilber E, Aynaci FM, Ahmetoglu A. Pontocerebellar hypoplasia in two siblings with dysmorphic features. J. Child Neurol. 2002;17:64–66. doi: 10.1177/088307380201700119. [DOI] [PubMed] [Google Scholar]
- 24.Budde BS, Namavar Y, Barth PG, Poll-The BT, Nürnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, Aronica E, van der Knaap MS, Hohne W, Toliat MR, Crow YJ, Steinling M, Voit T, Roelenso F, Brussel W, Brockmann K, Kyllerman M, Boltshauser E, Hammersen G, Willemsen M, Basel-Vanagaite L, Krageloh-Mann I, de Vries LS, Sztriha L, Muntoni F, Ferrie CD, Battini R, Hennekam RC, Grillo E, Beemer FA, Stoets LM, Wollnik B, Nurnberg P, Baas F. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 25.Barkovich AJ, Millen KJ, Dobyns WB. A developmental and genetic classification for midbrain-hindbrain malformations. Brain. 2009;132:3199–3230. doi: 10.1093/brain/awp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolehmainen J, Wilkinson R, Lehesjoki AE, Chandler K, Kivitie-Kallio S, Clayton-Smith J, Traskelin AL, Waris L, Saarinen A, Khan J, Gross-Tsur V, Traboulsi EI, Warburg M, Fryns JP, Norio R, Black GC, Manson FD. Delineation of Cohen syndrome following a large-scale genotype-phenotype screen. Am. J. Hum. Genet. 2004;75:122–127. doi: 10.1086/422197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotoda Y, Wakamatsu N, Kawai H, Nishida Y, Matsumoto T. Missense and nonsense mutations in the lysosomal α-mannosidase gene (MANB) in severe and mild forms of α-mannosidosis. Am. J. Hum. Genet. 1998;63:1015–1024. doi: 10.1086/302048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg T, Riise HM, Hansen GM, Malm D, Tranebjaerg L, Tollersrud OK, Nilssen O. Spectrum of mutations in a-mannosidosis. Am. J. Hum. Genet. 1999;64:77–88. doi: 10.1086/302183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewal SS, Shapiro EG, Krivit W, Charnas L, Lockman LA, Delaney KA, Davies SM, Wenger DA, Rimell FL, Abel S, Grovas AC, Orchard PJ, Wagner JE, Peters C. Effective treatment of α-mannosidosis by allogeneic hematopoietic stem cell transplantation. J. Pediatr. 2004;144:569–573. doi: 10.1016/j.jpeds.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Marzouki N, Benomar A, Yahyaoui M, Birouk N, Elouazzani M, Chkili T, Benlemlih M. Vitamin E deficiency ataxia with (744 del A) mutation on a-TTP gene: Genetic and clinical peculiarities in Moroccan patients. Eur. J. Med. Genet. 2005;48:21–28. doi: 10.1016/j.ejmg.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, Martin E, Ouvrard-Hernandez AM, Tessa A, Bouslam N, Lossos A, Charles P, Loureiro JL, Elleuch N, Confavreux C, Cruz VT, Ruberg M, Leguern E, Grid D, Tazir M, Fontaine B, Filla A, Bertini E, Durr A, Brice A. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat. Genet. 2007;39:366–372. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- 32.Uhlenberg B, Schuelke M, Rüschendorf F, Ruf N, Kaindl AM, Henneke M, Thiele H, Stoltenburg-Didinger G, Aksu F, Topaloğlu H, Nurnberg P, Hübner C, Weschke B, Gärtner J. Mutations in the gene encoding gap junction protein α12 (connexin 46.6) cause Pelizaeus-Merzbacher–like disease. Am. J. Hum. Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 36.Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, Buja A, Krieger A, Yoon S, Troge J, Rodgers L, Iossifov I, Wigler M. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O'Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Gunel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH, Jr., Geschwind D, Roeder K, Devlin B, State MW. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal MM. Mobile medical computing driven by the complexity of neurologic diagnosis. J. Child Neurol. 2006;21:595–599. doi: 10.1177/08830738060210071601. [DOI] [PubMed] [Google Scholar]
- 39.Lifton RP. Individual genomes on the horizon. N. Engl. J. Med. 2010;362:1235–1236. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 40.Murray SS, Oliphant A, Shen R, McBride C, Steeke RJ, Shannon SG, Rubano T, Kermani BG, Fan JB, Chee MS, Hansen MS. A highly informative SNP linkage panel for human genetic studies. Nat. Methods. 2004;1:113–117. doi: 10.1038/nmeth712. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann K, Lindner TH. easyLINKAGE-Plus—Automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 42.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper—An interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, Nickerson DA. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.