Highlights
-
•
Uterine serous carcinoma (USC) is an aggressive subtype of endometrial cancer.
-
•
Mutations in DNA polymerase ε gene (POLE) are detected in a subset of USC and confer a strong mutator phenotype.
-
•
USC patients diagnosed with POLE mutations experience a significantly better prognosis.
Keywords: Endometrial cancer, Uterine serous carcinoma, Polymerase epsilon somatic mutations
Brief communication
Uterine serous carcinoma (USC) is a rare but highly aggressive subtype of endometrial cancer causing a disproportionate number of cancer-related deaths. The overall 5-year survival of USC is only 30 ± 9% for all stages and the recurrence rate after surgery is extremely high (50–80%) (Fader et al., 2013).
We have recently sequenced the exomes of a large cohort of comprehensively staged USC all uniformly treated per Institutional guidelines with adjuvant brachytherapy and/or pelvic radiation followed by six cycles of carboplatin/paclitaxel (Fader et al., 2013, Zhao et al., 2013). Within the 57 USC patients we identified a subset of patients (5/57 = 8.7%) demonstrating a hypermutator phenotype (i.e., greater than 4000 protein-altering somatic mutations per tumor) (Zhao et al., 2013). This was in strong contrast to the substantially reduced number of somatic mutations (i.e., less than 100) identified in the remaining 52 USC patients (Zhao et al., 2013). All hypermutator USC harbored somatic mutations in the DNA polymerase ε gene (Zhao et al., 2013), a primary guardian of DNA replication fidelity during cell division recently shown to confer a strong mutator phenotype and high incidence of spontaneous neoplasms in mice (Albertson et al., 2009) and humans (Palles et al., 2013). Examination of the POLE and MMR genes showed no germ-line mutations; however, somatic mutations in these genes were highly prevalent in these tumors (mean of 4 per tumor, including 4 premature termination mutations for MMR genes and a mean of 4.5 per tumor for POLE) and more frequent than expected by chance (P = 2.23 × 10 − 3) (Zhao et al., 2013).
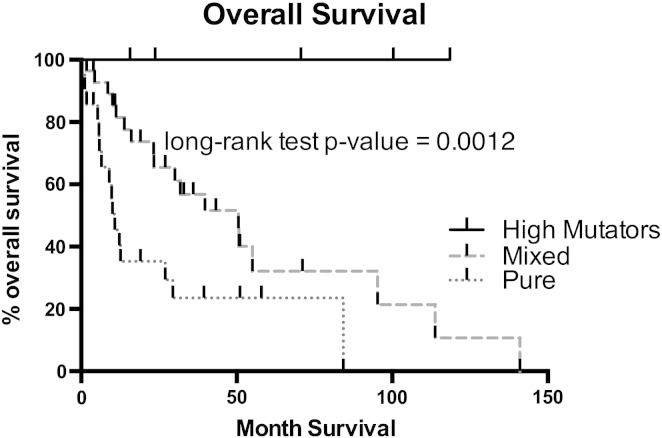
Here, we report the first evidence that patients diagnosed with hypermutator USC experience a significantly better prognosis when compared to the remaining USC patients (Fig. 1). These results were surprising considering that 2 of 5 of the long term USC survivors in our series harbored advanced stage disease (i.e., stage III) and the remaining 3 stage IB disease (risk of recurrence up to 80%) (Fader et al., 2013). Importantly, the survival results we report in patients harboring hypermutator Type II USC have recently been confirmed by the similarly excellent survival results found in 7% (17 out of 248) of the Type I endometrial cancer patients harboring proofreading POLE mutations reported by the TCGA group (The Cancer Genome Atlas Research Network, 2013) and in 15% (8 out of 53) grade 3 endometrioid adenocarcinoma of the endometrium reported by Meng et al. (2014).
Fig. 1.
Survival analysis of USC patients.
Kaplan–Meyers survival curves for 57 USC patients (5 harboring a hypermutator phenotype vs 52 moderately mutated) according to mixed and pure tumor histology. The median (range) duration of follow-up was 24 (1–141) months in the USC patients while the median (range) of follow up was 71 (16–119) months in the POLE patients. P value was calculated by the log-ranked test for survival differences. Secondary to the low number of patients in the POLE mutated subset, a comparison for stage is not feasible.
While it is currently not understood why patients developing hypermutator Type I (The Cancer Genome Atlas Research Network, 2013, Meng et al., 2014) and Type II (USC) endometrial cancers may experience such a good prognosis, it is possible that the high number of somatic mutations present in these tumors may render these cancers highly immunogenic for the host due to the large number of mutated epitopes. Alternatively, these cancers might be differentially sensitive to standard adjuvant treatments (i.e., radiation and/or chemotherapy).
These data provide novel information for the identification of a subset of biologically aggressive USC with unique prognostic features and biological properties. It will be of great interest to determine whether the POLE mutations we found in USC are of similar prognostic significance in the multiple additional types of human cancers recently found by the TCGA network to harbor POLE mutations (https://tcga-data.nci.nih.gov/tcga/).
Conflict of interest
There is no conflict interest.
References
- Albertson T.M., Ogawa M., Bugni J.M., Hays L.E., Chen Y., Wang Y., Treuting P.M., Heddle J.A., Goldsby R.E., Preston B.D. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106(40):17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader A.N., Santin A.D., Gehrig P.A. Early stage uterine serous carcinoma: management updates and genomic advances. Gynecol. Oncol. 2013;129(1):244–250. doi: 10.1016/j.ygyno.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Meng B., Hoang L.N., McIntyre J.B., Duggan M.A., Nelson G.S., Lee C.-H., Köbel M. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol. Oncol. 2014;134:15–19. doi: 10.1016/j.ygyno.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Palles C., Cazier J.B., Howarth K.M., Domingo E., Jones A.M., Broderick P., Kemp Z., Spain S.L., Almeida E.G., Salguero I., Sherborne A., Chubb D., Carvajal-Carmona L.G., Ma Y., Kaur K., Dobbins S., Barclay E., Gorman M., Martin L., Kovac M.B., Humphray S., CORGI Consortium, WGS500 Consortium, Lucassen A., Holmes C.C., Bentley D., Donnelly P., Taylor J., Petridis C., Roylance R., Sawyer E.J., Kerr D.J., Clark S., Grimes J., Kearsey S.E., Thomas H.J., McVean G., Houlston R.S., Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Choi M., Overton J.D., Bellone S., Roque D.M., Cocco E., Guzzo F., English D.P., Varughese J., Gasparrini S., Bortolomai I., Buza N., Hui P., Abu-Khalaf M., Ravaggi A., Bignotti E., Bandiera E., Romani C., Todeschini P., Tassi R., Zanotti L., Carrara L., Pecorelli S., Silasi D.A., Ratner E., Azodi M., Schwartz P.E., Rutherford T.J., Stiegler A.L., Mane S., Boggon T.J., Schlessinger J., Lifton R.P., Santin A.D. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]