Abstract
Helicobacter pylori causes peptic ulceration and gastric adenocarcinoma. The aims were to study the influence of dupA1 positivity upon interleukin-8 (IL-8) secretion from gastric mucosa and determine the prevalence of mutations responsible for clarithromycin and fluoroquinolone resistance. DNA was extracted from 74 biopsies and the virulence factors were studied. Levels of IL-8 in gastric mucosa were measured using ELISA and the mutations responsible for clarithromycin and fluoroquinolone resistance were determined using a GenoType-HelicoDR assay. The prevalence of cagA in strains isolated from gastric ulcer (GU) and duodenal ulcer (DU) was significantly higher than those isolated from non-ulcer disease (NUD) (90% and 57.9% versus 33.3%; p 0.01). The vacA s1m1 genotype was more prevalent in patients with DU (73.7%) and GU (70%) than in those with NUD (13.3%) (p 0.01). The prevalence of dupA1 was higher in DU patients (36.8%) than those with GU (10%) and NUD (8.9%) (p 0.01). Multivariate analysis showed that a cagA+/vacA s1i1m2 virulence gene combination was independently associated with the developing peptic ulcer disease (PUD) with increased odds of developing PUD (p 0.03; OR = 2.1). We found no significant difference in the levels of IL-8 secretion in gastric mucosa infected with H. pylori dupA-negative and H. pylori dupA1-positive strains (dupA-negative: mean ± median: 28 ± 26 versus 30 ± 27.1 for dupA1; p 0.6). While 12 strains were clarithromycin resistant, only three isolates were levofloxacin resistant. A significant association was found between dupA1 genotype and A2147G clarithromycin resistance mutation (p <0.01). Further study is needed to explore the relationship between virulence factors and disease process and treatment failure.
Keywords: Antibiotic sensitivity, clarithromycin, dupA, fluoroquinolones, Iraq
Introduction
Helicobacter pylori infection is associated with peptic ulcer disease (PUD) and gastric cancer. Virulence factors such as cagA, vacA and dupA play an important role in the pathogenesis of H. pylori-related diseases. Strains of H. pylori possessing cagA have been shown to associate with a significantly increased risk for the development of atrophic gastritis, peptic ulcer disease and gastric cancer [1]. The vacA and its polymorphism contribute significantly to the disease development process. According to the segment region (s) and middle region (m) the status of vacA can be classified into four subtypes: s1m1, s1m2, s2m1 and s2m2. In vitro experiments showed that s1m1 strains are the most cytotoxic, followed by s1m2 strains, whereas s2m2 strains have no cytotoxic activity, and s2m1 strains are rare [1]. Research showed that subjects infected with vacA s1 or m1 isolates have an increased risk of peptic ulcer and/or gastric cancer compared with those with s2 or m2 isolates [1–3]. Rhead et al. have recently described a novel determinant of VacA toxicity, the intermediate (i) region [4]. The duodenal ulcer promoting gene A (dupA) is another virulence determinant that has been shown to associate with duodenal ulcer (DU) in some countries. Also, the presence of dupA was found to be related to neutrophil infiltration and a high level of interleukin-8 (IL-8) production by epithelial cells [5,6]. Furthermore, dupA was previously classified into two main subtypes: the functional dupA with an extended open reading frame (ORF) within jhp0917-19 (dupA1), and non-functional dupA with an early stop codon to truncate the ORF (dupA2) [5].
Since the discovery of H. pylori, the mainstream treatment of PUD has changed significantly [7]. The use of antibiotics to eradicate H. pylori infection is usually not chosen on the basis of routine susceptibility testing because of the difficulty of growing H. pylori. The eradication regimen should include a proton pump inhibitor and more than one antibiotic [7]. Historically, clarithromycin has been used as a first-line treatment for H. pylori. The eradication rate drops from 88% in the case of a clarithromycin-susceptible strain to <20% in the case of clarithromycin resistance [7]. With fluoroquinolones, which can be used as a second line of treatment, resistance was also shown to be correlated with treatment failure [8,9]. Because resistance rates vary according to the country and patient characteristics, the choice of antibiotics on the basis of susceptibility pattern in the region might be an effective strategy to improve H. pylori eradication [7]. Resistance to clarithromycin and fluoroquinolone is the result of mutations and these mutations are now well known [9–12]. Clarithromycin resistance is the result of point mutations in the rrl gene encoding the 23S rRNA. Three major mutations have been described: A2146C, A2146G and A2147G [10]. The resistance of H. pylori to quinolones results from point mutations in the gyrA gene encoding the A subunit of the DNA gyrase, mainly at codons 87 and 91 [9,11].
Helicobacter pylori virulence factors and their relationship with disease outcomes in Iraq were studied in this paper. In addition, the association of dupA polymorphisms with IL-8 secretion from gastric mucosa was studied. Also, molecular techniques were used to study the sensitivity pattern of H. pylori to clarithromycin and fluoroquinolone.
Materials and methods
Gastric biopsies
Gastric biopsies were obtained from 74 H. pylori+ patients from Iraq undergoing routine upper gastrointestinal endoscopy to investigate dyspepsia. Mean age ± standard deviation was 37 ± 13 years. Endoscopic diagnoses were: DU 19, gastric ulcer (GU) 10 and non-ulcer disease (NUD) 45. During gastroscopy, biopsy samples were taken and placed in 1 mL of isosensitest broth (Oxoid, Basingstoke, UK) containing 15% (volume/volume) glycerol and stored in liquid nitrogen. DNA was extracted directly from the biopsy specimens and used for PCR-based H. pylori typing.
The study protocol was approved by the Ethics and Research Committees of the individual hospitals and all patients gave informed consent to the study.
Antibiotic susceptibility
Helicobacter pylori antimicrobial susceptibility was investigated using mutational analysis to clarithromycin and fluoroquinolone using the GenoType HelicoDR kit (Hain Lifescience, Nehren, Germany) according to the manufacturer's instructions. To detect fluoroquinolone resistance, four gyr87 wild-type probes (gyr87WT1–gyr87WT4) and one mutant probe (gyr87MUT), and one gyr91 wild-type probe (gyr91WT1) and three mutant probes (gyr91MUT1–gyr91MUT3) were used to detect fluoroquinolone resistance at positions 87 and 91, respectively. To detect clarithromycin resistance, one wild-type probe (23SWT) and three mutant probes (23SMUT1–23SMUT3) were used with the presence of three controls: conjugate control (CC), amplification control (AC) and H. pylori (HP). When one of the wild-type probes stained positive together with the gyr91WT as well as 23SWT and no mutation band formed, the results were interpreted as susceptible to the respective antibiotic. The presence of a band at CC and AC meant that the CC and AC were in the right frame whereas a band at HP implied the presence of H. pylori according to the manufacturer's instruction.
Virulence factors genotyping
Thermal cycling for amplifying cagA was 95°C for 30 s, 50°C for 1 min, and 72°C for 2 min, for a total of 35 cycles. PCR amplification of cagA used previously described primers cag2 (5′-GGAACCCTAGTCGGTAATG-3′) and cag4 (5′-ATCTTTGAGCTTGTCTATCG-3′) to amplify about 500-bp product from the middle of cagA (PAI) [13]. For vacA signal and middle region, thermal cycle conditions were 30 s at 95°C, 60 s at 56°C and 90 s at 72°C, performed for 35 cycles. Primers used for the middle region were vag-F (5′-CAATCTGTCCAATCAAGCGAG-3′), and vag-R (5′-GCGTCTAAATAATTCCAAGG-3′). For the signal region, A3436 (5′-ATGGAAATACAACAAACACAC-3′) and C1226 (5′-CTGCTTGAATGCGCCAAAC-3′) primers were used [14]. The PCR conditions for vacA i region were as follows: the template mixtures were amplified 35 cycles with primers VacF1 (5′-GTTGGGATTGGGGGAATGCCG-3′) and C1R (5′-TTAATTTAACGCTGTTTGAAG-3′) for i1 region and VacF1 and C2R (5′-GATCAACGCTCTGATTTGA-3′) for i2 region, at 95°C for 30 s, 53°C for 1 min, and 72°C for 30 s [4]. The dupA was amplified using three sets of primers: DupA-WXF (5′-GATATACCATGGATGAGTTCYRTAYTAACAGAC) and DAR1 (5′-TTAAATACTCTTCCTTATAAGTTTCTTGG); JHP0919R2 (5′-GCCCACCAGTTGCAAAAACAAATGAAC) and DupA918F (5′-CCTATATCGCTAACGCGCTC); and dupA-F0 (5′-TGG CGT GTG GCA GTC TAA TGC) and dupA-R1 (5′-GCT CAA CAA AAT GCC CAC CAG TCG C) then, the amplified genes were sequenced on both strands using different primers (Table 1).
Table 1.
Oligonucleotide primers used for DNA sequencing of Helicobacter pylori dupA
| Primers | Sequences |
|---|---|
| DupA918F | CCTATATCGCTAACGCGCTC(9) |
| JHP0919R2 | GCCCACCAGTTGCAAAAACAAATGAAC(9) |
| DAR1 | TTAAATACTCTTCCTTATAAGTTTCTTGG(9) |
| DupA-WXF | GATATACCATGGATGAGTTCYRTAYTAACAGAC(9) |
| dupA-F0 | TGG CGT GTG GCA GTC TAA TGC(26) |
| dupA-F1 | GCA AAC GCT CAA ACT ATT GCC(26) |
| dupA-F2 | ATG TTT CTT GGT TTA GAG GG(26) |
| dupA-F3 | CAG AAC ACA AGC TTT AAA TGA ATT G(26) |
| dupA-F4 | ATG AGT TCT ATA CTA ACA GAC TTTGAG CC(26) |
| dupA-F5 | GGT TTC TAC TGA CAG AGC AC(26) |
| dupA-F6 | GAT AAT TGG TAG CAC AGG AAG CGG(26) |
| dupA-F7 | GGC TCT AGC GAA CAA GAT TTT AAT GAG(26) |
| dupA-R1 | GCT CAA CAA AAT GCC CAC CAG TCG C(26) |
Mucosal IL-8 level
ELISA was used to measure the IL-8 levels in the antral biopsy specimens (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Briefly, samples were homogenized first and then aliquots of supernatant tissues were obtained by centrifugation. The total protein in the supernatants was measured by using the Bradford assay (Bio-Rad, Richmond, CA). ELISA sensitivities were approximately 5 pg/mL for IL-8.
Data analysis
Statistical analysis of data was performed by using stepwise logistic regression and the chi-squared test with significance set at a p < 0.05 using Minitab 15 software (Minitab Ltd., Coventry, UK).
Results
Association between virulence factors and clinical outcomes cagA, vacA, vacAs, m, combinations and dupA status
The prevalence of cagA+ strains in the study was 47.3% (35/74). Regarding the vacA genotypes, 70/74 (94.6%) of the studied strains typed as vacAs1. The vacAm2 was found in 47 (63.51%) of our 74 samples. In addition, the vacA i1 genotype was found in 42 of the 74 (56.76%). Regarding the combination of the s region, i region and m region of vacA, the prevalence of the genotype vacA s1i2m2 was highest (37.8%) (28/74), the prevalences of genotypes vacA s1i1m1 and s1i2m1 were 36.49% (27/74) and 20% (15/74), respectively. The genotype vacA s212m2 was found in only four patients, all of them with NUD (Table 2).
Table 2.
Association of vacA polymorphisms with clinical outcomes
| Outcome (n) | s1i1m1, n (%) | s1i2m1, n (%) | s1i2m2, n (%) | s2i2m2, n (%) |
|---|---|---|---|---|
| DU (19) | 14 (73.7)∗ | 3 (15.8) | 2 (10.5) | 0 (0) |
| GU (10) | 7 (70)∗ | 3 (30) | 0 (0) | 0 (0) |
| NUD (45) | 6 (13.3) | 9 (20) | 26 (57.8) | 4 (8.9) |
| Total (74) | 27 (36.4) | 15 (20.2) | 28 (37.8) | 4 (5.34) |
Significant association.
dupA was previously classified into two main groups: the common extended ORF within jhp0917-19 (dupA1), and dupA with an early stop codon to truncate the ORF (dupA2) [15]. In our study the dupA1 genotype was found in 12/74 (16.2%) and only 5/74 (6.7%) patients had the dupA2 genotype.
The effect of the virulence factors on the disease outcome
DNA was extracted from gastric mucosa of 74 H. pylori-positive patients (19 DU, 10 GU and 45 NUD). The prevalence of cagA+ strains isolated from GU and DU patients was found to be significantly higher than those isolated from patients with NUD (90% and 57.9% versus 33.3%; p 0.01 for both).
All H. pylori strains isolated from patients with DU and GU had the vacAs1 genotype, whereas 94.6% of the strains isolated from patients with NUD had the vacAs1 genotype (p >0.05). The vacAm1 genotype was found to be more prevalent in strains isolated from DU (73.7%) and GU (70%) than those from NUD (14%) (p <0.01). The prevalences of vacAi1 strains isolated from DU and GU were 89.47% and 90%, respectively, and were significantly higher than that found in NUD (33.3%) (p 0.01 for both). The vacA s1i1m1 genotype was more prevalent in patients with DU (73.7%) and GU (70%) than those with NUD (13.3%) (p 0.01). The vacA s2i2m2 genotype was only found in patients with NUD (8.9%) (Table 2). Seventeen (22.9%) of our 74 strains typed as dupA-positive, among which 12 strains typed as dupA1. The prevalence of dupA1 was higher in DU (36.8%) than in GU (10%) and NUD (8.9%) (p 0.01).
Multivariate analysis showed that cagA+/ vacA s1i1m2 strains were independently associated with the developing PUD with increased odds of developing PUD (p 0.03; OR = 2.1).
The prevalence of clarithromycin and levofloxacin resistance and its association with virulence factors
Two mutations were associated with clarithromycin resistance: A2147G and A2146G. Levofloxacin resistance is associated with D91N mutation. The prevalence of these mutations was studied. Overall in 74 patients, 12/72 (16.2%) strains were clarithromycin resistant, of which ten carried the A2147G mutation (Table 3). Only three patients were levofloxacin resistant. The A2147G mutation was found in 8.1% in patients with NUD, 4.1% in patients with DU and 1.4% in patients with GU. The other mutation, A2146G, was found in only 2.7% of patients. 1.4% of patients with NUD and 1.4% of patients with DU tested positive for the presence of this mutation. The levofloxacin-resistance mutation (D91N) was found in three of the 74 patients (4.1%; one DU, one GU and one NUD).
Table 3.
The relation between antibiotic resistance and virulence factors
| Clarithromycin resistance |
Levofluxacin resistance |
||
|---|---|---|---|
| A2147G | A2146G | D91N | |
| cagA | 27% | 0% | 6% |
| dupA1 | 50%∗ | 0% | 0% |
| dupA2 | 0% | 0% | 0% |
| vacA s1 | 13% | 3% | 4% |
| vacA s2 | 25% | 0% | 0% |
| vacA m1 | 15% | 0% | 7% |
| vacA m2 | 13% | 4% | 2% |
| vacA i1 | 15% | 0% | 5% |
| vacA i2 | 12% | 6% | 3% |
| vacA s1m1 | 15% | 0% | 8% |
| vacA s1m2 | 12% | 5% | 2% |
| vacA s2m2 | 25% | 0% | 0% |
Significant association.
A significant association was found between dupA1 genotype and A2147G clarithromycin resistance mutation (p <0.01) (Table 3). No significant association was found between A2146G or D91N and virulence factors.
The association between IL-8 and dupA status
To compare the levels of gastric IL-8 secretions, we classified our samples into three groups: dupA-negative, dupA1 and dupA2. We found no significant difference between the levels of IL-8 secretion in gastric mucosa infected with H. pylori dupA-negative mucosa and H. pylori dupA1-positive strains (dupA-negative: mean ± median: 28 ± 26 versus 30 ± 27.1 for dupA1; p 0.6). Also, no significant difference was found between H. pylori dupA-negative mucosa and H. pylori dupA2-positive strains (28 ± 26 versus 35 ± 26; p 0.7) (Fig. 1). Also, we classified our samples into two groups: samples infected with cagA-positive strains and those infected with cagA-negative strains. It was found that the same levels of IL-8 were elicited in biopsies infected with cagA-positive strains and cagA-negative strains (cagA-negative: 29 ± 25 pg/mL; cagA-positive: 28.4 ± 27 pg/ml) (Fig. 2).
Fig. 1.
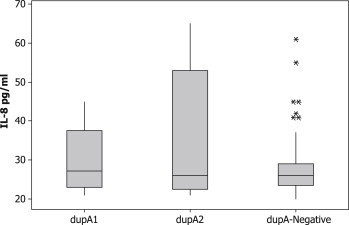
Interleukin-8 levels in gastric biopsies from cagA-negative Helicobacter pylori-infected tissues, dupA1 H. pylori-infected tissues and dupA2 H. pylori-infected tissues. No significant differences were found; * indicates outlier.
Fig. 2.
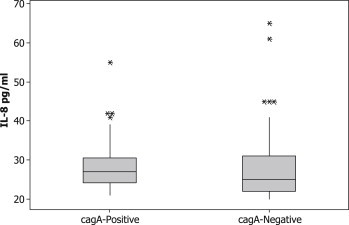
Interleukin-8 levels in gastric biopsies from cagA-negative Helicobacter pylori-infected tissues and cagA-positive H. pylori-infected tissues. No significant differences were found; * indicates outlier.
Discussion
Helicobacter pylori infection is associated with peptic ulceration and gastric adenocarcinoma, the second highest cause of cancer deaths worldwide [7]. Clarithromycin and fluoroquinolone are two drugs used widely in the eradication of H. pylori. Resistance to these drugs is emerging and presents a challenge. Studies all over the world have reported resistance to clarithromycin and fluoroquinolone [16–18]. Proton pump inhibitor–clarithromycin-containing triple therapy without previous susceptibility testing should be abandoned when the clarithromycin-resistance rate in the region is between 15 and 20% [7]. No study has been conducted in Iraq to examine the resistance rate to these medications. It was found that 16.2% of the strains used in this study were clarithromycin resistant. These results need to be confirmed by culture and sensitivity and if true, then clarithromycin-containing triple therapy should not be considered for the eradication of H. pylori because of the high failure rate. In addition, only three (4%) patients were levofloxacin resistant. This is a low resistance rate in comparison to isolates from Belgium, France, Italy and Germany where higher resistance rates to ciprofloxacin or levofloxacin ranging between 16.8% and 23% [16,17] were found. Also, higher resistance rates (ciprofloxacin 33.8%; levofloxacin 21.5%) have been observed in Japan [18]. These differences in resistance rate could be attributed to geographical region and drug usage differences.
The study of H. pylori virulence factors in populations is important because they contribute to disease risk. For example, in Iran, where gastric cancer is common, more than 75% of H. pylori strains are cagA positive [2]. We looked within our populations for the associations between virulence factors and PUD. In agreement with previous reports from Iraq [19–21], a significant association between cagA status and PUD was observed. Also, research from a neighbouring country, Turkey, has shown results similar to those from Iraq [3]. Next, we examined the association between vacA polymorphisms and clinical outcomes. The vacA s1, m1 and s1m1 genotypes were significantly associated with PUD. This is in agreement with other reports from Iraq and other countries [2,22]. The vacA i region polymorphisms are a relatively a new virulence determinant and have not been examined thoroughly. It was previously shown that the vacA i1 genotype is associated with gastric cancer in Iran [4]. We studied the association between vacA i region polymorphisms and clinical outcomes. It was found that vacA i1 is significantly associated with GU and DU. A previous report from Iraq showed a significant relationship between vacA i1 and GU but not DU [2]. However, the sample size used in the previous study was small and more study was suggested to investigate such a relationship.
Multivariate analysis showed that cagA+/ vacA s1i1m2 strains were independently associated with the developing PUD. Other virulence factors, solely or in combination, could not show any independent association with clinical outcome. Therefore we suggest that single bacterial factors could not solely explain the outcomes of gastroduodenal diseases.
The dupA is a virulence factor that comprises both jhp0917 and jhp0918. Lu et al. found a significant relationship between dupA and DU, and the presence of dupA was related to neutrophil infiltration and a high level of IL-8 production by epithelial cells [6]. Since then, contradicting results reported about the relationships of dupA and clinical outcomes [5]. Then, dupA was classified into dupA1 (functional) and dupA2 (non-functional, including the originally described form, in which the the ORF was broken by a stop codon) [5]. Such a classification helped the explanation of contradicting results. In agreement with a previous report from Iraq, we found a significant relationship between dupA1 and DU [15]. In addition, a previous report showed an association between dupA status and failure of H. pylori eradication [23]. No explanation was given in that report about how dupA could prevent the eradication of this bacteria. We found a novel association between dupA1 and A2147G, a mutation responsible for clarithromycin resistance. Such an association is difficult to explain and warrants further study. However, this relationship might be because dupA1 may increase the pro-inflammatory factors including reactive oxygen species and these factors may help to induce mutations in the bacteria. Further study is needed to confirm such a result and explore how these genotypes may have become associated.
Lu et al. found that the presence of dupA was related to neutrophil infiltration and a high level of IL-8 production [6]. We also previously reported that dupA1 correlates with clinical outcome and gastric IL-8 levels in Iraqi H. pylori-infected samples [15]. However, such a relationship could not be obtained in this report. This contradiction is difficult to explain and warrants further studies.
To conclude, there is a high resistance rate to clarithromycin in our region. Such results need to be confirmed and, if true, it should be discussed whether clarithromycin should be dropped from the first-line eradication regimen. Univariate analysis showed that cagA, vacA s1, m1, and s1m1 genotypes were significantly associated with PUD. However, multivariate study showed that only cagA+/ vacA s1i1m2 combination is an independent marker for disease outcome. dupA1 was shown to be associated with DU and clarithromycin resistance but not IL-8 secretion. The association between dupA1 and clarithromycin may help to explain the previous report of the association between dupA and treatment failure. Further study is needed to explore the relationship between dupA polymorphisms and clinical outcomes and drug resistance.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Atherton J.C. The pathogenesis of Helicobacter pylori induced gastro-duodenal diseases. Annu Rev Pathol Mech Dis. 2006;1(1):63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 2.Hussein N.R., Mohammadi M., Talebkhan Y., Doraghi M., Letley D.P., Muhammad M.K. Differences in virulence markers between Helicobacter pylori strains from Iraq and those from Iran: potential importance of regional differences in H. pylori-associated disease. J Clin Microbiol. 2008;46(5):1774–1779. doi: 10.1128/JCM.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saribasak H., Salih B.A., Yamaoka Y., Sander E. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J Clin Microbiol. 2004;42:1648–1651. doi: 10.1128/JCM.42.4.1648-1651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhead J.L., Letley D.P., Mohammadi M., Hussein N., Mohagheghi M.A., Eshagh Hosseini M. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133(3):926–936. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 5.Hussein N.R., Argent R.H., Marx C.K., Patel S.R., Robinson K., Atherton J.C. Helicobacter pylori dupA is polymorphic, and its active form induces proinflammatory cytokine secretion by mononuclear cells. J Infect Dis. 2010;202(2):261–269. doi: 10.1086/653587. [DOI] [PubMed] [Google Scholar]
- 6.Lu H., Hsu P.I., Graham D.Y., Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128(4):833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malfertheiner P., Megraud F., O'Morain C.A., Atherton J., Axon T.R., Bazzoli F. Management of Helicobacter pylori infection the Maastricht IV/Florence Consensus report. Gut. 2012;61(5):646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 8.Nishizawa T., Suzuki H., Kurabayashi K., Masaoka T., Muraoka H., Mori M. Gatifloxacin resistance and mutations in gyrA after unsuccessful Helicobacter pylori eradication in Japan. Antimicrob Agents Chemother. 2006;50(4):1538–1540. doi: 10.1128/AAC.50.4.1538-1540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishizawa T., Suzuki H., Umezawa A., Muraoka H., Iwasaki E., Masaoka T. Rapid detection of point mutations conferring resistance to fluoroquinolone in gyrA of Helicobacter pylori by allele-specific PCR. J Clin Microbiol. 2007;45(2):303–305. doi: 10.1128/JCM.01997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oleastro M., Ménard A., Santos A., Lamouliatte H., Monteiro L., Barthélémy P. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41(1):397–402. doi: 10.1128/JCM.41.1.397-402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Doorn L.J., Glupczynski Y., Kusters J.G., Mégraud F., Midolo P., Maggi-Solcà N. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR Line Probe Assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45(5):1500–1504. doi: 10.1128/AAC.45.5.1500-1504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cambau E., Allerheiligen V., Coulon C., Corbel C., Lascols C., Deforges L. Evaluation of a New Test, GenoType HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47(11):3600–3607. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudi J., Kolb C., Maiwald M., Kuck D., Sieg A., Galle P.R. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36(4):944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atherton J.C., Cao P., Peek R.M., Jr., Tummuru M.K., Blaser M.J., Cover T.L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 15.Hussein N.R., Abdullah S.M., Salih A.M., Assafi M.A. dupA1 Is associated with duodenal ulcer and high interleukin-8 secretion from the gastric mucosa. Infect Immun. 2012;80(8):2971–2972. doi: 10.1128/IAI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung K.H., Sheu B.S., Chang W.L., Wu H.M., Liu C.C., Wu J.J. Prevalence of primary fluoroquinolone resistance among clinical isolates of Helicobacter pylori at a University Hospital in Southern Taiwan. Helicobacter. 2009;14(1):61–65. doi: 10.1111/j.1523-5378.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- 17.Megraud F., Coenen S., Versporten A., Kist M., Lopez-Brea M., Hirschl A.M. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62(1):34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 18.Miyachi H., Miki I., Aoyama N., Shirasaka D., Matsumoto Y., Toyoda M. Primary levofloxacin resistance and gyrA/B mutations among Helicobacter pylori in Japan. Helicobacter. 2006;11(4):243–249. doi: 10.1111/j.1523-5378.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 19.Abdullah S.M., Hussein N.R., Salih A.M., Merza M.A., Goreal A.A., Odeesh O.Y. Infection with Helicobacter pylori strains carrying babA2 and cagA is associated with an increased risk of peptic ulcer disease development in Iraq. Arab J Gastroenterol. 2012;13(4):166–169. doi: 10.1016/j.ajg.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Hussein N.R., Napaki S.M., Atherton J.C. A study of Helicobacter pylori-associated gastritis patterns in Iraq and their association with strain virulence. Saudi J Gastroenterol. 2009;15(2):125. doi: 10.4103/1319-3767.48971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salih A.M., Goreal A., Hussein N.R., Abdullah S.M., Hawrami K., Assafi M. The distribution of cagA and dupA genes in Helicobacter pylori strains in Kurdistan region, northern Iraq. Ann Saudi Med. 2012;33(3):290–293. doi: 10.5144/0256-4947.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsunari O., Shiota S., Suzuki R., Watada M., Kinjo N., Murakami K. Association between Helicobacter pylori virulence factors and gastroduodenal diseases in Okinawa, Japan. J Clin Microbiol. 2012;50(3):876–883. doi: 10.1128/JCM.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiota S., Nguyen L.T., Murakami K., Kuroda A., Mizukami K., Okimoto T. Association of Helicobacter pylori dupA with the failure of primary eradication. J Clin Gastroenterol. 2012;46(4):297–301. doi: 10.1097/MCG.0b013e318243201c. [DOI] [PMC free article] [PubMed] [Google Scholar]


