Abstract
Microscopy reveals complex patterns of cellular heterogeneity that can be biologically informative. However, a limitation of microscopy is that only a small number of biomarkers can typically be monitored simultaneously. Thus, a natural question is whether additional biomarkers provide a deeper characterization of the distribution of cellular states in a population. How much information about a cell’s phenotypic state in one biomarker is gained by knowing its state in another biomarker?
Here, we describe a framework for comparing phenotypic states across biomarkers. Our approach overcomes the current limitation of microscopy by not requiring co-staining biomarkers on the same cells; instead we require staining of biomarkers (possibly separately) on a common collection of phenotypically diverse cell lines.
We evaluate our approach on two image datasets: 33 oncogenically diverse lung cancer cell lines stained with 7 biomarkers, and 49 less diverse subclones of one lung cancer cell line stained with 12 biomarkers. We first validate our method by comparing it to the “gold standard” of co-staining. We then apply our approach to all pairs of biomarkers and use it to identify biomarkers that yield similar patterns of heterogeneity. The results presented in this work suggest that many biomarkers provide redundant information about heterogeneity. Thus, our approach provides a practical guide for selecting independently informative biomarkers and, more generally, will yield insights into both the connectivity of biological networks and the complexity of the state space of biological systems.
Keywords: Heterogeneity, Biomarker Selection, Systems Biology, Microscopy, Bioimage Informatics, Biological Networks, Single-Cell Variability, Information Theory
Introduction
Single-cell studies are revealing wide-ranging differences from one cell to another, even within presumably isogenic populations. While this variability has traditionally been viewed as “noise”, a growing body of evidence suggests that analysis of this variability can reveal novel biological information (1–4). For example, previous work has shown that studies of variability can be used to infer network topology (5–9), predict responses of cancer populations to drugs (10–13), impute mechanisms of drug action (14), and identify new cellular states during differentiation (15,16).
There are a growing number of approaches for measuring heterogeneity within populations. Sequencing allows for analysis of heterogeneity in mutational and transcriptional state at the genomic scale (17–19). Flow cytometry allows for measurements of dozens of total and phosphorylated protein concentrations within single cells (20,21). And microscopy—the focus of our current study, and the most classical of all approaches for observing cellular heterogeneity—allows simultaneous measurement of cell morphology and biomarker expression and localization patterns for cells in situ (22,23).
A natural way of investigating heterogeneity using microscopy is to co-stain a population of cells with biomarkers of interest. However, a limitation of microscopy is that only a small number of biomarkers can typically be monitored simultaneously. (Though, new approaches are always being developed to increase these numbers (24–29).) With a strict economy on the number of readouts that can be selected in microscopy, a fundamental and practical question is whether subpopulations of cells that appear to be in the same phenotypic state as assessed by one biomarker are in the same phenotypic state as assessed by another biomarker. That is, will additional biomarkers provide a deeper characterization of heterogeneity?
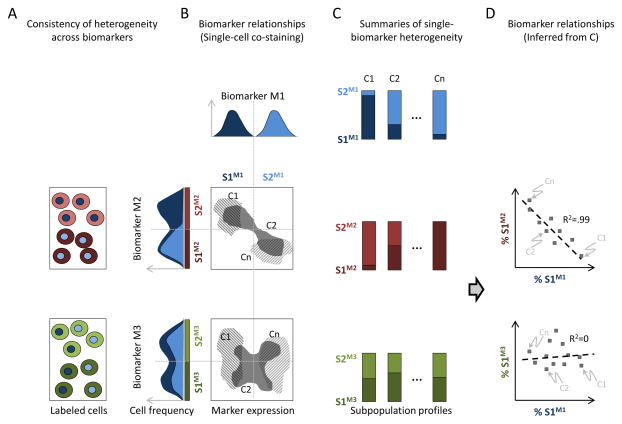
Here, we describe a framework for assessing the extent to which different biomarkers reveal different structures, or “decompositions” of heterogeneity. First, we model heterogeneity in our panels of cell populations, one biomarker at a time. Here, cellular heterogeneity is described by mixtures of phenotypically distinct subpopulations, identified using automated image analysis (Methods) (10,14). Second, we develop a regression-based approach to compare our decompositions of heterogeneity across biomarkers (Fig. 1). Third, we demonstrate interpretability of our approach using a simple 2-state (low vs. high expression) model of heterogeneity (Figs. 2, 3Aii). Finally, we evaluate (Fig. 3) and apply (Fig. 4) our regression-based approach to identify biomarkers that have similar subpopulation-based decompositions of heterogeneity.
Figure 1.
Figure 2.
Figure 3.
Figure 4.
We make use of two image datasets: 1) a panel of 33 Lung Cancer Cell populations (termed the LCC dataset (Methods)) that captures a wide variety of cancer oncogenotypes (Supp. Fig. S1) stained with seven biomarkers, and 2) a phenotypically less diverse panel of 49 Clonal Populations of the H460 lung cancer cell line (termed the CP dataset (Methods)) stained with twelve biomarkers. The use of diverse cell populations addresses a general limitation in all studies of phenotypic variability that a single population of cells may not reveal the entire spectrum of phenotypic states (30). For example, the expected negative correlation between opposing EMT biomarkers β-catenin and vimentin may not be apparent within a single cell population, yet emerges after considering a panel of diverse cell populations (Fig. 3Aii).
Our framework for relating subpopulations across biomarkers will yield insights into the connectivity of biological networks, the complexity of the state space of a biological system, and provide practical guides for selecting biomarkers in studies of heterogeneity.
Materials and Methods
Experimental Methods
In our study, we made use of two different imaging datasets:
CP
Panel of 49 low-passage Clonal Populations from the non-small cell lung cancer cell line H460 (previously published (10)).
LCC
Panel of 33 non-small cell Lung Cancer Cell lines (developed specifically for this study). These lines are: HCC95, H596, EKVX, H2073, HCC78, H1355, H157, HCC4011, H226, HCC193, H1650, H358, H2009, H292, H322, PC9, H460, A549, HCC4017, HCC827, H1993, H1648, HCC1359, HCC515, H1395, H1299, H1264, H1819, H2126, H1693, HCC2935, H2887, and HCC4006. All lung cancer cell lines used in this study were obtained from the Hamon Cancer Center Collection (University of Texas Southwestern Medical Center). This set of cell lines well represents the diversity of mutational states observed across a larger panel of 134 lung cancer cell lines (Supp. Fig. S1).
Cell Culture
CP
Clones were all cultured in supplemented RPMI 1640 medium and seeded in imaging plates at a density of 10,000 cells per well as previously published (10).
LCC
Cell lines were all maintained in RPMI 1640 medium (Fisher Scientific) supplemented with 2 mM L-glutamine, 10% fetal bovine serum (FBS), and penicillin-streptomycin at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. All cell lines were mycoplasma tested by EZ-PCR Mycoplasma Test kit (Fisher Scientific). Cells were plated in four plates on 96-well BD imaging plates. The majority of cell lines were seeded at a density of 10,000 cells per well. To optimize for image segmentation, a few cell lines were seeded at different densities: EKVX: 15k/well, HCC1359: 5k/well, PC9: 20k/well, HCC827: 5k/well, H1648: 5k/well, H1264: 15k/well, HCC2935: 5k/well. H460 and A549 cell lines were seeded and assayed in each of the four plates as controls for intensity normalization.
Biomarkers
CP
Clones were stained with 6 different biomarker sets as previously published (10) and in Supp. Table S1. MS1: DNA/pSTAT3/pPTEN, MS2: DNA/pERK/pP38, MS3: DNA/E-cadherin/GSK3-β/β-catenin, MS4: DNA/pAKT/H3K9Ac, MS5: DNA/Actin/Tubulin, and MS6: DNA/GAPDH)
LCC
Cells were fixed with 4% paraformaldehyde for 5 minutes, permeabilized with 0.2% Triton X-100 solution in TBS for 3 minutes, then blocked with 5% IgG-free BSA solution in TBST at 4°C overnight. Cells were then washed with PBS three times and stored in PBS at 4°C before staining. 5% BSA in TBST was used for primary and secondary antibody dilutions. Primary antibody staining occurred overnight at 4°C. Secondary staining occurred for two hours at room temperature in the dark. PBS was used to wash in between and after staining steps and for storage. The four lung cancer cell line plates were stained together after all had been seeded, fixed, permeabilized and blocked. Four biomarker sets, each containing Hoechst 33342 to stain DNA, were selected for the lung cancer data: (Supp. Table S2, MS1: DNA/β-catenin/vimentin, MS2: DNA/pSTAT3/pPTEN, MS3: DNA/pAKT/H3K9Ac, and MS4: DNA/E-cadherin).
Image acquisition, preprocessing, cellular region segmentation, and quality control
CP
Images were acquired at 20x magnification, rolling-ball background subtraction was applied, cellular regions were identified using a watershed-based algorithm, images were manually inspected and poor quality images were dropped as previously published (10) Approximately 100,000 cells per biomarker were identified in the CP dataset.
LCC
All fluorescence images in the LCC dataset were acquired using a Nikon epifluorescence microscope, with a 20x objective lens and 1×1 camera binning. Image acquisition was controlled by Nikon Elements software. For consistency with the CP dataset, background correction was performed using National Institute of Health ImageJ rolling-ball background subtraction software (31). Cellular regions were determined using a watershed-based segmentation algorithm (32) which uses Hoechst staining to identify nuclear regions and cytosolic biomarkers in the images to identify cellular boundaries. Each individual image was visually inspected and those with focus issues, imaging artifacts or poor segmentation were discarded from analysis. Approximately 20,000 cells per biomarker were identified in the LCC dataset.
Plate-to-plate fluorescence intensity normalization
CP
Images were normalized from plate to plate as previously published (10) to give equal median intensity to the control cell line that was present on all plates.
LCC
Images were normalized from plate to plate to give fixed median intensity of two pooled control cell lines: H460 and A549. We use two cell lines to provide a more robust control for normalization. Grey scale values of each image’s pixels were normalized against the merged distribution of pixels from images of both control lines. Pixels in each plate, p, and each fluorescence channel, m, were rescaled by a plate specific normalization factor, . The normalization factor was chosen such that the median of the pooled cell lines had a fixed intensity, I0.
The median intensity of the pooled control lines, , was used to transform each pixel intensity, , of all images from channel m in plate p to a new value, . I0 was chosen to be the mean of ’s across all p’s. This normalization reduces variation across plates allowing for cell line comparison.
Analytical Methods
The following methods were applied independently to the two datasets described above.
Feature extraction
To computationally describe cellular phenotypes, we selected biomarker-specific phenotypic descriptors (based on intensity and localization) that we believed are relevant to the biomarkers’ biology (Supp. Tables S3 and S4). For example, we characterized the nuclear biomarker Acetylated Histone 3 Lysine 9 (H3K9-Ac) by its nuclear intensity, while we used the cytoplasmic intensity of β-catenin as a readout of the epithelial state of the cell (as opposed to β-catenin’s nuclear intensity, which is commonly used as a readout of wnt signaling (33)).
Identifying subpopulations based on a single biomarker
For each biomarker, we identified subpopulations representing local high-density clusters of cells in the feature space defined by these phenotypic descriptors. To capture the spectrum of heterogeneity in our datasets, subpopulations were identified on a pooled population of 10,000 cells sampled equally from each cell population in the dataset. For each biomarker, we used a Gaussian mixture model (GMM) (14,34) to identify phenotypically distinct cellular subpopulations based on the features described above. (Because GMM approaches may give different results for each run, end results, such as model-fit criteria or mutual information scores, were averaged when appropriate across multiple runs, as described below.) We tested models with a range from 2 to 10 subpopulations for each biomarker (Supp. Fig. S2). To select the number of subpopulations presented in the main paper, we computed a model-fit criteria (BIC, averaged over 10 GMM runs). We then selected the smallest subpopulation number for each biomarker that was within 5% of its minimum observed fit criteria within our scanned range (this was used to avoid the situation when increased numbers of subpopulations only marginally improved model fit) (Supp. Tables S3 and S4). Given these models, we then determined, for each cell, a posterior probability of its belonging to each subpopulation. Cells were then assigned the subpopulation that they most likely belonged to, based on their posterior probabilities.
Using mutual information to quantify co-stained biomarker relations
We used the mutual information to quantify the extent to which the subpopulation state of one biomarker is predictive of the subpopulation state of another biomarker. We note that such a direct quantification was only possible in case when individual cells were co-stained with biomarkers. For every pair of co-stained biomarkers m1 and m2, we subsampled 10,000 cells (distributed uniformly across all cell populations) to construct the co-occurrence matrix, Pm1m2 of their respective subpopulations. An element, Pm1m2 (i, j), of this matrix thus measures the fraction of cells that are in both subpopulation i, based on biomarker m1 and also in subpopulation j based on biomarker m2. The mutual information between m1 and m2 was then calculated as:
where , are the marginal probabilities of biomarker m1’s subpopulations, and k1, k2 are the number of subpopulations for m1, m2 (respectively). Finally, mutual information values were averaged across multiple GMM runs (10 per biomarker).
Subpopulation Profiles
For each biomarker, M, a cell population C was described by a subpopulation profile (10):
where N is the total number of cells, k is the number of subpopulations, and nsj is the number of cells in the jth subpopulation.
Using subpopulation profiles to quantify biomarker relationships
We used a regression-based approach to estimate the probability, Tm1m2 (si, sj), for cells in subpopulation si of biomarker m1 to be in subpopulation sj of biomarker m2. That is, for any cell line C, we tried to relate the subpopulation profiles and between two biomarkers by . We estimated Tm1m2 as the matrix that minimizes the differences between the predicted profiles and the observed profiles across all cell lines:
Here, we used the Kullback-Leibler divergence, , to measure the similarity between pairs of subpopulation profiles. The minimization to determine Tm1m2 was performed using an interior point solver, subject to the constraints that ensure Tm1m2’s probabilistic interpretation (elements lie between zero and one and rows sum up to one). We then calculated a raw score
which reported on the difference between the observed subpopulation profiles for biomarker m2 and the ones predicted from transforming m1: the higher the score (i.e. closer to 0), the more similar are the actual and predicted profiles.
(We note that our choice of the similarity measure above provided consistency with the mutual information, described above, which is simply the Kullback-Leibler divergence between the joint distribution and the product of the marginal distributions. We also note that the T matrix need not be square; if we identified 3 and 5 subpopulations for biomarkers m1 and m2, then the resulting Tm1m2 would be a 3×5 matrix.)
Finally, we performed 3 steps of normalization designed to improve the robustness of our approach:
-
We converted raw similarity scores into z-scores
where μRandom and σRandom are the mean and standard deviation respectively of SRaw calculated with the cell line identities randomized. The higher the z-score, the more similar are the actual and predicted profiles (i.e. the more similar are the subpopulation structures between the two biomarkers).
We accounted for the randomness inherent to the process of sub-population construction z(m1, m2) by averaging across runs of subpopulation construction to give . takes into account all 100 possible combinations of generated replicate models.
-
Finally, we generated the Subpopulation Structure Similarity (S3) score:
First, we note S3(m1, m1) = 1 and S3(m1, m2) = 0 when m1 and m2 are perfectly unrelated. Second, in theory z(m1, m1) ≥ 0, but for our dataset we found z(m1, m1) > 0. Third, in general S3(m1, m2) need not be the same as S3(m2, m1) due to the asymmetric nature of DKL, which reflects the asymmetric nature of biological networks. For example, the state of an upstream biomarker is more likely to be predictive of the states of its downstream targets, while the reverse is less likely to be true, as the state of a downstream target may be influenced by multiple upstream effectors. However, when comparing to the standard symmetric measure of mutual information (Fig. 3), a symmetrized version of the S3 score, , was used.
Code and Data
Raw feature and subpopulation profile data along with the MATLAB code to generate the figures in the main text are included in the Supplementary Material. More up-to-date versions can be downloaded from our GitHub repository located at: https://github.com/AltschulerWu-Lab/On_Comparing_Heterogeneity_Across_Biomarkers
Results
In this paper, we develop methodology to determine the extent to which decompositions of heterogeneity are preserved from one biomarker to another. Specifically, we focus on the question of how much information about a cell state in one biomarker is gained by knowing its state in another biomarker (Fig. 1A). At one extreme, cells in the same subpopulation for one biomarker are also in the same subpopulation in another biomarker (Fig. 1B top density plot; compare low/hi subpopulations for biomarkers 1 vs. 2). In this case, the two biomarkers clearly identify the same underlying subpopulation structures. On the other extreme, cells in the same subpopulation from one biomarker are randomly spread across subpopulations of the other biomarker (Fig. 1B bottom density plot; compare low/high subpopulations for biomarkers 1 vs. 3). In this case, information about the state of a cell based on one biomarker gives no information about the state of a cell based on the other biomarker, and subpopulations identified from the two biomarkers are considered to be “unrelated”. Our goal is to assess how deep or consistent heterogeneity is across a set of biomarkers.
In principle, co-staining of biomarkers will allow for the determination of whether the biomarkers re-identify the same underlying subpopulations (Figs. 1A,B). However, a primary challenge in relating heterogeneity across a large panel of biomarkers using microscopy is the experimental difficulty of simultaneously assaying multiple biomarkers. Here, we propose an experimental-theoretical framework to compare biomarker heterogeneity that does not require biomarkers to be co-stained. Instead, we require only that the biomarkers of interest be stained (possibly separately) on a common collection of cell lines. Then, for each cell line, we calculate the fractions of cells belonging to the different subpopulations for each biomarker (Fig. 1C). (We note that our results produce multiplexed biomarker information at the subpopulation level; we do not determine the states of multiple biomarkers in individual cells.) We show here that the relationship between subpopulation profiles of two biomarkers on the same collection of cell lines can be used to infer a relationship between their respective subpopulations (Fig. 1D).
How can we relate subpopulations across biomarkers when these biomarkers are never simultaneously measured on the same cells? Our approach is to look for co-variation of subpopulation profiles determined from different biomarkers across a common collection of cell lines. As with any regression, an underlying assumption is that the true variation is large enough to overcome experimental noise. This translates to the requirement that the cell lines considered have diverse subpopulation profiles. In practice, we relate subpopulations from two biomarkers by performing a multivariate regression between two sets of profiles. We then quantify the strength of the relationship between the two biomarkers based on the goodness of fit of this regression (described in detail in Materials and Methods). In the simplified case of two subpopulations per biomarker (Fig. 1D), the regression is between the fraction of cells in one subpopulation of one biomarker against a corresponding subpopulation fraction for another biomarker. When the two biomarkers re-identify the same subpopulation structure, a clear regression trend is seen (Fig. 1D, top panel), while when the subpopulations are unrelated, the quality of regression is poor (Fig. 1D, bottom panel).
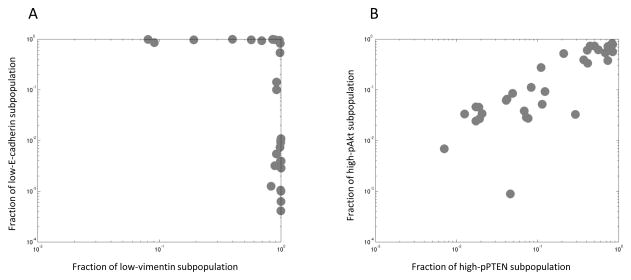
To give a concrete demonstration of these concepts, we examine two cases of non-co-stained biomarkers with known relationships (Fig. 2). For ease of interpretation, we use a two-subpopulation model to characterize each biomarker. Because our features were chosen to quantify cellular biomarker expression, the two subpopulations reflect either high or low biomarker levels. We observe how the occurrence of these subpopulations co-varied across our collection of lung cancer cell lines. First, we look at E-cadherin and vimentin (Fig. 2A), whose expression levels are well-known to report on opposing epithelial or mesenchymal cellular programs (33). Consistently, our analysis reveals a striking switch-like relationship between the two biomarkers: cell lines either have a significant fraction of cells that were low in vimentin and high in E-cadherin (epithelial-like), or the other way around (mesenchymal-like). Next, we look at pAkt and pPTEN (Fig. 2B). Here, we find a striking correlation between the proportions of high-expressing pAkt and pPTEN subpopulations in our cell lines, reflecting a well-known relationship between these signaling molecules (35,36). These results suggest that studies of co-variation can reveal subpopulation-level relationships among biomarkers. Below, we allow model-fit criteria to select the number of subpopulations (between two to eight; Materials and Methods) for each biomarker when performing analytical studies of co-variation.
The Subpopulation Structure Similarity (S3) score
We compute an information theoretic measure for goodness of fit to quantify the relationship between biomarkers, which we term the Subpopulation Structure Similarity (S3) score (described in detail in Materials and Methods). The S3 score is normalized such that 0 corresponds to the case when biomarkers are completely unrelated and 1 corresponds to the comparison of a biomarker to itself. To better understand if these bounds could be reached in practice, we use the DNA (Hoechst) channel of the LCC dataset as a positive control. In principle, DNA-based subpopulations constructed using replicate data (by keeping the DNA channel and ignoring the other co-stained biomarkers) should be perfectly related, giving an S3 score of 1. Thus, any deviation from this score should reflect degradation in signal due to experimental noise. In practice, across-biomarker-set comparisons of DNA-based subpopulations yielded scores of 0.9 or higher, indicating that scores close to 1 are nearly achievable (Supp. Fig. S4). Ideally, we would also have liked to test whether artifactual covariation inflated the relatedness of un-related biomarkers to be greater than 0. However, identifying the appropriate negative control of perfectly un-related biomarkers (apart from the randomization used to set our bounds) is challenging. Here, we simply note that subpopulations identified using EMT biomarkers such as E-cadherin showed little or no relation to those identified using DNA, with S3 scores as low as 0.05. Taken together, these observations suggest that biomarkers whose subpopulations are unrelated will give a low S3 score (close to 0), while biomarkers that nearly perfectly rediscover each other’s subpopulations will give a high S3 score (close to 1.0).
Validation of method on co-stained biomarkers
Our method is designed to determine the extent to which different biomarkers identify similar subpopulation structures, though without the need to co-stain these biomarkers. For validation, we therefore compare our results to the more direct case when biomarkers are, in fact, co-stained in the same cells. With co-staining, we are able to use the mutual information between the subpopulation assignments across biomarkers (whose joint probability can now be calculated) as our gold standard quantification of the relationship between the biomarkers.
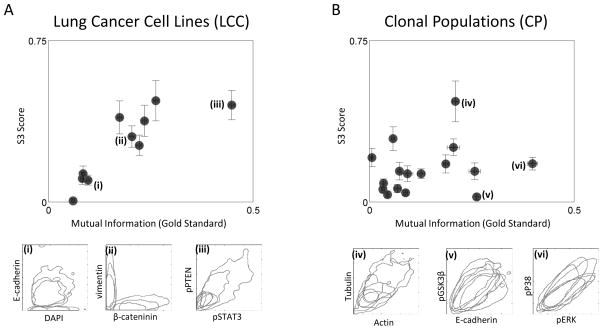
We first test our method on nine pairs of co-stained biomarkers assayed in the LCC dataset of 33 diverse lung cancer cell lines. While some pairs of biomarkers show little relationship at both a single-cell and a cell-line level (e.g. Fig. 3A(i)), others show clear relationships (Fig. 3A(ii–iii)); both the mutual information and the S3 score capture these properties. Overall, we find a clear and strong positive correlation between the S3 score and the co-stained mutual information (Fig. 3A), thereby providing confirmation of our methodology. Next, we explore how reduced variation among cell lines affects the performance of our approach. To this end we apply our method to 15 pairs of co-stained biomarkers in the CP dataset of 49 clonal populations derived from single cells of the same H460 lung cancer cell line. We observe a positive trend between the S3 score and the mutual information from co-staining. However, for the case of our clonal populations, the strength of biomarker-biomarker relationships reported by the S3 score is diminished for all biomarker pairs, and increased mutual information from co-stained biomarker pairs does not necessarily imply increased similarity of subpopulation structure based on our methodology (Fig. 3B). A closer examination of these biomarkers indicates that, while there is extensive heterogeneity and correlation of levels of the biomarkers within a cell line, the “phenotype space” occupied by our clonal cell lines is nearly identical (Fig. 3B(v–vi)), with Actin vs. Tubulin staining being the exception (Fig. 3B(iv)).
Taken together, our results suggest that when cell lines are phenotypically distinct, biomarker-biomarker relations inferred using our methods agree with those obtained by the “gold-standard” of direct co-staining. As the cell lines become more similar, the ability to detect biomarker-biomarker relationships is diminished in a biomarker-specific fashion.
Comparison of non-co-stained biomarkers
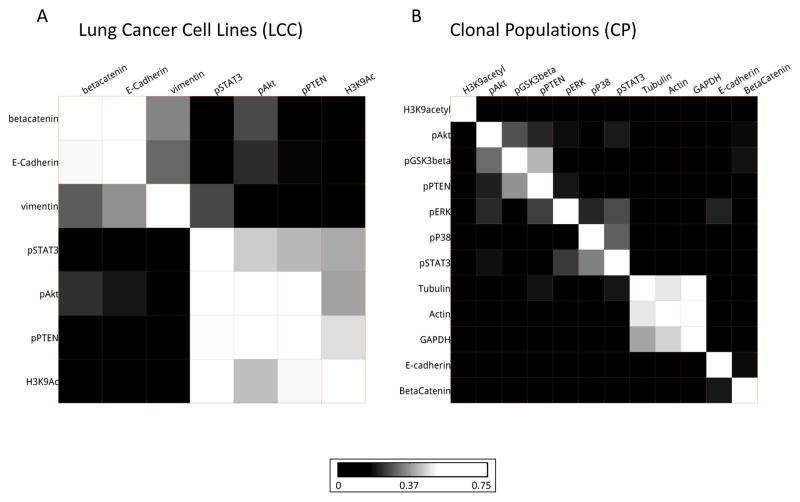
We next examine the relationships between all pairs of biomarkers in the LCC dataset (Fig. 4A, Supp. Figs. S3 and S4). We observe two pronounced clusters of biomarkers. The first captures the well-known relationships between the EMT biomarkers vimentin, E-cadherin, and β-catenin. The second groups the biomarkers pAkt, H3K9Ac, pPTEN, and pSTAT3, which have known roles in promoting cell growth. pAkt and pPTEN have particularly high scores in this cluster, potentially reflecting their known pathway relationship previously mentioned (35).
In the CP dataset, far fewer biomarker pairs exhibit similar subpopulation structures than in the LCC dataset (Fig. 4B). Nonetheless, we do observe clusters of strongly related biomarkers. For example, the cytoskeletal biomarkers actin and tubulin are found to be strongly related to one another and to the housekeeping gene GAPDH. The insulation from other pathways seen in our results is consistent with their frequent use as control biomarkers in a variety of experiments (and also suggests that they are not freely “independent” as controls) (37). We also observe relationships between groups of signaling biomarkers. For this dataset, pAkt and pPTEN are weakly connected via pGSK3β in a cluster. Additionally, we see that pP38, pERK and pSTAT3 cluster together. These three proteins have been implicated together in lung cancer response to potential chemotherapy (38).
As expected, some of the interactions that are detected in the LCC data are missed in the H460 clones. This is particularly evident when considering EMT related biomarkers. As with E-cadherin and vimentin discussed previously, β-catenin and vimentin are also negatively correlated EMT biomarkers: if membrane/cytosolic β-catenin is high in a cell, cells are considered to be more epithelial and, conversely, vimentin-high cells are more mesenchymal (33). However, the epithelial or mesenchymal nature is a cell-line level property: within any given cell line these biomarkers’ expressions are in fact slightly positively correlated (Fig. 3A(ii)). The LCC dataset contains a mixture of epithelial and mesenchymal lines, and hence enough variation to detect the relationship between the EMT biomarkers. On the other hand all the clones of the H460 are epithelial, making it far more difficult to relate the EMT biomarkers. Together, these results suggest the importance of diversity among cell lines for inferring biomarker relationships.
Discussion
Microscopy, through its ability to capture subtle differences in cellular phenotypes, reveals the immense complexity of cellular populations. This complexity can be conceptually broken up into (at least) two aspects: the breadth of phenotypic differences between cells, and the depth of characterization of single cells via multiple biomarkers. Past work has shown that the breadth of heterogeneity can be made tractable (39) by modeling cellular populations as mixtures of a small number of so-called subpopulations (10,14). Here, we investigate the extent to which more biomarkers would give deeper coverage of phenotypic states. If additional biomarkers simply re-identify previously identified subpopulations, then they add little new information and complexity. On the other hand, if each biomarker leads to an unrelated decomposition of heterogeneity, then the complexity of the systems blows up with the number of biomarkers studied.
As a first step towards answering such questions, we built an experimental-theoretical framework to compare subpopulations identified by different biomarkers. A fundamental challenge is that traditional microscopy setups allow at most 5 biomarkers to be simultaneously assayed. We attempted to overcome this limitation using a novel approach, which requires only that the biomarkers be stained (but not necessarily co-stained) on a common set of cell lines. The approach we took for comparing heterogeneity was general and could be applied to different choices of cellular features or approaches for defining subpopulations (that may be motivated by specific biological questions). The extent to which biomarkers identify the same subpopulations can then be estimated by the ability of one biomarker’s subpopulation profiles to predict those of another biomarker across the set of cell lines. Our preliminary results using this methodology suggest an intermediate level of complexity: although there are a number of biomarker groups within which subpopulations are re-identified, subpopulations identified by biomarkers that do not belong to the same group are largely unrelated. The groupings of biomarkers we obtain largely reflect established signaling relationships, with groups consisting of EMT biomarkers, housekeeping readouts and particularly strong relations between biomarkers in the same signaling pathway.
The major advantage of our proposed method is experimental extensibility. To relate a new biomarker to a panel of biomarkers that has already been assayed, only the new biomarker (but not previously assayed biomarkers) need be stained across the collection of cell lines. Additionally, imputing biomarker relations using our methods can offer advantages over studying natural variation or perturbation experiments. Using natural variation runs the risk that the amount of true variation within a population is so small that it gets swamped by non-biological signals (40). For example, as we saw, the expression levels of β-catenin and vimentin can be positively correlated in a single cell line; the well-established negative correlation only emerges when we look across multiple cell lines. Although the use of perturbations can resolve this issue, perturbations often push cells far outside their usual operating regime, thereby undermining the biological relevance of any results discovered. For example, protein interactions found from overexpression studies may not reflect the behavior of cells in their basal state. Thus, with judiciously chosen cell lines, our approach offers the prospect of a middle ground: with increased variation, but cells still operating within their normal parameters.
The dependence of our results on our choice of cell lines raises interesting questions about the biology revealed at different scales of variation. When the cell lines are closely related, relationships discovered can be subtle and specific to this system. However, as the cell lines become more diverse, only coarse global relations that are supported by the entire panel will be discovered. Ultimately, once the cell lines become fundamentally different, each may support a different set of relationships between biomarkers, leaving no preserved relationships to discover. Anecdotal evidence suggests that a panel of cancer cell lines from similar tissue and sub-type provides a sweet spot in inter-cell-line-variation (data not shown). This suggests that a systematic study of how the degree of variation affects the biology that can be discovered is an interesting avenue to explore in the future. Previous work has suggested that crosstalk between network components can be phenotype specific (41). Our methods will allow us to evaluate whether different phenotypes extracted from the same biomarker identify the same subpopulation structures and how different phenotypes affect identified relationships between biomarkers.
Although our motivations were primarily conceptual, there are a number of practical applications of the method proposed here. The most obvious application is to the selection of a panel of biomarkers to investigate cellular heterogeneity. Since there is a premium on the number of biomarkers that can be simultaneously assayed in microscopy, it is crucial to select the least redundant set possible. Our method provides a way of determining which biomarkers share information, and thus a means of selecting those that provide largely independent views of cellular states (42). Another possible application, made possible by the extensibility of this method, is to infer novel biomarker interactions. A new biomarker need only be assayed on the common set of cell lines, and relations to any previously assayed biomarkers could be tested. Being based on statistical correlation, such hypotheses would, of course, need to be rigorously tested with more specific experiments. Finally, although we have not explored the specific subpopulations that were deemed as being related, this mapping can itself be of biological interest. For example one could answer questions such as: How does the subpopulation of cells with activated JAK/STAT signaling vary with respect to EMT, MAPK, Akt, and cell cycle signaling states? Or, given a prognostic biomarker, what additional biomarkers could be used to confirm the presence of a disease-related subpopulation?
In conclusion, the apparent heterogeneity of a cellular population is dependent on the assay used for phenotypic characterization. Microscopy facilitates highly detailed characterization using a few biomarkers, but there are experimental limits to how many biomarkers can be simultaneously assayed. It has therefore been challenging to determine the effect, on heterogeneity, of assaying a large number using microscopy. To this end, we have attempted to build an experimental-theoretical framework that allows us to determine if different biomarkers yield similar decompositions of heterogeneity. By staining biomarkers on a common set of cell lines, we have been able to overcome microscopy’s limitations on the number of simultaneous biomarkers. Our results, using this framework, suggest that biomarkers can be partitioned into different groups that each result in a similar decomposition of heterogeneity. The groupings found in this way are consistent with known signaling pathways. We believe this work represents a useful first step in deeper profiling of heterogeneity using microscopy.
Supplementary Material
Acknowledgments
We would like to thank Jason Altschuler for his useful comments on the manuscript. This work was supported by the National Cancer Institute grants NCI R01CA133253 (SA) and NCI SPORE P50CA70907 (JM), the National Institute of Health grant NIH R01 GM081549 (LW), the Welch Foundation I-1619 (SA) and I-1644 (LW), and Rita Allen Foundation (SA).
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–63. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelkmans L. Cell Biology. Using cell-to-cell variability--a new era in molecular biology. Science. 2012;336:425–6. doi: 10.1126/science.1222161. [DOI] [PubMed] [Google Scholar]
- 3.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–34. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 4.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–37. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farkash-Amar S, Zimmer A, Eden E, Cohen A, Geva-Zatorsky N, Cohen L, Milo R, Sigal A, Danon T, Alon U. Noise genetics: inferring protein function by correlating phenotype with protein levels and localization in individual human cells. PLoS Genet. 2014;10:e1004176. doi: 10.1371/journal.pgen.1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Ku CJ, Zhang ER, Artyukhin AB, Weiner OD, Wu LF, Altschuler SJ. Identifying network motifs that buffer front-to-back signaling in polarized neutrophils. Cell Rep. 2013;3:1607–16. doi: 10.1016/j.celrep.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janes KA, Wang CC, Holmberg KJ, Cabral K, Brugge JS. Identifying single-cell molecular programs by stochastic profiling. Nat Methods. 2010;7:311–7. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–8. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh DK, Ku CJ, Wichaidit C, Steininger RJ, 3rd, Wu LF, Altschuler SJ. Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol Syst Biol. 2010;6:369. doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyson DR, Garbett SP, Frick PL, Quaranta V. Fractional proliferation: a method to deconvolve cell population dynamics from single-cell data. Nat Methods. 2012;9:923–8. doi: 10.1038/nmeth.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–15. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 13.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–22. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Slack MD, Martinez ED, Wu LF, Altschuler SJ. Characterizing heterogeneous cellular responses to perturbations. Proc Natl Acad Sci U S A. 2008;105:19306–11. doi: 10.1073/pnas.0807038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo LH, Lin HJ, Singh DK, Lyons KM, Altschuler SJ, Wu LF. Heterogeneity in the physiological states and pharmacological responses of differentiating 3T3-L1 preadipocytes. J Cell Biol. 2009;187:375–84. doi: 10.1083/jcb.200904140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–7. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–8. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz RF, Trinh A, Sipos B, Brenton JD, Goldman N, Markowetz F. Phylogenetic quantification of intra-tumour heterogeneity. PLoS Comput Biol. 2014;10:e1003535. doi: 10.1371/journal.pcbi.1003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–9. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 21.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 22.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Li F, Yin Z, Jin G, Zhao H, Wong ST. Chapter 17: bioimage informatics for systems pharmacology. PLoS Comput Biol. 2013;9:e1003043. doi: 10.1371/journal.pcbi.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–7. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–42. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–22. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 27.Gustafsdottir SM, Ljosa V, Sokolnicki KL, Anthony Wilson J, Walpita D, Kemp MM, Petri Seiler K, Carrel HA, Golub TR, Schreiber SL, et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS One. 2013;8:e80999. doi: 10.1371/journal.pone.0080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo LH, Lin HJ, Steininger RJ, 3rd, Wang Y, Wu LF, Altschuler SJ. An approach for extensibly profiling the molecular states of cellular subpopulations. Nat Methods. 2009;6:759–65. doi: 10.1038/nmeth.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AW. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24:1270–8. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 30.Keller PJ, Lin AF, Arendt LM, Klebba I, Jones AD, Rudnick JA, DiMeo TA, Gilmore H, Jefferson DM, Graham RA, et al. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res. 2010;12:R87. doi: 10.1186/bcr2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasband WS. ImageJ. National Institutes of Health; Bethesda, Maryland, USA: 1997–2008. http://rsb.info.nih.gov/ij/ [Google Scholar]
- 32.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–53. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Yin Z, Zhou X, Bakal C, Li F, Sun Y, Perrimon N, Wong ST. Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics. 2008;9:264. doi: 10.1186/1471-2105-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 36.Georgakis GV, Li Y, Rassidakis GZ, Medeiros LJ, Mills GB, Younes A. Inhibition of the phosphatidylinositol-3 kinase/Akt promotes G1 cell cycle arrest and apoptosis in Hodgkin lymphoma. Br J Haematol. 2006;132:503–11. doi: 10.1111/j.1365-2141.2005.05881.x. [DOI] [PubMed] [Google Scholar]
- 37.Sturzenbaum SR, Kille P. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:281–9. doi: 10.1016/s1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- 38.Xue P, Zhao Y, Liu Y, Yuan Q, Xiong C, Ruan J. A novel compound RY10-4 induces apoptosis and inhibits invasion via inhibiting STAT3 through ERK-, p38-dependent pathways in human lung adenocarcinoma A549 cells. Chem Biol Interact. 2014;209:25–34. doi: 10.1016/j.cbi.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Candia J, Maunu R, Driscoll M, Biancotto A, Dagur P, McCoy JP, Jr, Sen HN, Wei L, Maritan A, Cao K, et al. From cellular characteristics to disease diagnosis: uncovering phenotypes with supercells. PLoS Comput Biol. 2013;9:e1003215. doi: 10.1371/journal.pcbi.1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padovan-Merhar O, Raj A. Using variability in gene expression as a tool for studying gene regulation. Wiley Interdiscip Rev Syst Biol Med. 2013;5:751–9. doi: 10.1002/wsbm.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ku CJ, Wang Y, Weiner OD, Altschuler SJ, Wu LF. Network crosstalk dynamically changes during neutrophil polarization. Cell. 2012;149:1073–83. doi: 10.1016/j.cell.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005;3:185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.