Abstract
Orthogonal aminoacyl-tRNA synthetase/tRNA pairs from archaea have been evolved to facilitate site specific in vivo incorporation of unnatural amino acids into proteins in Escherichia coli. Using this approach, unnatural amino acids have been successfully incorporated with high translational efficiency and fidelity. In this study, CHARMM-based molecular docking and free energy calculations were used to evaluate rational design of specific protein-ligand interactions for aminoacyl-tRNA synthetases. A series of novel unnatural amino acid ligands were docked into the p-benzoyl-L-phenylalanine tRNA synthetase, which revealed that the binding pocket of the enzyme does not provide sufficient space for significantly larger ligands. Specific binding site residues were mutated to alanine to create additional space to accommodate larger target ligands, and then mutations were introduced to improve binding free energy. This approach was used to redesign binding sites for several different target ligands, which were then tested against the standard 20 amino acids to verify target specificity. Only the synthetase designed to bind Man-α-O-Tyr was predicted to be sufficiently selective for the target ligand and also thermodynamically stable. Our study suggests that extensive redesign of the tRNA synthatase binding pocket for large bulky ligands may be quite thermodynamically unfavorable.
Keywords: TyrRS, CDOCKER, Protein Design, Binding Pocket
1. Introduction
A general approach to genetically encode unnatural amino acids (AA) with diverse chemical, biophysical and biological properties into prokaryotes and eukaryotes was developed recently.1–4 Prior to protein synthesis, each of the 20 standard amino acids (AA) must be attached to their specific tRNA molecule by a specific aminoacyl-tRNA synthetase (AA tRNA-RS). During protein synthesis, mRNA codons are recognized by a specific tRNA anticodon resulting in selective AA incorporation into the elongating protein chain in the ribosome. Aminoacyl-tRNA synthetase/tRNA pairs from archaea have been shown to be orthogonal to the endogenous AA tRNA-RS/AA tRNA pairs in E. coli, which means they do not interfere with any of the host pairs. In vivo incorporation of unnatural AA into proteins was facilitated in response to the amber codon TAG by AA tRNA-RS selectively charging the orthogonal tRNA with a specific unnatural AA. Using this approach, more than 30 unnatural AA have been cotranslationally incorporated into proteins with high fidelity and efficiency in vivo.1–4 The identity of the AA is determined by the AA tRNA-RS specificity to covalently link a specific AA exclusively to its designated tRNA. This reaction involves several steps: selective binding of the AA and ATP to the synthetase, AA adenylation to activate the AA, selective binding of tRNA, and finally transfer of the adenylated AA to the 3′ end of the tRNA forming the aminoacyl-tRNA via a covalent ester linkage between AA and tRNA. Experimentally, a directed evolution approach is used to alter the specificity of the orthogonal synthetase enzyme for the target unnatural AA. This is accomplished by randomizing the AA identity of 4–6 positions in the binding pocket. Libraries of enzyme variants comprising on the order of 109 mutants are passed through a series of positive and negative selection steps. Repeated rounds of positive and negative selection may result in the isolation of specific enzyme mutants that successfully incorporate target unnatural AA but not endogenous AA.
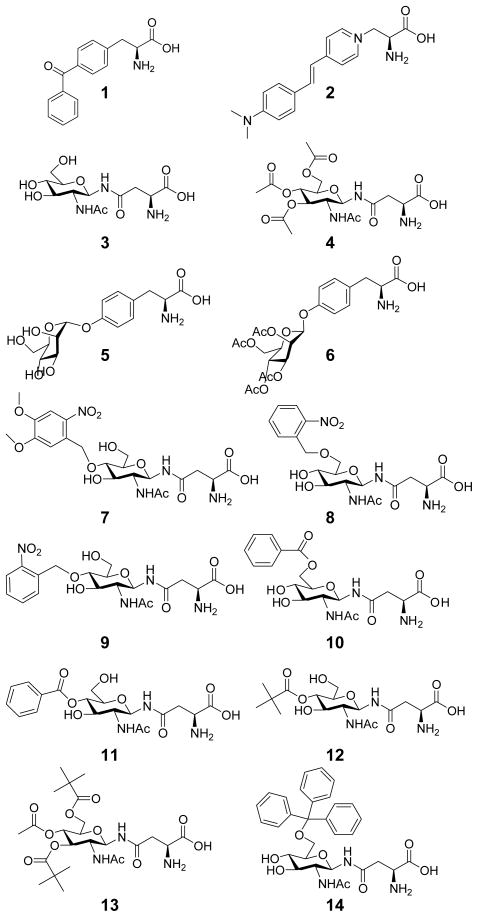
The objectives for this study are to demonstrate that CHARMM-based molecular docking and free energy calculations can augment the experimental approach in three ways: 1. by predicting reasonable binding geometries for target ligands and 2. by evaluating rationally designed mutants that improve the binding affinity for specific target ligands. 3. evaluating the change in thermodyanmic stability of the designs compared to the template synthatase. We designed and rebuilt AA tRNA-RS binding sites to accommodate new unnatural AA comprising significantly larger and/or more polar moieties. Our design effort focused on different N-linked GlucNAc AA with various protecting groups, Man-α-O-Tyr, as well as a styryl dye based AA as examples (Figure 1). Incorporation of these AA or their partially protected versions may have widespread use in addressing chemical and biological questions, and in the development of vaccines that require site-specific protein modification.5,6 Due to its high fidelity and relatively large AA binding site, the pBpa synthetase (specific for 1) was used as a template to design new AA tRNA-RS.7–9 Our study was aimed at identifying synthetase binding site variants with favorable thermodynamic ligand binding and protein stability.
Figure 1.
A series of 14 ligands that were docked into the pBpa synthetase. 1. p-benzoyl-Phe (pBpa) 2. Styryl Dye 3. GlucNAc-β-Asn 4. Ac4GlucNAc-β-Asn 5. Man-α-O-Tyr 6. Ac4Man-β-O-Tyr 7. 4-(2-Nitro-4,5-DimethoxyPh)GlucNAc-β-Asn 8. 6-(2-NitroPh) GlucNAc-β-Asn 9. 4-(2-NitroPh)GlucNAc-β-Asn 10. 6-BenzoylGlucNAc-β-Asn 11. 4-BenzoylGlucNAc-β-Asn 12. 4-PivGlucNAc-β-Asn 13. 4-Ac-3,6-DiPivGlucNAc-β-Asn 14. 6-Trt-GlucNAc-β-Asn
Our approach to designing specific protein-ligand interactions is based on the assumption that the AA binding pocket is rigid and that successful binding of the AA to the AA binding pocket is the most crucial aspect of the reaction. We assume that if the AA is capable of binding in the correct geometry with a reasonable affinity, adenylation will proceed forming an adenylated AA followed by efficient transfer to the tRNA. The crystal structures of E. coli TyrRS-Tyr (1x8x) and TyrRS-TyrAMS (1vbm) (TyrAMS is a L-tyrosyladenylate analogue) show that there are indeed induced fit conformational changes upon binding the AA and formation of the adenylated AA.10 These conformational changes are predominantly in the region involved in ATP binding (res:44–53) and the loops for tRNA recognition (res:226–245). However, there are minimal changes to the main chain of the AA binding pocket, and very slight deviations in side chain conformations. We conclude from these observations that it is reasonable to assume that the AA side chain binding pocket is rigid for designing specificity for an unnatural AA.
There have been a few previous attempts to use computational design methodology to suggest binding site mutations for AA tRNA-RS enzymes to increase the binding specificity of a target unnatural AA. Mayo and co-workers designed a PheRS variant that allows efficient incorporation of p-acetyl-L-phenylalanine.11 Two mutations, T261G and A314G, were designed for improved selectivity for p-acetyl-L-phenylalanine; these were subsequently verified experimentally.11 Goddard and co-workers used the docking methodology HierDock to predict the relative free energies of binding of various Phe analogues to PheRS, which were shown to correlate well with experimental translational activities.12,13 This approach was also used to study MetRS and SerRS, and the predicted binding affinities of AA variants correlated with experimental translational activities.14,15 Goddard and co-workers also used protein design to predict a double mutant Y32Q/D158A for Methanococcus jannaschii TyrRS that would preferentially bind O-methyl-L-Tyrosine.16 These two mutations were also identified experimentally from a previous library selection experiment by Schultz and co-workers. However, it has not yet been verified experimentally that these two mutations alone provide sufficient incorporation of O-methyl-L-Tyrosine.17,18 In the previous attempts to design specificity for p-acetyl-Phe and O-methyl-Tyr, both analogs were in the range of 4–6 atoms different than the natural AA, and the mutations (no more than 2 residues) were on the order of 12–20 atoms different than the natural binding pocket. Several of our target ligands are significantly different than the starting AA 1 (on the order of 20–45 atoms different), and therefore these ligands require many more mutations to accommodate the steric requirements.
Our basic approach to re-designing the binding site is similar to previous approaches, but our sampling of sequence space is limited to only a few binding site residues that have been used in experimental selection experiments. Our design goals aim to identify a set of possible binding site mutations for significantly larger ligands. In these cases, an experimental selection experiment of only 4–5 binding site residues may not provide sufficient steric space for target ligands, even if all residues were alanine. As these ligands are also much larger and more flexible than p-acetyl-Phe and O-methyl-Tyr, we allow the ligands to be flexible in rounds of design by placing harmonic restraints on the peptide backbone atoms: allowing the rest of the ligand to be flexible in a simulated annealing search for favorable conformations. Rather than selecting side chain positions that exhibit atom clashes with a specific ligand, 8 side chains were selected to be mutated to Ala to create an empty binding pocket for all of the ligands. The empty binding pocket was optimally rebuilt for each target ligand to optimize the binding free energy and minimize the number of mutations from the template synthatase. The change in thermodynamic stability compared to the pBpa synthetase was also calculated for each design.
2. Results and Discussion
2.1. Free Docking to pBpa Synthetase
A series of fourteen possible ligands were docked into the crystal structure of the pBpa synthetase. Docking this series established that there was not sufficient space in the current binding pocket for all of the larger sugar derivatives with protecting groups. For the reference ligand 1 the three lowest energy conformations were very close to the crystal structure: 1.52, 1.08 and 0.14 Å (heavy atom) RMSD. The accuracy of this docking protocol has been previously validated by predicting native-like ligand geometries on a diverse series of protein-ligand complexes from the LPDB.19,20
2.2. Comparison with 20 Natural AA
Assuming that all AA bind with the same peptide conformation, we used the Harmonic Restraint Simulated Anealing (HRSA) procedure to compare binding of 1 to the standard natural AA (Table 1). Using HRSA, the lowest two energy conformations for 1 were 1.0 and 0.7 Å (heavy atom) RMSD from the crystallographic conformation, which is similar to the results from free docking. As expected, Phe, Leu, and Tyr all found low energy conformations similar to the crystallographic conformation of the first phenyl group of 1. The calculated ΔGbinding for 1 was −8.9 kcal/mol from the lowest energy conformation from the HRSA search and −11.9 kcal/mol using the conformation from the crystal structure. After performing HRSA for the 20 standard AA, the four most favorable were Leu (−6.1 kcal/mol), Phe, (−5.5) Ile (−5.2), and Tyr (−4.9). Based on the conformations predicted from the HRSA search and our rough approximation to ΔGbinding, this would suggest a Kd ≥ 3×10−7 for 1 and a range of Kd from 4×10−5 to 9×10−5 for the best natural amino acids. This difference of 2 orders of magnitude is consistent with the experimental observation that the pBpa synthetase incorporates 1 with a high degree of selectivity.7,9 Expression experiments have not observed any evidence of incorporation of natural amino acids, but demonstrate specific incorporation of 1. These predictions for ΔGbinding provide a metric as to how specific a cognate protein-ligand interaction needs to be to design specificity for novel protein-ligand pairs. Our aim is to design a variant that is sufficiently selective in ligand binding and also thermodynamically stable. In the literature, this type of comparison has only been performed for native synthetase enzymes (PheRS, TyrRS, SerRS, MetRS),12,14–16 but has never been performed for a synthetase variant from selection experiments.
Table 1.
Calculated free energy of binding for target and standard amino acids to their cognate synthetase enzymes (ΔGbinding ≤ −6.0 kcal/mol shown in bold). The target ligand for the pBpa synthetase is ligand 1, and the target ligand for the 8Ala synthetase is ligand 4. The six synthetase enzymes shown in Figure 2 correspond to the six columns of this table.
| pBpa Synthetase | 8Ala Synthetase | Synthetase Design for Ligand 4 | Synthetase Design for Ligand 5 | Synthetase Design for Ligand 7 | Synthetase Design for Ligand 2 | |
|---|---|---|---|---|---|---|
| Target Ligand | −8.9 | −9.2 | −12.8 | −11.2 | −12.7 | −7.4 |
| Ala | −1.6 | −0.9 | −1.4 | 0.1 | −2.4 | −2.0 |
| Arg | 3.8 | 3.6 | 2.2 | 5.6 | −2.4 | 5.5 |
| Asn | −0.2 | −3.1 | −3.0 | −4.6 | −5.2 | −3.2 |
| Asp | 8.1 | 3.9 | 2.0 | 6.8 | 1.0 | 2.0 |
| Cys | −0.3 | 0.8 | 0.4 | −1.5 | −3.3 | −1.6 |
| Gln | −4.6 | −0.9 | −3.9 | −4.6 | −1.5 | −3.4 |
| Glu | 9.0 | −3.5 | 7.0 | 7.9 | 7.0 | 5.6 |
| Gly | 0.6 | 2.0 | 1.0 | 0.4 | −0.8 | −0.6 |
| His | −2.5 | −0.4 | −2.0 | −4.0 | −4.7 | −2.5 |
| Ile | −5.2 | −2.5 | −3.3 | −5.7 | −3.1 | −8.3 |
| Leu | −6.1 | −3.5 | −6.3 | −8.5 | −4.5 | −8.0 |
| Lys | 5.6 | 1.2 | 0.7 | −0.2 | 3.7 | 1.8 |
| Met | −0.3 | −1.6 | −3.6 | −0.7 | −1.5 | −0.7 |
| Phe | −5.5 | −5.0 | −5.1 | −4.5 | −5.5 | −5.9 |
| Pro | −4.4 | −3.0 | −2.6 | −4.9 | −4.4 | −2.9 |
| Ser | 0.6 | 1.8 | 0.3 | 0.0 | −3.1 | 0.5 |
| Thr | 1.0 | 1.1 | −0.9 | 0.5 | −1.9 | −0.9 |
| Trp | 0.3 | 2.2 | 0.8 | −0.3 | −0.8 | 0.0 |
| Tyr | −4.9 | −2.7 | −4.4 | −6.5 | −5.3 | −5.3 |
| Val | −3.4 | −0.5 | −2.7 | −3.0 | −2.5 | −3.4 |
2.3. Protein Design for Specific Target Ligands
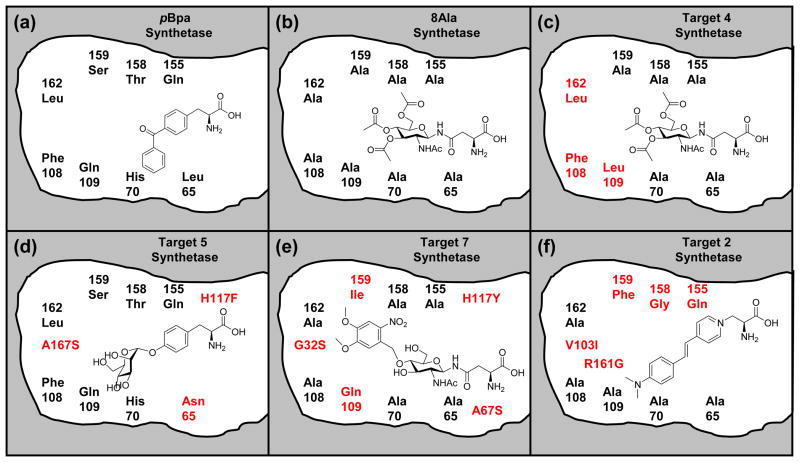
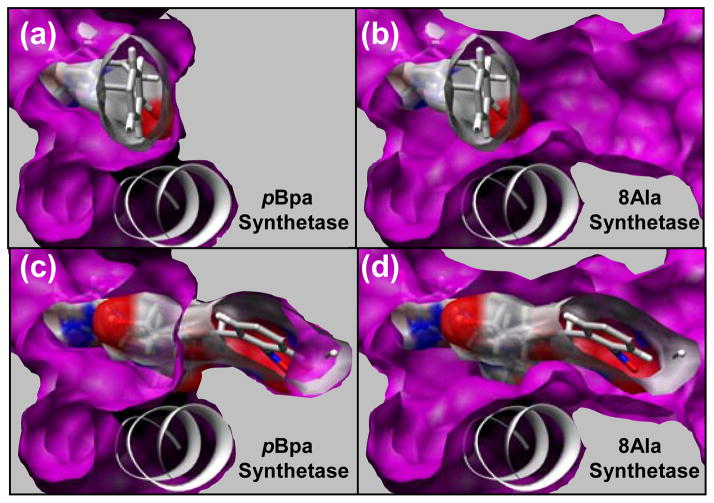
As the pBpa synthetase binding pocket did not have enough space to accommodate the large target ligands, 8 amino acids that caused steric clashes were mutated to Gly. Once HRSA was performed with 4 in the 8 Gly variant, it was clear that all 8 positions could tolerate an Ala residue. We denote this variant the “8Ala synthetase” (L65A, H70A, F108A, Q109A, Q155A, T158A, S159A, L162A) (Figure 2). Another iteration of HRSA with the 8Ala synthetase demonstrated that most of our target ligands (2–11) were now able to find low energy conformations in this redesigned binding pocket (Figure 3). For 4, the fit is fairly tight and only a few of the 8 residues could be mutated to anything other than Ala. The predicted ΔGbinding for 4 and 6 was −9.2 and −7.4 kcal/mol respectively. The predicted ΔG for 4 is as favorable as 1 to its specific synthetase (−8.9 kcal/mol). Non-bonded van der Waals interactions were primarily responsible for the favorable ΔGbinding. The fit to the binding site is quite tight for the fully acetylated sugar derivatives (4 and 6), and there was very little free volume remaining. The less favorable predicted ΔGbinding for 5 (−4.4 kcal/mol) compared to the larger acetylated derivatives is predominantly due to missing interactions and empty space in the binding pocket.
Figure 2.
Cartoon representation of the binding site of pBpa synthetase (A) and designs for target ligands. The position of residues in this cartoon attempt to represent the relative positions of the residues with respect to the peptide binding site and the back of the binding pocket defined by residues L162 and F108. However, proximity of mutations to functional groups shown in the ligands do not necessarily indicate close molecular interactions. In the 8Ala synthetase (B) all of the 8 residues shown were mutated to alanine. The mutations for the best designs for (C) 4, (D) 5, (E) 7, and (F) 2 are shown. The three mutations shown in red in (D) are relative to the pBpa synthetase, and mutations shown in red for (C), (E), and (F) are relative to the 8Ala synthetase.
Figure 3.
Comparison of the AA binding site for the pBpa synthetase (A, C) and the 8Ala synthetase (B, D). This view is from the perspective of the back of the binding site (L162, F108) looking towards the peptide binding site in the front of the binding site. Helix (res:150–163) is also shown, which is the predominant secondary structure element defining the bottom of the binding site. Solid gray is shown next to the surface of the binding pocket to indicate that this area is occupied by well packed side chains. The crystallographic conformation of 1 is shown in the AA binding pocket of the pBpa synthetase in (A, C) and in the 8Ala synthetase in (B, D). Surface of the ligand is shown to illustrate the free volume between the surface of the ligand and the binding pocket. The lowest energy conformation of 7 in the AA binding pocket of the 8Ala synthetase is shown in (C, D). The surface of this ligand is shown in (C, D) to illustrate the free volume between the surface of the ligand and the binding pocket. This lowest energy conformation of 7 has significant atom clashes with the surface of the pBpa synthetase, which is shown in (C) where the ligand penetrates into the solid gray area that is occupied by side chains.
A recent theoretical study of attractive interactions in carbohydrate binding sites showed that CHARMM performed well at reproducing interaction energies compared to ab initio calculations at the MP2 level of theory, suggesting that these molecular mechanics approaches are good approximations for aromatic-sugar interaction energies.21 It is possible that our approximation of ΔGbinding with the GBMV implicit solvent model is overestimating ΔGbinding because explicit dispersion forces are included in the calculation of the interaction energy in the protein-ligand complex (from explicit protein atoms), while there are no explicit solvent dispersion forces. This difference could lead to an overestimation of ΔGbinding for larger ligands compared to smaller ligands, but this difference should still remain a minor contribution to the free energy. In addition, our approximation of ΔGbinding does not include an entropic penalty for more flexible degrees of freedom, for example in 4 compared to ligand 1. However, this too should be a minor contributor to ΔGbinding.
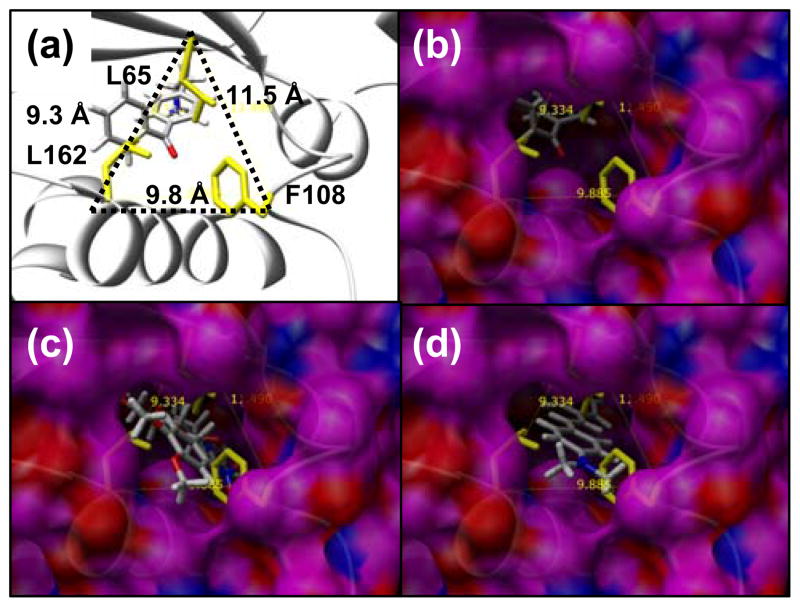
Several of the ligands had functional groups that bound in a newly created deep pocket near the back of the 8 Ala synthetase AA binding site. Two residues that were mutated to Ala (L162 and F108) define the back of the binding pocket at the C-terminus of a helix (res:50–63), such that mutating them to alanine opens up the back of the binding site to solvent (Figure 4). We denote these two residues L162A and F108A the “back door” residues, and the mutations L65A and Q109A are the next two closest mutations to the back door. Several of the target ligands had binding geometries that required the back door to be open, including 2 and 7–11. In comparing the benzoyl protecting group at the 4- or at the 6- position, it was found that the 4-benzoyl derivative 11 (−7.3 kcal/mol) was more favorable than the 6-benzoyl derivative 10 (−5.8 kcal/mol). The 6-benzoyl derivative 10 bound in a similar geometry to the 4-dimethoxy derivative 7, such that it requires the back door to be open (L162A, F108A). In contrast, the 4-benzoyl group of 11 bound in a slightly different conformation which was less solvent exposed. It was quite evident that building mutations for this geometry of 11 would allow position 109 to be Phe instead of alanine, which would likely result in a more stable protein in that it is more similar to the pBpa synthetase. Therefore, we conclude that further design for the 4-benzoyl group would likely be more promising than for the 6-benzoyl group.
Figure 4.
Mutations L162A, F108A, and L65A open up the back of the binding site to be exposed to solvent - the “back door”. The position between these residues relative to helix (res:150–163), the crystallographic conformation of pBpa, and the β-sheet that covers the top of the binding site is shown in (A). The distance between the Cα atom of each of these three residues is shown in (A). A surface of the outside of the 8Ala synthetase is shown in (B, C, D) where the view through the solvent exposed back door shows ligands in the binding site. Residues L162, F108 and L65 are shown in yellow, but alanine residues at these positions contribute to the surface shown for the 8Ala synthetase. In (B) the crystallographic conformation of pBpa is shown, in (C) the lowest energy conformation of 7 in the 8Ala synthetase is shown, and in (D) the lowest energy conformation of the 2 is shown in the 8Ala synthetase. Successful designs for 7 and 2 required that the back door remained open and exposed to solvent. The best designs for 4 and 5 were successful in closing the back door so no part of the binding site is exposed to solvent in this region.
We predict that opening up the back door to solvent in the 8 Ala synthetase AA binding site significantly destabilizes the protein by 8.4 Kcal/mol (Table 2). This may represent a physical limit to the size of a ligand that can be accommodated into the binding pocket. It would be very interesting if the protein could be stable with the back door open, or if an unstable protein could be stabilized by ligand binding during protein expression. If this was possible and the synthetase reaction was still viable, longer substituents could be designed to hang out of the back door of the binding site and into solvent. An example of such a molecule may be the alkyl chain of a fatty acid. Any viable design using an open back door will need to be stabilized by many other compensating mutations elsewhere in the sequence that optimize stability but are not perturbing to structure and folding. However, we are limiting our study to only binding site mutations.
Table 2.
Calculated change in the free energy of stability for each designed synthatase compared to the reference pBpa synthetase (ΔG shown in kcal/mol). The five synthetase enzymes designed to bind ligands shown in Figure 2 correspond to the five rows of this table. Only the design for 5 (ManOTyr) is predicted to have a thermally stable native state and the others are predicted to be unstable.
| Synthatase Name | ΔΔG | N Mutations |
|---|---|---|
| 8 Ala | 8.4 | 8 |
| design for 4 | 9.8 | 6 |
| design for 5 | −0.2 | 3 |
| design for 7 | 4.9 | 10 |
| design for 2 | 13.8 | 9 |
2.3.1. Protein Design for Target 4
Recent experimental studies have shown that E. coli cells for the expression system uptake 4 (Schiller & Schultz unpublished data), and cell lysates have been shown to contain 4, which is not significantly de-acetylated by enzyme activity in the cell lysate (Schiller & Schultz unpublished data). This fully acetylated version of 3 was therefore the most likely species to be able to diffuse to the variant synthetase. For the 8Ala synthetase, the predicted ΔGbinding for 4, (−9.2 kcal/mol) is on the order of the predicted ΔGbinding for 1 and its cognate synthetase. The binding of 20 standard AA were compared to the 8Ala enzyme, and the most favorable natural amino acid was Phe (−5.0 kcal/mol). From the binding geometry of 4 into the 8Ala synthetase, 5 rounds of building and HRSA refinement were performed. It was obvious from the initial binding geometry that the mutations from the back door of the enzyme did not need to be Ala, and they were restored to their native identity (L162A, F108A). Closing the back door further enhanced the predicted ΔG binding for 4 from −9.2 kcal/mol to −11.2 kcal/mol. The only other residue of the original 8 positions mutated to Ala that could tolerate a non-Ala residue was 109. The A109L mutation introduces a close hydrophobic interaction with the acetyl methyl group protecting the 6-hydroxyl group, and further decreases the ΔGbinding for 4 from −11.2 to −12.8 kcal/mol. These three mutations from the 8Ala protein (A162L, A108F, (should be 108) and A109L) completely close the back door of the binding site from solvent, and are more favorable for binding, but do not sufficiently improve protein stability (Table 2). The binding of 20 standard AA were compared to this best design for 4, and the most favorable natural AA was Phe (−6.3 kcal/mol), again suggesting that selectivity would be sufficient.
2.3.2. Protein Design for Target 5
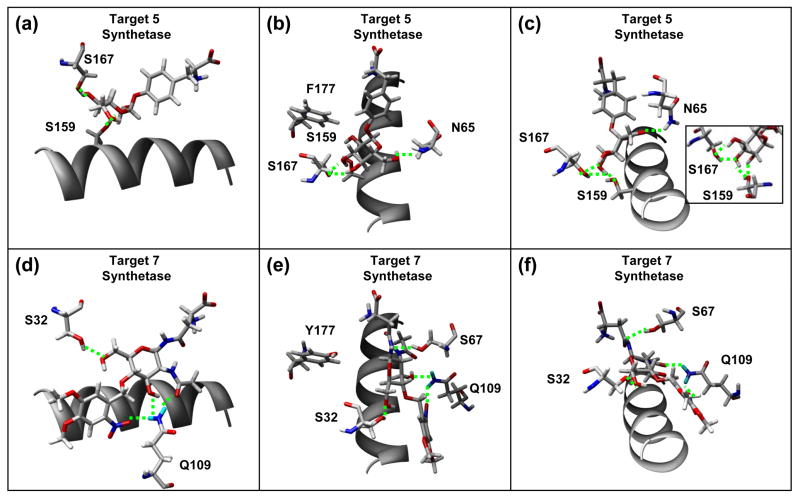
Recent experimental studies have shown that E. coli uptakes 5, and cell lysates have been shown to contain 5 (Schiller & Schultz unpublished data). Compound 5 bound in a reasonable conformation into the 8Ala synthetase (−4.4 kcal/mol), but it was clear that most of the Ala mutations could be replaced with native pBpa residues. HRSA also yielded a very similar conformation of 5 in the binding site of the unmodified pBpa synthetase (−3.3 kcal/mol). This is interesting considering that initial free docking was unsuccessful at finding this conformation. Since 5 was able to bind to the pBpa biding site, 13 rounds of building and HRSA were performed from the pBpa synthetase. The best design was 3 mutations from the native pBpa synthetase (L65N, A167S, and H177F), which had a predicted ΔGbinding of −11.2 kcal/mol, and also has a thermostability that is similar to the native pBpa synthetase (Table 2). This best design incorporates 4 specific hydrogen bonding interactions to the 3, 4 and 6-hydroxyl groups of the sugar (Figure 5). The mutation at position 65 to Asn incorporates a hydrogen bond from the NH2 group to the 6-hydroxyl group of the sugar. The mutation at position 167 to Ser introduces two hydrogen bonds to the 3-hydroxyl and the 4-hydroxyl group of the sugar. The native Ser159 provides hydrogen bonds to the 3-hydroxyl of the sugar. The H177F mutation replaces a less favorable polar - nonpolar aromatic interaction with a more favorable nonpolar aromatic interaction. However, this mutation, which is closer to the peptide binding site, also increased the affinity for Leu, Ile, Phe and Tyr. Compared to native pBpa, the H177F mutation decreases the predicted ΔG binding for Ile and Tyr by −2.4 and −1.6 kcal/mol respectively making them much more favorable. If the two mutations L65N and A167S are sufficient experimentally for specificity, the H177F mutation may not be necessary. It is interesting to consider this theoretical variant in a selection experiment. Is it possible that many successful variants from positive selection that successfully incorporate the target AA would be eliminated in multiple rounds of negative selection due to a slightly increased background.
Figure 5.
Specific hydrogen bonding interactions of the best designs for 5 (A, B, C) and 7 (D, E, F). Helix (res:150–163) is shown for reference. For 5, specific hydrogen bonds are made from S159, S167 and N65. For 7, specific hydrogen bonds are made by S32 and Q109, and favorable close polar-polar interactions are also made from S67 and Y177.
2.3.3. Protein Design for Target 7
This sugar variant contains a photocleavable protection group. Even though this target had a reasonable predicted binding affinity to the 8Ala synthetase (−8.1 kcal/mol), mutations were introduced to improve binding affinity in 67 rounds of sequential protein design. Every possible residue that could contribute a Lys or Arg residue to interact with the nitro group was examined, and very few had favorable Dunbrack library rotamers. The two main possibilities became Y114R and A109K. As Gln is the native residue at position 109 in the pBpa synthetase, the A109K and A109Q mutations were both found to be favorable. The best design for this ligand (A109Q, G32S, H117Y, A159I, A67S) has a predicted ΔGbinding of −12.7 kcal/mol, but was still predicted to be thermodynamically unstable by 4.9 kcal/mol (Table 2). This best design is 5 mutations away from the 8Ala synthetase, and only 7 mutations away from the pBpa synthetase (Figure 2e). Some specific hydrogen bonds and electrostatic interactions are shown for the best design in Figure 5. In the binding geometry of this best design, the A109Q mutation introduces a hydrogen bond to the 4-hydroxyl group and the nitro group. The G23S mutation introduces a hydrogen bond with the 6-hydroxyl group, and the A67S mutation introduces a close polar interaction between the Ser hydroxyl group and the N-linked nitrogen close to the peptide binding site. The H117Y mutation introduces a closer polar interaction between the Tyr hydroxyl and the ether oxygen in the sugar ring. For this best design for 7, the binding of the standard 20 AA were compared, and Phe, Tyr, and Asn were found to be the best (−5.5, −5.3, −5.2 kcal/mol respectively), which are much less favorable than that predicted for the target. The mutation H177Y makes the binding of Asn more favorable, as the endogenous Asn side chain interacts in a similar way to the region of the N-linkage in the ligand with the Y177 hydroxyl group.
This cycle of rational protein design may also be useful to predict mutations in a directed evolution library experiment. For these experiments, it must be determined which positions will be variable and which positions will be fixed at a specific AA identity. The best design for 7, which is five mutations from the 8Ala synthetase includes the mutations A109Q, G32S, H117Y, A159L, and A67S. The best design with only two mutations from the 8Ala synthetase is (A109K, G32S), and the best design with only three mutations is (A109Q, G32S, H117Y). All of the best designs incorporate combinations of the following mutations (A109A, A109K, Y114R, R161S, G32S, H117F, H117Y, A159I, A159L, A67S, and A155S). From this information it is possible to propose suggestions for library experiments for this specific ligand. The consensus from these rounds of building show that the best positions to fix as Ala are (65, 70, 108, 155, 158, 162) and the best positions to randomize are: 159, 177, 154, 176, and possibly 109. The best positions to fix as a non-Ala residue are Ser32 and Gln109.
2.3.4. Protein Design for Target 2
For ligand 2 we were unable to find conformations with a favorable predicted binding affinity for the 8Ala synthetase (+0.9 kcal/mol). The resonance delocalization of the positively charged ligand makes the ring in 2 rigidly planar. Very slight deviations from planarity are observed in crystal structures and simulations of stilbene bound to proteins,22,23 but it is expected that large deviations in planar geometry are extremely unfavorable. Several cycles of rational protein design demonstrated that the lowest energy conformations always had significantly unfavorable deviations from the planar structure. The most important mutations to minimize deviations in planar structure were R161G and A158G, which are both on the helix (res:150–163). These two mutations were necessary for 2 to lay down flat into the deep pocket along the helix formed by the opening of the back door (L162A, and F108A).
In 16 rounds of building, the best design was 5 mutations from the 8Ala synthetase (R161G, A158G, V103I, A159F, A155Q). This best design had the least deviations in planar structure and a predicted ΔGbinding of −7.41 kcal/mol (Figure 6). This protein-ligand interaction is predicted to be much weaker than the interaction between the benchmark 1 and its cognate synthetase. In addition, compared to the 3 other designed binding sites this one is not sufficient for selectivity. Out of all of the synthatase designs to improve binding affinity, this one was the most destabilizing to the protein (13.8 Kcal/mol), Ligand 2 was the most difficult of the ligand targets for computational design because of the planarity of its ring system. Because of problems associated with keeping this ring system planer when bound to the protein, we conclude that the design for 2 is the least reliable prediction. It was also interesting that the best design for 2 is less less selective, allowing several endogenous AA to bind more favorably, including Ile (−8.3 kcal/mol) and Leu (−8.0 kcal/mol) (Table 1). As with the design for 5, this increased affinity for the natural AAs is due to the mutation H177F, which is a close favorable hydrophobic interaction for Ile, Leu, Phe and Tyr. This observation may have interesting consequences for the design of selection experiments, as it is possible to have increased affinity for the target, but also have measurable background incorporation of standard AA. Negative selection rounds may eliminate some of the very favorable variants from positive selection that still have a reasonable affinity for natural AA.
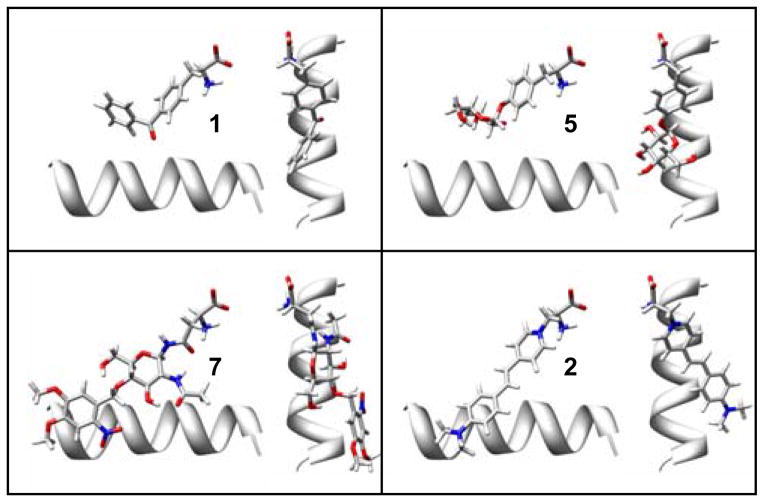
Figure 6.
Predicted ligand binding geometries of the best designs for 5, 7, and 2 compared to the crystallographic conformation of 1. The helix (res:150–163) is shown for reference. The binding geometry of 5 is very similar to 1. The 2-Nitro-4,5-DimethoxyPh protecting group of 7 can lay along the helix in an unstrained conformation. However, the requirement that 2 bound in a planar conformation was a significant complication that lead to less favorable designs.
2.4. Free Docking of Target Ligands Back into their Designed Proteins
All of our protein designs were constructed with the restraint that the peptide group of the ligand binds like the peptide group of 1. Therefore as an additional check on the design, the target ligands were re-docked without any restraints back into the designed enzymes. Free docking showed that each ligand could dock close to the designed geometry. In each case, the heavy atoms of the peptide groups come reasonably close to the native conformation (1–3 Å). This is in sharp contrast to free docking of the original 14 potential ligands into the pBpa synthetase, where most of the ligands bound in the ATP binding pocket instead of the AA specificity pocket. For docking into the 8Ala synthetase, ligand 4 was 4.5 Å (heavy atom RMSD) from the binding geometry after HRSA. In both of these cases the peptide groups are also very close to the native conformation, although the rest of the molecule exhibited some slight deviations.
For the more specific designs, better binding accuracy is achieved for target ligands compared to the 8Ala synthetase, as expected. For the best design for 4, the lowest energy pose was 5.0 Å RMSD, but the second lowest energy pose was 1.3 Å RMSD from the designed geometry. For the best design for 5, 7, and 2 the lowest energy pose was 1.6, 3.5 and 0.8 Å from the designed geometry respectively. For all of these ligands, the lowest energy pose from free docking also had very reasonable geometries for the peptide group, such that we may reasonably expect that the reaction could take place and form the covalent adenylated unnatural AA. Free docking tests the specificity of the designed protein-ligand interactions (binding geometry), and provided a good independent check on the selectivity of the design for the target ligand.
3. Conclusions
The original pBpa synthetase binding site was not of sufficient size to accommodate large sugar derivative target ligands, so the binding site was rebuilt with 8 mutations to Ala. Mutations to this binding site were designed for 4 specific targets: 2, 4, 5, and 7. Our approach has predicted a design for 4 that is only 3 mutations away from the 8Ala synthetase construct, and a design for 5 that is only 3 mutations away from the original pBpa synthetase. The designs for 7 and 2 require that the back door of the binding site is open, and are predicted to be thermodynamically unstable proteins. This highlights potential steric and thermodynamic limitations of synthetase AA binding pockets for incorporating large bulky ligands. The cycles of design are also useful in predicting which positions should be variable and which positions should be fixed in library selection experiments. The best protein designs for 4, 5, and 7 show a significantly more favorable ΔGbinding for the target ligand that any of the 20 endogenous AA. These predicted ΔG binding are on the order of the specificity predicted for 1 compared to the standard 20 AA in its own cognate pBpa synthetase from selection experiments. However, only the design for 5 (Man-α-O-Tyr) was predicted to have sufficient ligand selectivity and native-like thermostability.
4. Computational Details
4.1. Docking with CHARMM (CDOCKER)
A 1.0 Å grid is used to describe the static protein conformation of the protein binding site, where the interaction energy of 20 types of probe atoms is calculated for every point on the grid. The grid is calculated to extend 8 Å in all directions from any atom in the ligand. This is large enough to describe the AA binding site as well as the ATP binding site. The flexible ligands are modeled with an all atom representation including hydrogen atoms. The 3D coordinates were built using ChemOffice 2006 and CS Chem3D Ultra (CambridgeSoft, Version 10.0) from chemdraw files. The CHARMm force field,24–26 originally parameterized by Momany and Rone, has been extended to describe ligands in the ligand-protein databank (LPDB) and was used to build the potential energy function for all ligands.19
During the docking procedure, a random configuration of the flexible ligand is generated by running 1000 steps (1 fs) of molecular dynamics (MD) at 1000 K in vacuo. During this generation of novel conformations, electrostatic interactions are turned off to facilitate conformational randomization. For each random conformation generated, 10 random rigid body rotations about its center of mass are used as the initial conformation in the vicinity of the binding site. For the native ligand 1, this results in starting positions that are as far away as 33 Å heavy atom RMSD from the native conformation. The ligand interacts with the potential energy for the protein mapped to a grid using a soft-core repulsion term for both van der Waals and electrostatics interactions. An MD simulated annealing is used starting from these initial binding site conformations to search for low energy conformations of the ligand in the 3D grid. The heating phase consists of 4000 MD steps (1 fs) heating from a temperature of 300 K to 700 K. The cooling phase consists of 10000 MD steps (1 fs) from 700 K back to 300 K. The potential energy of the ligand on the grid is minimized with a steepest descent minimization of 1000 steps. Finally, the all atom representation of the rigid protein is restored and the all atom protein-ligand representation is minimized fixing the coordinates of the protein, using the standard all atom potential function with a distant dependent dielectric. This interaction energy is taken as the score for the final ligand pose. Other details of the CDOCKER setup and protocol have been published previously.20,27–32 A series of 14 ligands (Figure 1) were docked into the 2.5 Å crystal structure of the pBpa synthetase (specific for 1).33 For each free docking run a total of 2000 docking attempts were performed from 200 generated random conformations (each with 10 different random rotations).
4.2. Harmonic Restraint Simulated Annealing (HRSA)
Given that the position of the binding site for the AA peptide group is known (this position is relatively conserved in all known TyrRS mutants, a requirement for activation and transfer to the tRNA), harmonic restraints were used to reduce the conformational space for the simulated annealing search for low energy conformations of the amino acid side chain in the binding pocket. It was assumed that the peptide groups of all possible amino acids (both unnatural and endogenous) would bind exactly like 1. Harmonic restraints of 1000 kcal/mol/Å2 were applied to the coordinates of five heavy atoms of the peptide backbone (N, CA, C, O1, O2). Multiple cycles of restrained simulated annealing were performed with a ligand. To randomize the backbone restrained position of the AA ligand 5000 steps of MD were performed in vacuo (with the electrostatic interactions turned off). During this search the peptide group remained restrained based on its known binding conformation. After the search, the grid representation of the protein was restored and the all-atom model of the ligand interacts with the potential energy for the protein mapped to a grid using a soft-core repulsion term for both van der Waals and electrostatic interactions. Five sequential cycles of simulated annealing were performed for each random conformation generated. A heating cycle, 4000 MD steps (1 fs) for heating (300 to 700 K), and a cooling cycle, 10000 MD steps (1 fs) for cooling (700 K to 300 K) were performed. For each restrained refinement trial, 25 initial conformations were generated and each was refined with sequential simulated annealing cycles.
The final structures were subjected to minimization, first on the grid and then with the all-atom representation of the protein as described above for free docking. The CHARMM energies from the minimization with the all-atom representation of the protein-ligand complex was used to rank ligand poses. The conformational space of this search in the binding pocket is reduced sufficiently such that the searches converged rapidly to a small number of, 10–20, unique low energy conformations (depending on ligand complexity). The three lowest energy conformations from all trials were selected as solutions. These conformations were then re-scored with an implicit solvent model to calculate ΔG binding. The Generalized-Born Molecular Volume (GBMV) model is used to represent the solvent.34–36 Each model was minimized with 200 steps of steepest descent minimization using the GBMV implicit solvent and the rest of the CHARMM all atom potential function. This minimization was performed separately on the complex, free protein and free ligand. After minimization, the total potential energy was calculated as the sum of all components including the GBMV energy. The potential energy difference between the products and the reactants [complex - (protein + ligand)] was used as our estimate of ΔGbinding. This approximation includes the desolvation of the ligand in going from a solvated state to the bound state via the potential energy term from the GBMV implicit solvent model. In using this approach to score the native conformations of crystal structures from the Ligand Protein Database (LPDB),19 scaling the difference by a constant results in magnitudes that are approximately on the order of experimental Δ Gbinding values (Armen & Brooks unpublished data). In this paper we report these scaled ΔGbinding values from HRSA refinement. However, they are not intended to represent a quantitative prediction of binding free energy but are rather a computationally inexpensive “heuristic score” to rank protein designs.
4.3. Protein Design for Specific Ligand Targets
The HRSA procedure was used to design specific protein-ligand interactions for target unnatural amino acids, assuming that the peptide groups bind in the correct orientation for the adenylation reaction to proceed. As most of the target unnatural AA were much larger than 1, our approach was first to identify all large side chains that may result in atom clashes with target ligands. Eight amino acid positions in the binding pocket were mutated to glycine and later to alanine (“8Ala synthetase”). HRSA was then used to search for low energy conformations of the target AA in the binding pocket. Rational mutations were designed based on these low energy conformations, and binding pocket side chains were built back from Ala using the backbone (ϕ,ψ) specific Dunbrack rotamer library (2002 version).37–39 In building side chains only the most favorable 1–3 rotamers were considered. While placing the most favorable rotamers at a specific residue position, it was assumed that all other residues remained rigid.
For each round of building and HRSA, the ligand remained flexible, but the side chain rotamers built from the Dunbrack library were rigid as was the rest of the protein. For each round of building, the three lowest energy conformations were used to calculate ΔG, and the lowest ΔG was used to rank protein designs. Designs for target ligands 2, 4, and 7 were built starting from the 8Ala synthetase. For target ligands 2, 4, and 7, multiple rounds of building (15, 5, and 67, respectively) and HRSA refinement were performed starting from 8Ala synthetase. For target ligand 5, 11 rounds of building were performed starting from pBpa synthetase.
4.3. Calculation of Change in Free Energy of Stability
The GBMV solvent model was used for solvation energies for the calculation of changes in thermodynamic stability. A linear interaction energy (LIE) model using the GBMV implicit solvent model was fit to describe an experimental dataset of 183 mutations and corresponding change in stability (ΔΔG) for 7 small proteins with well characterized protein folding (ubiquitin, barnase, interleukin-1 beta, C-MYB, protein G, Histidine-containing Phosphocarrier protein HPR, and CI2).40 The LIE model perfomed as well as results published for a method based on machine learning with vector support machines40, where both models had a similar perfomance over all 183 mutations (R2=0.31, AUE=0.9 Kcal/mol, RMS=1.1 kcal/mol). Using the LIE model, differences in potential energy components Δ(vdw) and Δ(elec+gbe) upon mutation were calculated for both the folded state (modeled from the crystal strucure) and the unfolded state (modeled by extended AAXAA peptides). An LIE model of the form [ΔG=(α*(Δ(VDW))+β(Δ(elec+gbe))] was used to calculate the various legs [ ΔΔGN-D =ΔGN−ΔGD ] of the free energy cycle. The values of the coefficients for the best fit (α=0.279 β=−0.001) to 105 barnase mutants (R2=0.34, AUE=0.9 Kcal/mol) are similar in sign and magnitude of other recent fits for protein-ligand interactions using LPDB Charmm forcefield which provides confidence in using these LIE weights. For each designed synthatase variant, the LIE model fit to the barnase mutants was used to predict the change in thermostability.
Acknowledgments
This work computational/theoretical work presented here was supported by grants from the National Institutes of Health to C.L.B. (GM037554) and R.S.A. (GM076836). The experimental work by S.M.S. has been supported by the National Institutes of Health (GM62159) to P.G. Schultz. Finally, we acknowledge Professor Pete Schultz for discussing aspects of their ongoing experiments.
Abbreviations
- AA
amino acid
- ATP
adenosine triphosphate
- AA tRNA-RS
aminoacyl-tRNA synthetase
- E. coli
Escherichia coli
- MetRS
methionine-tRNA synthetase
- PheRS
phenylalanine-tRNA synthetase
- TyrRS
tyrosine-tRNA synthetase
- pBpa
p-benzoyl-L-phenylalanine
- MD
molecular dynamics
- LPDB
Ligand-Protein Database
- fs
femtosecond
References
- 1.Wang L, Schultz PG. Expanding the genetic code. Angew Chem Int Ed Engl. 2005;44(1):34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 3.Xie J, Schultz PG. Adding amino acids to the genetic repertoire. Curr Opin Chem Biol. 2005;9(6):548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, Schultz PG. An expanding genetic code. Methods. 2005;36(3):227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Xu R, Hanson SR, Zhang ZW, Yang YY, Schultz PG, Wong CH. Site-specific incorporation of the mucin-type N-acetylgalactosamine-alpha-O-threonine into protein in Escherichia coli. J Amer Chem Soc. 2004;126(48):15654–15655. doi: 10.1021/ja044711z. [DOI] [PubMed] [Google Scholar]
- 6.Zhang ZW, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. A new strategy for the synthesis of glycoproteins. Science. 2004;303(5656):371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 7.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci, USA. 2002;99(17):11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin JW, Schultz PG. In vivo photocrosslinking with unnatural amino acid mutagenesis. Chembiochem. 2002;3(11):1135–1137. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Ryu YH, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3(4):263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Takimura T, Sekine R, Kelly VP, Kamata K, Sakamoto K, Nishimura S, Yokoyama S. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase (vol 346, pg 105, 2005) J Mol Biol. 2005;354(3):739–739. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 11.Datta D, Wang P, Carrico IS, Mayo SL, Tirrell DA. A designed phenylalanyl-tRNA synthetase variant allows efficient in vivo incorporation of aryl ketone functionality into proteins. J Amer Chem Soc. 2002;124(20):5652–5653. doi: 10.1021/ja0177096. [DOI] [PubMed] [Google Scholar]
- 12.Kekenes-Huskey PM, Vaidehi N, Floriano WB, Goddard WA. Fidelity of phenylalanyl-tRNA synthetase in binding the natural amino acids. J Phys Chem B. 2003;107(41):11549–11557. [Google Scholar]
- 13.Wang P, Vaidehi N, Tirrell DA, Goddard WA., III Virtual screening for binding of phenylalanine analogues to phenylalanyl-tRNA synthetase. J Amer Chem Soc. 2002;124(48):14442–14449. doi: 10.1021/ja0175441. [DOI] [PubMed] [Google Scholar]
- 14.Datta D, Vaidehi N, Zhang DQ, Goddard WA., III Selectivity and specificity of substrate binding in methionyl-tRNA synthetase. Prot Sci. 2004;13(10):2693–2705. doi: 10.1110/ps.04792204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClendon CL, Vaidehi N, Kam VWT, Zhang DQ, Goddard WA., III Fidelity of seryl-tRNA synthetase to binding of natural amino acids from HierDock first principles computations. Prot Engn Des & Sel. 2006;19(5):195–203. doi: 10.1093/protein/gzl001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang DQ, Vaidehi N, Goddard WA, Danzer JF, Debe D. Structure-based design of mutant Methanococcus jannaschii tyrosyl-tRNA synthetase for incorporation of O-methyl-L-tyrosine. Proc Natl Acad Sci U S A. 2002;99(10):6579–6584. doi: 10.1073/pnas.052150499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292(5516):498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang L, Schultz PG, Wilson IA. Crystal structures of apo wild-type M-jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Prot Sci. 2005;14(5):1340–1349. doi: 10.1110/ps.041239305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roche O, Kiyama R, Brooks CL. Ligand-Protein DataBase: Linking protein-ligand complex structures to binding data. J Med Chem. 2001;44(22):3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 20.Taufer M, Crowley M, Price DJ, Chien AA, Brooks CL. Study of a highly accurate and fast protein-ligand docking method based on molecular dynamics. Concurrency and Computation-Practice & Experience. 2005;17(14):1627–1641. [Google Scholar]
- 21.Spiwok V, Lipovova P, Skalova T, Vondrackova E, Dohnalek J, Hasek J, Kralova B. Modelling of carbohydrate-aromatic interactions: ab initio energetics and force field performance. J Comput-Aided Mol Des. 2005;19(12):887–901. doi: 10.1007/s10822-005-9033-z. [DOI] [PubMed] [Google Scholar]
- 22.Salsbury FR, Jr, Han WG, Noodleman L, Brooks CL., III Temperature-dependent behavior of protein-chromophore interactions: a theoretical study of a blue fluorescent antibody. Chemphyschem. 2003;4(8):848–855. doi: 10.1002/cphc.200300694. [DOI] [PubMed] [Google Scholar]
- 23.Simeonov A, Matsushita M, Juban EA, Thompson EHZ, Hoffman TZ, Beuscher AE, Taylor MJ, Wirsching P, Rettig W, McCusker JK, Stevens RC, Millar DP, Schultz PG, Lerner RA, Janda KD. Blue-fluorescent antibodies. Science. 2000;290(5490):307–313. doi: 10.1126/science.290.5490.307. [DOI] [PubMed] [Google Scholar]
- 24.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM - a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 25.Momany FA, Rone R. Validation of the general-purpose QUANTA(R)3. 2/CHARMM(R) force-field. J Comput Chem. 1992;13(7):888–900. [Google Scholar]
- 26.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., III Comparative study of several algorithms for flexible ligand docking. J Comp Mol Des. 2003;17(11):755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 28.Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: The effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47(1):45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- 29.Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL., III Assessing scoring functions for protein-ligand interactions. J Med Chem. 2004;47(12):3032–3047. doi: 10.1021/jm030489h. [DOI] [PubMed] [Google Scholar]
- 30.Vieth M, Hirst JD, Dominy BN, Daigler H, Brooks CL. Assessing search strategies for flexible docking. J Comput Chem. 1998;19(14):1623–1631. [Google Scholar]
- 31.Vieth M, Hirst JD, Kolinski A, Brooks CL., III Assessing energy functions for flexible docking. J Comp Chem. 1998;19(14):1612–1622. [Google Scholar]
- 32.Wu GS, Robertson DH, Brooks CL, III, Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER - A CHARMm-based MD docking algorithm. J Comp Chem. 2003;24(13):1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 33.Liu W, Alfonta L, Mack AV, Schultz PG. Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases. Angew Chem Int Ed Engl. 2007;46(32):6073–6075. doi: 10.1002/anie.200701990. [DOI] [PubMed] [Google Scholar]
- 34.Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL., III Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J Comp Chem. 2004;25(2):265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 35.Lee MS, Feig M, Salsbury FR, Brooks CL., III New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J Comp Chem. 2003;24(11):1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 36.Lee MS, Salsbury FR, Brooks CL., III Novel generalized Born methods. J Chem Phys. 2002;116(24):10606–10614. [Google Scholar]
- 37.Bower MJ, Cohen FE, Dunbrack RL. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: A new homology modeling tool. J Mol Biol. 1997;267(5):1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 38.Dunbrack RL. Rotamer libraries in the 21(st) century. Curr Opin Struct Biol. 2002;12(4):431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 39.Dunbrack RL, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Prot Sci. 1997;6(8):1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capriotti E, Fariselli P, Calabrese R, Casadio R. Predicting protein stability changes from sequences using support vector machines. Bioinformatics. 2005;21(Suppl 2):ii54–58. doi: 10.1093/bioinformatics/bti1109. [DOI] [PubMed] [Google Scholar]