Abstract
Background
We previously identified a correlation between increased expression of the PI3K regulatory subunit p85α and improved survival in human pancreatic ductal adenocarcinoma (PDAC). The purpose of this study was to investigate the impact of changes in p85α expression on response to chemotherapy as well as the regulation of p85α by microRNA-21 (miR-21).
Materials and Methods
PDAC tumor cells overexpressing p85α were generated by viral transduction, and the effect of p85α overexpression on sensitivity to gemcitabine was tested by MTT assay. Primary human PDAC tumors were stained for p85α and miR-21 via immunohistochemistry and in situ hybridization, respectively. Additionally, PDAC cells were treated with miR-21 mimic, and changes in p85α as well as phospho-AKT were assessed by western blot. Finally, a luciferase reporter assay system was used to test direct regulation of p85α by miR-21.
Results
Higher p85α expression resulted in increased sensitivity to gemcitabine (p<0.01), which correlated with decreased PI3K-AKT activation. Human tumors demonstrated an inverse correlation between miR-21 and p85α expression levels (R=−0.353, p<0.001). In vitro, overexpression of miR-21 resulted in decreased levels of p85α and increased phosphorylation of AKT. Luciferase reporter assays confirmed the direct regulation of p85α by miR-21 (p<0.01).
Conclusions
Our results demonstrate that p85α expression is a determinant of chemosensitivity in PDAC. Additionally, we provide novel evidence that miR-21 can influence PI3K-AKT signaling via its direct regulation of p85α. This data provides insight into potential mechanisms for the known relationship between increased p85α expression and improved survival in PDAC.
Keywords: p85α, PIK3R1, miR-21, pancreatic cancer, chemoresistance
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related deaths in the United States. As a result of late diagnosis and frequent resistance to chemotherapy, the prognosis for PDAC is dismal, with an overall 5-year survival rate of 6% [1]. Chemoresistance in PDAC is multifactorial and results from both tumor cell (TC) intrinsic and extrinsic mechanisms involving a variety of signaling pathways [2]. A better understanding of the molecular alterations driving PDAC progression and treatment resistance is necessary to improve the prognosis of this fatal disease.
The phosphoinositide 3-kinase (PI3K) and AKT signaling pathway is activated by diverse stimuli, including receptor tyrosine kinases and RAS [3]. Because of its role in cell proliferation and survival, alterations in the PI3K-AKT pathway have been described in various cancers [4]. In addition to its contribution to cancer development and progression, aberrant PI3K-AKT signaling is involved in resistance to various chemotherapeutic agents [5]. PI3K class IA molecules, the only members of this kinase family involved in cancer, consist of a heterodimer comprised of the p110 catalytic and p85 regulatory subunits [3,4]. The major regulatory subunit, p85α, has diverse functions. As a positive regulator, upon receptor activation, p85α participates in trafficking of p110 to the cell membrane as the initial step in the PI3K-AKT signaling cascade. As a negative regulator, p85α mediates basal inhibition of the p110 subunit. Furthermore, monomeric p85α competes with p85-p110 dimers for activated receptor binding [6,7]. Therefore, proper regulation of p85α expression is necessary to maintain physiologic flux through the PI3K-AKT signaling pathway.
The importance of PI3K-AKT signaling is highlighted by the fact that approximately 60% of human PDAC tumors exhibit increased AKT activity [8,9]. PI3K-AKT signaling has been implicated in PDAC tumorigenesis [9-13] as well as patient outcomes [8,14,15]. Increased AKT activation in human PDAC tumors is associated with higher histologic grade [8] as well as decreased patient survival [14]. Decreased expression of phosphatase and tensin homolog (PTEN), an important negative regulator of PI3K signaling, is the best described mechanism of PI3K-AKT activation in PDAC [9,16]. We recently showed that lower expression of p85α in resected pancreatic tumors correlates with worse patient survival [15], suggesting another potential mechanism for PI3K-AKT dysregulation in PDAC.
While genetic modifications of p85α have been observed in several cancers [17-22], this has not been the case in PDAC [23], indicating another mechanism of altered expression. MicroRNAs (miR) are small, non-coding RNAs that regulate gene expression by binding the 3′ untranslated region (3′UTR) of target mRNAs [24]. Changes in miR expression have been implicated in a wide range of cancers [25]. In our previous work, we have shown that miRs are extensively involved in regulating the expression of survival-associated genes [15]. Of particular interest in PDAC is miR-21, which is frequently over-expressed in TCs as compared to benign ductal cells and has been linked to proliferation, invasion and chemoresistance [26-28]. Additionally, increased miR-21 is correlated with worse patient survival [27-29]. MiR-21 is a known regulator of the PI3K-AKT pathway via inhibition of PTEN [30-32]. In addition, p85α is a predicted miR-21 target by sequence analysis, although this has not yet been experimentally validated.
The objective of this study was to investigate the impact of p85α expression on response to chemotherapy as well as the regulation of p85α by miR-21. We hypothesized that negative regulation of PI3K-AKT signaling by p85α in PDAC TCs results in increased chemosensitivity. We also hypothesized that miR-21 enhances PI3K-AKT signaling by downregulating p85α.
2. Materials and Methods
2.1. Cell culture, transfections and transductions
TCs were grown in DMEM (MiaPaCa-2 and PANC-1) or RPMI (Hs766T) supplemented with 10% FBS + 1X Penicillin-Streptomycin. PANC-1 cells were used for p85α overexpression studies because of their relatively low baseline expression of p85α. MiaPaCa-2 and Hs766T were used in miR-21 overexpression studies because of their relatively high baseline expression of p85α. For p85α overexpression, lentivirus was produced by transfecting HEK 293T packaging cells in polyethylenimine (Polysciences) with a 3-plasmid system. DNA for transfections was prepared by mixing pCMV-Δ8.9, pCMV-VSVG and pLenti4/V5-DEST. The plasmid pLenti4/V5-DEST-P85 was purchased from Addgene (#40219), and empty vector was generated by removing the p85α coding sequence. Lentiviral supernatants were harvested at 24h post-transfection, filtered, and frozen at −80°C for long-term storage. PANC-1 cells were transduced with lentivirus in the presence of 8μg/ml polybrene (Sigma) for 24h. Cells were then selected for 7 days in 2.5μg/mL puromycin. Overexpression of p85α was confirmed by western blot. For miR-21 overexpression experiments, 10nM miScript miR-21 mimic (Qiagen) was utilized. Cells were transfected with HiPerfect Transfection reagent (Qiagen), and miR-21 overexpression was confirmed by qRT-PCR.
2.2. Western blots
After 72h treatment with or without miR-21 mimic, cells were lysed with 2% SDS buffer containing protease and phosphatase inhibitors. Lysates were then sonicated for 15 seconds and BCA assays (Thermo Scientific) were performed to measure protein concentrations for each sample. 35μg of protein was loaded into each well of a 10% SDS-PAGE gel. Following resolution on the gel, samples were transferred to a PVDF membrane, blocked with 5% BSA in TBS + 0.1% Tween-20 (TBST), washed ×3 in TBST, then incubated overnight at 4°C in 5% BSA in TBS containing primary antibody: p85α 1:500 (Abcam, 54586), p-AKT 1:500 (Epitomics, 2118-1), AKT 1:1000 (Cell Signaling, 4691). β-actin 1:5000 (Sigma, A5441) was used as a loading control. Chemiluminescent imaging was performed by incubation with anti-mouse-1:10,000 or anti-rabbit- 1:5,000 HRP-conjugated secondary antibody (Jackson Labs) and Amersham ECL prime (GE Healthcare Life Sciences) followed by visualization on a ChemiDoc XRS+ (BioRad). All experiments were repeated a minimum of 2 times, and data displayed is from one representative experiment.
2.3. Viability assays
TCs were plated in 96-well plates at a density of 5×103 cells/well in serum containing media. The following day, gemcitabine was added at indicated concentrations. After 72h incubation, MTT reagent (Molecular Probes) was added to each well and incubated for 4h, followed by lysis with 10% SDS in 0.01M HCl. After overnight incubation, absorbance was read at 560nm on a microplate reader. All conditions were plated in quadruplicate. All experiments were repeated a minimum of 2 times, and data displayed is from one representative experiment.
2.4. Immunohistochemistry and in-situ hybridization
For immunohistochemistry (IHC), formalin-fixed, paraffin-embedded tumor samples were incubated at 60°C for 1h, deparaffinized in xylene, and rehydrated with graded alcohol washes. Slides were then boiled in 0.01M sodium citrate buffer for 15 min followed by quenching of endogenous peroxidase with 3% hydrogen peroxide for antigen retrieval. After 1h of blocking with 5% donkey serum at room temperature, primary antibodies were added (p85α 1:100, Epitomics 1675-1; PTEN 1:100, Cell Signaling 9559) and incubated overnight at 4°C. Biotin-conjugated anti-rabbit secondary antibody (1:500 Jackson Labs) was added and developed using Elite Vectastain ABC kit (Vector Laboratories).
For in-situ hybridization (ISH), slides were deparaffinized and rehydrated as described above then washed 3 times with diethyl pyrocarbonate-treated PBS, digested with 5μg/mL proteinase K at 37°C for 30 min, washed again twice with dieth yl pyrocarbonate-treated PBS, submerged in graded alcohol for 1 min each, and air-dried completely. Slides were then hybridized at 55°C for 2h with 50nM locked nucleic acid-modified digoxigenin-labeled probes for miR-21 (Exiqon). After hybridization, stringency washes were performed at 55°C with 5×, 1× and 0.2× saline-sodium citrate buffer respectively. The slides were then placed in blocking solution for 1h at room temperature, followed by incubation overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin Fab fragment in blocking solution. After three washes in PBS/0.1% Tween-20, the slides were incubated for 4–48h with 4-nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate substrate (Roche) for signal amplification and then counterstained with Nuclear Fast Red (Vector) for 1 min. The slides were washed with water, dehydrated in alcohol solutions and mounted with Eukitt mounting medium (Electron Microscopy Sciences).
14 resected PDAC tumors were stained as described above for p85α and PTEN (IHC) as well as miR-21 (ISH). Each tumor was divided into 25-30 regions which were individually scored for staining intensity: 0- negative, 1- low, 2- moderate, 3- high. IHC or ISH scores were then grouped into negative (0), low (1) or high (2 or 3) categories. Two independent, blinded scorers were used. Any discrepancies in scoring were reviewed by both scorers and a consensus score was agreed upon. Scores for p85α or PTEN were compared to those for miR-21 to assess for any correlation. The clinical data from the patients whose tumors were used for this analysis are presented in Table S1.
2.5. Luciferase reporter assay
Luciferase reporter constructs were generated using the pMIR-REPORT Firefly reporter system (Ambion) and a portion of the p85α 3′UTR amplified from human genomic DNA. In addition to wild type p85α 3′UTR, a mutant construct was generated via site-directed mutagenesis to introduce mutations into the two predicted miR-21 binding sites in the 3′ UTR (TargetScan). The AUAAGCU binding sites at position 861-867 and 1107-1113 were mutated to AGCGGCU using PfuUltra DNA polymerase (Stratagene) and the following primer sequences: site 1- 5′-CTG GGG TGG GGA AGC GGC TTC TGA AGG TAC ATT-3′ and 5′-AAT GTA CCT TCA GAA GCC GCT TCC CCA CCC CAG-3′; site 2- 5′-AAG CAG CTT TGA AGC GGC TCA GGC ACT GCC CAA-3′ and 5′-TTG GGC AGT GCC TGA GCC GCT TCA AAG CTG CTT-3′.
HEK 293T cells were plated in 24-well plates (5×104 cells/well) and incubated overnight. The following day, cells were co-transfected with 0.05μg p85α 3′UTR reporter construct (wild type or mutant) and miR-21 mimic or miScript AllStars Negative Control using attractene (Qiagen). The Renilla luciferase construct pRL-TK (Promega) was also included in all conditions (0.25μg). Luciferase assays were performed 48h after transfection using the Dual-Luciferase Reporter Assay System (Promega) and a microplate reader. Firefly luciferase was normalized to Renilla luciferase for each well. Experiments were repeated a minimum of 2 times, and data displayed is from one representative experiment.
2.6. Array analysis
Gene and miR expression array data (GEO accession GSE32688) from our previous study [15] were compiled to identify potential miR regulators of p85α. Array analyses are detailed in our prior study. Briefly, miR and gene expression arrays were performed on 25 PDAC samples with high tumor cell content and compared to 7 benign samples. Predicted miR binding sites in the p85α 3′UTR were identified using TargetScan (version 6.2). For our analysis, the expression levels of those miRs with predicted p85α binding sites were analyzed to identify inverse correlations between candidate miR and p85α as well as to compare the expression level of each miR in tumor vs. benign samples. Expression levels in tumor vs. benign samples were quantified using the log10 transformed p-value of Student’s T-test.
2.7. Statistical Analysis
Statistical analysis was performed with SPSS (version 22, IBM) and GraphPad Prism (version 6, GraphPad Software). Student’s T-test was used to compare means between groups. Chi-square and Pearson’s correlation were used to identify associations between miR-21 and p85α or PTEN histoscores. Image Lab software (BioRad) was used for densitometry analysis of western blots. For all experiments, statistical significance was defined as P≤0.05. Error bars indicate ± SD.
3. Results
3.1. Increased p85α expression in PDAC TCs results in decreased PI3K-AKT signaling and increased gemcitabine sensitivity
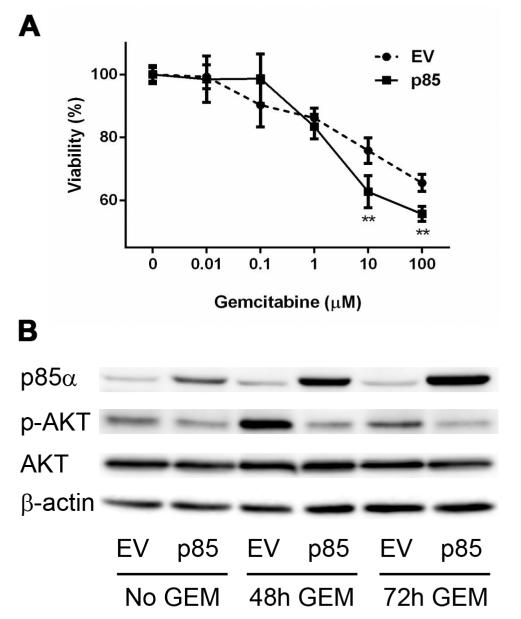
Given the positive correlation between p85α expression in PDAC TCs and patient survival [15], we sought to investigate the role of p85α in response to chemotherapy. PANC-1 cells, which have low p85α expression at baseline, were transduced with p85α or empty expression vectors, and response to gemcitabine was assayed via MTT. Overexpression of p85α resulted in increased sensitivity to gemcitabine (p<0.01; Fig. 1A). Additionally, PANC-1 cells overexpressing p85α exhibited decreased expression of phospho-AKT (p-AKT), both at baseline and after gemcitabine treatment (Fig. 1B).
Fig. 1. Overexpression of p85α increases sensitivity to gemcitabine and decreases PI3K-AKT activation in PDAC TCs.
(A) MTT assay comparing the response of p85α overexpressing (p85) and empty vector (EV) PANC-1 cells to various doses of gemcitabine. (B) Western blots for p85α and p-AKT/AKT in EV or p85 PANC-1 cells treated with or without gemcitabine (GEM) for the indicated times. **p<0.01.
3.2. Expression of p85α inversely correlates with miR-21 levels in human PDAC
Because no alterations in copy number were detected to explain differences in p85α expression in our previous study [15], we hypothesized that p85α could undergo miR-dependent regulation. Therefore, the expression data (GEO accession GSE32688) from our study was re-analyzed to identify potential miR regulators of p85α. First, miRs with predicted binding sites in the p85α 3′UTR were identified using the TargetScan database. Next, the array-based expression level of each miR was compared to that of p85α from our analysis of high tumor cell content PDAC samples. The 24 candidate miR regulators of p85α, based on predicted binding sites and inversely correlated expression levels, are summarized in Table 1. Of all the candidates, only miR-21 was found to be significantly elevated (p=0.04) in PDAC TCs compared to benign ductal epithelium. Furthermore, miR-21 was the only candidate with 2 predicted binding sites within the p85α 3′UTR.
Table 1.
Potential miR regulators of p85α
| miR | Tumor vs. Benigna | Conserved Sitesb | p85α Correlationc |
|---|---|---|---|
| miR-21 | 1.42 | 2 | −0.09 |
| miR-617 | −1.13 | 1 | −0.46 |
| miR-525-5p | −0.94 | 1 | −0.45 |
| miR-936 | −1.52 | 1 | −0.43 |
| miR-488 | −3.97 | 1 | −0.41 |
| miR-519 | −0.37 | 1 | −0.40 |
| miR-635 | −1.28 | 1 | −0.40 |
| miR-486-5p | −0.40 | 1 | −0.38 |
| miR-634 | 0.24 | 1 | −0.34 |
| miR-127-5p | −0.84 | 1 | −0.26 |
| miR-498 | 0.71 | 1 | −0.25 |
| miR-656 | 0.96 | 1 | −0.24 |
| miR-650 | 0.03 | 1 | −0.24 |
| miR-1285 | 0.01 | 1 | −0.22 |
| miR-1229 | 1.02 | 1 | −0.21 |
| miR-554 | −0.31 | 1 | −0.17 |
| miR-548o | 0.45 | 1 | −0.16 |
| miR-330-3p | −0.53 | 1 | −0.14 |
| miR-455-3p | 0.50 | 1 | −0.14 |
| miR-767-5p | −1.40 | 1 | −0.10 |
| miR-495 | 0.38 | 1 | −0.06 |
| miR-221 | 1.01 | 1 | −0.05 |
| miR-15a | 0.23 | 1 | −0.01 |
| miR-548g | 0.19 | 1 | −0.01 |
Log-transformed p-value of Student’s T-test comparing expression in PDAC tumor cells vs. normal ductal epithelium based on miR array. A positive (negative) value indicates higher (lower) expression in tumor as compared to normal cells.
Number of predicted binding sites in p85α 3′UTR for each miR.
Correlation coefficient between miR and p85α (mRNA) expression levels in PDAC tumor cells.
To further investigate the association between miR-21 expression and p85α as well as PTEN, we performed IHC and ISH on resected human PDAC tumor samples. The histoscore distributions are shown in Figure S1. Representative images are displayed in Figures 2A and S2. The majority of tumors with negative or low p85α expression exhibited high miR-21 staining. Conversely, the majority of tumors with high p85α expression had negative or low miR-21 staining (Fig. 2B). Chi-square test showed an association between p85α and miR-21 expression (p<0.001). Pearson correlation analysis confirmed the inverse relationship (R=−0.353, p<0.001). Interestingly, PTEN, a negative regulator of the PI3K-AKT pathway and a known miR-21 target, did not correlate well with miR-21 expression in our analysis (R=0.037, p=0.46; Fig. 2C). There was a modest positive correlation between p85α and PTEN expression (R=0.144, p=0.003).
Fig. 2. The expression levels of p85α and miR-21 are inversely correlated in human PDAC.
(A) Representative images demonstrating the inverse relationship between p85α and miR-21 in resected tumors. The distribution of p85α (B) and PTEN (C) staining along with the corresponding miR-21 level for the entire cohort is displayed. Each marker was categorized as negative, low or high based on histoscores of 0, 1 or 2-3 respectively. 20× magnification.
3.3. Overexpression of miR-21 results in decreased levels of p85α and increased PI3K-AKT activation
Based on the correlation between miR-21 and p85α expression in human samples, we sought to test the effect of direct manipulation of miR-21 on p85α levels. Two PDAC TC lines (MiaPaCa-2 and Hs766T) with high baseline p85α expression were treated with or without miR-21 mimic for 72h followed by lysis and protein harvest. The basal expression of miR-21 for each cell type used as well as the change in miR-21 levels upon mimic transfection are reported in Supplemental Figure S3. Overexpression of miR-21 resulted in decreased expression of p85α in both MiaPaCa-2 and Hs766T (Fig. 3). PTEN expression decreased in response to miR-21 overexpression in Hs766T (Fig. 3B) but not MiaPaCa-2 (Fig. 3A). Concomitant with the decrease in p85α seen in both cell lines, an increase in PI3K pathway activation was observed, as evidenced by increased p-AKT expression (Fig. 3). To test the functional effect of miR-21 overexpression, PANC-1 cells treated with or without miR-21 mimic were tested for gemcitabine sensitivity. Overexpression of miR-21 led to gemcitabine resistance (Fig. S4), the opposite of the effect of p85α overexpression (Fig. 1A).
Fig. 3. Overexpression of miR-21 in PDAC TCs causes decreased expression of p85α and decreased PI3K-AKT activation.
After treatment with control transfection reagent (NC) or miR-21 mimic (miR-21), western blots were performed for p85α, PTEN and p-AKT/AKT in MiaPaCa-2 (A) and Hs766T (B) cells. Expression of p85α, PTEN and p-AKT/AKT were quantified by densitometry.
3.4. p85α is a direct target of miR-21
Having demonstrated an inverse correlation between p85α expression and miR-21 levels in both human PDAC tumors and cell lines, we next sought to determine if p85α is regulated by miR-21 directly. A luciferase reporter construct was produced containing the 3′UTR of p85α and transfected into HEK 293T cells with or without miR-21 mimic. Treatment with miR-21 mimic resulted in a reduction of luciferase activity compared to negative control (p=0.009; Fig. 4). To more rigorously confirm direct regulation, a construct was produced with mutations introduced into the 2 predicted miR-21 binding sites within the p85α 3′UTR. Although luciferase activity decreased after treatment with miR-21 mimic in the mutant construct, it was significantly higher than the wild type construct (p=0.005; Fig. 4). The lack of complete reversal of luciferase activity in the mutant construct may indicate residual binding at mutant sites or the presence of additional miR-21 binding sites not identified by sequence-based prediction methods.
Fig. 4. P85α is a direct target of miR-21.
HEK 293T cells were transfected with luciferase reporter constructs containing wild-type (WT) or mutant (Mut) p85α 3′UTR sequences along with miR-21 mimic or AllStars negative control (NC). **p<0.01.
4. Discussion
In this study, we expand on our previous findings that p85α mRNA expression is correlated with improved survival by demonstrating that increased expression of p85α in PDAC TCs results in decreased PI3K-AKT signaling and increased sensitivity to gemcitabine. Additionally, we show an inverse correlation between miR-21 and p85α expression levels in PDAC TCs as well as human tumors. Finally, we present novel evidence that p85α is a direct miR-21 target. Taken together, our data provide novel insight into the regulation of p85α expression in PDAC and its potential impact on response to chemotherapy.
While the importance of the PI3K-AKT pathway in PDAC is well known, the role of p85α in PDAC initiation and progression has not been widely studied. Although p85α is necessary for stabilization and membrane recruitment of the p110 subunit, it also functions to inhibit PI3K activity [6]. In most cells, the p85 subunit is present in greater quantities than the p110 subunit, resulting in net inhibition of the pathway. As a result, heterozygous loss of p85α causes increased PI3K-AKT activity, while homozygous loss results in decreased activity [33]. Our finding that p-AKT levels are inversely related to p85α expression in PDAC TCs is consistent with these previous observations. The concept that decreased p85α expression can be protumorigenic is supported by the fact that lower levels of p85α are seen in various cancers when compared to normal tissues [34]. Furthermore, increased p85α expression has been linked to a better prognosis not only in PDAC [15] but also breast [35], brain [36] and lung [37] cancers. In our study, p85α expression influenced sensitivity to chemotherapy, elucidating a potential mechanism for the observed associations with survival.
Given the prognostic significance of p85α, understanding its regulation in PDAC is important. Here, we show that p85α is a direct target of miR-21. This finding is consistent with the presence of predicted miR-21 binding sites in 3′UTR of p85α in miR databases such as TargetScan and miRanda. Although enhancement of PI3K-AKT signaling by miR-21 has been previously described, this effect has been attributed to the negative regulation of PTEN [30-32]. In our study, miR-21 expression was much more strongly linked to p85α than PTEN expression. While this finding does not refute the previously described regulation of PTEN by miR-21, it does identify an alternative mechanism by which miR-21 can regulate the PI3K-AKT pathway. The effect of miR-21 on p85α and PTEN may also be inter-related because p85α can act to facilitate PTEN function [38]. Furthermore, miR-21 has a wide variety of targets and can regulate PI3K-AKT signaling via multiple mechanisms. As an example, miR-21 mediated downregulation of PDCD4 has been shown to increase AKT signaling in renal cell carcinoma [39]. Our results support the concept that miR-21 can regulate the PI3K-AKT pathway by a variety of mechanisms in a tissue specific manner. The mechanistic findings in this study are limited by the fact that they were generated via in vitro experiments, and further experiments using in vivo models will be necessary to confirm our results. Nevertheless, our data highlight a novel mechanism by which miR-21 regulates PI3K-AKT signaling that may be applicable beyond PDAC given the frequent dysregulation of both p85α and miR-21 in other cancers.
The PI3K-AKT pathway is being extensively evaluated as a therapeutic target in various malignancies, including PDAC [40]. Preclinical studies in PDAC have demonstrated the potential utility of PI3K-AKT inhibition [41,42], and several clinical trials examining both PI3K and AKT inhibitors are ongoing [43]. Presumably, inhibition of PI3K-AKT signaling is most likely to be effective in tumors with aberrant upregulation of the pathway. We have shown that the cascade of miR-21 overexpression resulting in p85α downregulation has the potential to cause increased PI3K-AKT signaling and chemoresistance. The impact of miR-21 expression on gemcitabine resistance has been well described [26,27,44]. The influence of miR-21 on sensitivity to other chemotherapeutic agents, such as 5-FU, has also been described in PDAC [28,45,46] as well as other cancers [47,48]. Additional in vivo studies will be necessary to confirm the relationship between miR-21, p85α and chemosensitivity in a more clinically relevant model. However, our data combined with the existing literature raise the compelling possibility that elevated miR-21 and decreased p85α may identify a subgroup of PDAC which is resistant to standard chemotherapy and in which PI3K-AKT inhibition may be effective.
5. Conclusions
We have demonstrated that p85α expression impacts PI3K-AKT activation as well as sensitivity to gemcitabine, the most commonly used chemotherapeutic agent in PDAC. In addition, we have shown that miR-21 and p85α expression are inversely correlated in human tumors and cell lines, and that p85α is a direct miR-21 target. These observations provide new insight into the molecular mechanisms of enhanced PI3K-AKT signaling and chemoresistance in PDAC.
Supplementary Material
Acknowledgements
This work was supported by the following funding sources: PT and BK- California Institute for Regenerative Medicine TG2-01169. TD- Concern Foundation for Cancer Research, Hirshberg Foundation for Cancer Research, CURE: Pilot and Feasibility Study NIH/NIDDK P30DK41301, STOP Cancer Foundation, Association for Academic Surgery, Digestive Disease Research Center supported by NIH grant P30DK41301
Abbreviations
- 3′UTR
3′ untranslated region
- IHC
immunohistochemistry
- ISH
in situ hybridization
- miR
microRNA
- PDAC
pancreatic ductal adenocarcinoma
- PI3K
phosphoinositide 3-kinase
- PTEN
phosphatase and tensin homolog
- TC
tumor cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
PT assisted with the viability assays, was involved in study design, coordination and interpretation and was the major contributor to manuscript composition. LL was involved in performing all experiments. BK was involved in study design and assisted with the IHC and ISH. AN was involved in study design and coordination. LT was responsible for the candidate miR analysis. NW assisted with IHC and performed western blots. DM assisted with cell culture and western blots. SP was responsible for designing and optimizing the luciferase assay experiments. DD was involved in study design and data interpretation. TD conceived of the study and oversaw study performance and manuscript composition.
Disclosure
The authors declare that they have no competing interests.
Appendices
SupplementalMaterial.pdf: Figures S1-4. Table S1.
References
- [1].American Cancer Society [Accessed: April 12, 2014];Cancer Facts & Figures 2014. http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2014/index.
- [2].Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, et al. Pancreatic cancer: understanding and overcoming chemoresistance. Nat Rev Gastroenterol Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- [3].Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- [4].Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- [5].Hafsi S, Pezzino FM, Candido S, Ligresti G, Spandidos DA, et al. Gene alterations in the PI3K/PTEN/AKT pathway as a mechanism of drug-resistance (review) Int. J. Oncol. 2012;40:639–644. doi: 10.3892/ijo.2011.1312. [DOI] [PubMed] [Google Scholar]
- [6].Luo J, Cantley LC. The negative regulation of phosphoinositide 3-kinase signaling by p85 and it’s implication in cancer. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- [7].Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schlieman MG, Fahy BN, Ramsamooj R, Beckett L, Bold RJ. Incidence, mechanism and prognostic value of activated AKT in pancreas cancer. Br. J. Cancer. 2003;89:2110–2115. doi: 10.1038/sj.bjc.6601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ying H, Elpek KG, Vinjamoori A, Zimmerman SM, Chu GC, et al. PTEN is a major tumor suppressor in pancreatic ductal adenocarcinoma and regulates an NF-kappaB-cytokine network. Cancer Discov. 2011;1:158–169. doi: 10.1158/2159-8290.CD-11-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Elghazi L, Weiss AJ, Barker DJ, Callaghan J, Staloch L, et al. Regulation of pancreas plasticity and malignant transformation by Akt signaling. Gastroenterology. 2009;136:1091–1103. doi: 10.1053/j.gastro.2008.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- [12].Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70:7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- [14].Kennedy AL, Morton JP, Manoharan I, Nelson DM, Jamieson NB, et al. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol. Cell. 2011;42:36–49. doi: 10.1016/j.molcel.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clin. Cancer Res. 2012;18:1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, et al. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- [17].Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, et al. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- [19].Urick ME, Rudd ML, Godwin AK, Sgroi D, Merino M, et al. PIK3R1 (p85alpha) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 2011;71:4061–4067. doi: 10.1158/0008-5472.CAN-11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ross RL, Burns JE, Taylor CF, Mellor P, Anderson DH, et al. Identification of mutations in distinct regions of p85 alpha in urothelial cancer. PLoS One. 2013;8:e84411. doi: 10.1371/journal.pone.0084411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Quayle SN, Lee JY, Cheung LW, Ding L, Wiedemeyer R, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One. 2012;7:e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [25].Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- [26].Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 2009;8:1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- [27].Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- [28].Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J. Gastrointest. Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int. J. Oncol. 2013;42:219–228. doi: 10.3892/ijo.2012.1707. [DOI] [PubMed] [Google Scholar]
- [32].Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin. Chim. Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- [33].Ueki K, Fruman DA, Brachmann SM, Tseng YH, Cantley LC, et al. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell. Biol. 2002;22:965–977. doi: 10.1128/MCB.22.3.965-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 2010;70:5305–5315. doi: 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cizkova M, Vacher S, Meseure D, Trassard M, Susini A, et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer. 2013;13:545. doi: 10.1186/1471-2407-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fransson S, Abel F, Kogner P, Martinsson T, Ejeskar K. Stage-dependent expression of PI3K/Aktpathway genes in neuroblastoma. Int. J. Oncol. 2013;42:609–616. doi: 10.3892/ijo.2012.1732. [DOI] [PubMed] [Google Scholar]
- [37].Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Taniguchi CM, Tran TT, Kondo T, Luo J, Ueki K, et al. Phosphoinositide 3-kinase regulatory subunit p85alpha suppresses insulin action via positive regulation of PTEN. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12093–12097. doi: 10.1073/pnas.0604628103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bera A, Das F, Ghosh-Choudhury N, Kasinath BS, Abboud HE, et al. microRNA-21-induced dissociation of PDCD4 from rictor contributes to Akt-IKKbeta-mTORC1 axis to regulate renal cancer cell invasion. Exp. Cell Res. 2014;328:99–117. doi: 10.1016/j.yexcr.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Akinleye A, Avvaru P, Furqan M, Song Y, Liu D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J. Hematol. Oncol. 2013;6:88. doi: 10.1186/1756-8722-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cao P, Maira SM, Garcia-Echeverria C, Hedley DW. Activity of a novel, dual PI3-kinase/mTor inhibitor NVP-BEZ235 against primary human pancreatic cancers grown as orthotopic xenografts. Br. J. Cancer. 2009;100:1267–1276. doi: 10.1038/sj.bjc.6604995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Awasthi N, Yen PL, Schwarz MA, Schwarz RE. The efficacy of a novel, dual PI3K/mTOR inhibitor NVP-BEZ235 to enhance chemotherapy and antiangiogenic response in pancreatic cancer. J. Cell. Biochem. 2012;113:784–791. doi: 10.1002/jcb.23405. [DOI] [PubMed] [Google Scholar]
- [43].National Institutes of Health [Accessed: April 16, 2014]; ClinicalTrials.gov. http://www.clinicaltrials.gov/
- [44].Wang P, Zhuang L, Zhang J, Fan J, Luo J, et al. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Donahue TR, Nguyen AH, Moughan J, Li L, Tatishchev S, et al. Stromal MicroRNA-21 levels predict response to 5-fluorouracil in patients with pancreatic cancer. J. Surg. Oncol. 2014 doi: 10.1002/jso.23750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2012;18:534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- [47].Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br. J. Cancer. 2010;103:1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc. Natl. Acad. Sci. U. S. A. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.