Results from single-molecule and macroscopic electrophysiology and kinetic analysis provide a model for activation of the glutamate-bound NMDA receptor by glycine.
Abstract
N-methyl-d-aspartate (NMDA) receptors are the only neurotransmitter receptors whose activation requires two distinct agonists. Heterotetramers of two GluN1 and two GluN2 subunits, NMDA receptors are broadly distributed in the central nervous system, where they mediate excitatory currents in response to synaptic glutamate release. Pore opening depends on the concurrent presence of glycine, which modulates the amplitude and time course of the glutamate-elicited response. Gating schemes for fully glutamate- and glycine-bound NMDA receptors have been described in sufficient detail to bridge the gap between microscopic and macroscopic receptor behaviors; for several receptor isoforms, these schemes include glutamate-binding steps. We examined currents recorded from cell-attached patches containing one GluN1/GluN2A receptor in the presence of several glycine-site agonists and used kinetic modeling of these data to develop reaction schemes that include explicit glycine-binding steps. Based on the ability to match a series of experimentally observed macroscopic behaviors, we propose a model for activation of the glutamate-bound NMDA receptor by glycine that predicts apparent negative agonist cooperativity and glycine-dependent desensitization in the absence of changes in microscopic binding or desensitization rate constants. These results complete the basic steps of an NMDA receptor reaction scheme for the GluN1/GluN2A isoform and prompt a reevaluation of how glycine controls NMDA receptor activation. We anticipate that our model will provide a useful quantitative instrument to further probe mechanisms and structure–function relationships of NMDA receptors and to better understand the physiological and pathological implications of endogenous fluctuations in extracellular glycine concentrations.
INTRODUCTION
NMDA receptors are glutamate- and glycine-gated ion channels that mediate excitatory neurotransmission in the mammalian central nervous system (CNS). They are required for normal brain development and function throughout life but are also implicated with acute and chronic neurodegeneration, neuropathic pain syndromes, and developmental disorders such as schizophrenia and autism (Paoletti et al., 2013). NMDA receptors are heterotetramers of two GluN1 and two GluN2 subunits; throughout the CNS, the obligatory subunit GluN1 is expressed ubiquitously, and four GluN2 (A–D) isoforms are differentially represented according to neuronal type, developmental stage, and synaptic or extrasynaptic location. Glycine binding to GluN1 subunits and glutamate binding to GluN2 subunits are both required for receptor activation (Kleckner and Dingledine, 1988; Hirai et al., 1996; Laube et al., 1998).
Endogenous glycine was initially considered inhibitory to CNS neurotransmission (Curtis et al., 1967; Werman et al., 1967), until it was found to enhance neuronal NMDA-dependent currents (Johnson and Ascher, 1987), and later to be required for NMDA receptor activity in heterologous expression systems (Kleckner and Dingledine, 1988). Extracellular glycine concentrations vary with brain region and neuronal activity, and are controlled in an activity-dependent manner by uptake and release through GlyT1, a glycine transporter expressed in astrocytes and postsynaptic neurons (Zafra et al., 1995; Snyder and Ferris, 2000; Harsing and Matyus, 2013). Dynamic changes in endogenous glycine concentrations modulate NMDA receptor responses and have been successfully exploited as therapeutic interventions in schizophrenia (Coyle and Tsai, 2004).
Early studies in dissociated neurons revealed the potent and mechanistically complex effects of glycine on NMDA receptors. Glycine increases NMDA-elicited currents with submicromolar potency (EC50 of <1 µM) and also strongly slows their desensitization (Kleckner and Dingledine, 1988; Mayer et al., 1989; Johnson and Ascher, 1992). In oocytes expressing recombinant GluN1/GluN2A receptors, glycine has similarly high potency (EC50 of ∼1–3 µM) (Chen et al., 2008). With maximally effective glycine concentrations (>10 µM), macroscopic NMDA receptor currents elicited with seconds-long glutamate exposures relax slowly (τD of ∼1–2 s) to a steady-state current that is 20–40% lower than the peak current. However, when glycine concentrations are below its EC50 value, in addition to reduced peak currents, traces also display much faster and deeper relaxation, a biophysical property known in the literature as “glycine-dependent desensitization” (Benveniste et al., 1990; Lerma et al., 1990; Vyklický et al., 1990).
This phenomenon has been explained by postulating negative cooperativity between the glutamate- and glycine-binding sites (Benveniste et al., 1990). In brief, this hypothesis assumes that in concert with binding glutamate, the receptor’s affinity for glycine decreases approximately fivefold; thus, in subsaturating glycine concentrations, the initial glutamate-elicited peak current decreases quickly as glycine molecules dissociate to reach the new equilibrium dictated by the lowered glycine affinity of glutamate-bound receptors. The molecular basis for this phenomenon is unresolved. Structural models of NMDA receptors illustrate that the agonist-binding sites are located on distant, quasi-independent modules; however, substantial interactions exist between these domains (Furukawa et al., 2005; Karakas and Furukawa, 2014; Lee et al., 2014), thus providing a possible basis for a concerted conformational change. At odds with the hypothesis of allosteric coupling between binding sites, mutations in the glycine-binding site that decrease glycine affinity up to 100-fold have no effect on glutamate affinity (Hirai et al., 1996), and some allosteric modulators can selectively modulate the receptor’s affinity for glycine but not for glutamate (Nahum-Levy et al., 2002; Hansen et al., 2012). Therefore, the issue remains controversial.
At the single-channel level, increasing glycine concentration does not change the receptor’s mean open durations but increases their bursting frequency (Johnson and Ascher, 1987; Schorge et al., 2005). This observation is consistent with glycine dissociating from a preopen closed state (Mayer et al., 1989). For this reason, and also in accord with the strong evidence that both glutamate and glycine must bind before the receptor transitions into open conformations, previous state models of glycine binding assumed that both glutamate and glycine dissociate from the same closed kinetic state of NMDA receptors (Benveniste et al., 1990; Nahum-Levy et al., 2001, 2002; Schorge et al., 2005). Fitting these models to NMDA receptor responses estimated rate constants in agreement with the high potencies of both glutamate and glycine (Kd of 1–3 µM).
More recently, reaction mechanisms for glycine-bound GluN1/GluN2A receptors have been expanded to include multiple closed and open states, as required by single-channel observations. These models account for the full spectrum of microscopic receptor behaviors and have produced novel insights into previously unexplained macroscopic phenomena (Popescu et al., 2004; Zhang et al., 2008). We used these models as a starting point to investigate the role of glycine in NMDA receptor activation. Here, we describe results from kinetic analyses and modeling of one-channel current recordings obtained in the presence of glutamate and a series of glycine-site agonists at several concentrations. Based on these results, we propose a new reaction scheme that describes the principal kinetic steps during glycine activation of glutamate-bound NMDA receptors. This model expands the reach of quantitative modeling to the glycine-binding reaction and offers deeper insight into how glycine concentrations sculpt the NMDA receptor response.
MATERIALS AND METHODS
Cell culture and receptor expression
Plasmids encoding GluN1-1a (GenBank accession no. U08261), GluN2A (GenBank accession no. M91561), and GFP cloned in pcDNA3.1 (+) were transiently transfected into HEK 293 cells (CRL-1573; ATTC) at passage 16–34 in a 1:1:1 ratio using the calcium phosphate precipitation method (Chen and Okayama, 1987). Cells were used for electrophysiological measurements 24–48 h after transfection.
Managing glycine contamination
Levels of contaminating glycine can reach 40–50 nM (Kleckner and Dingledine, 1988; Lerma et al., 1990). At these concentrations, glycine supports a substantial level of NMDA receptor activity and may influence the observed results. Therefore, we took several precautions when handling and preparing solutions for the work described here. These included using ultrapure reagents (>99.99% purity; Sigma-Aldrich) and ultrapure double-distilled deionized water (Thermo Fisher Scientific) for the preparation of solutions. In addition, all glassware and equipment were acid washed and soaked in ultrapure water (>24 h) to leech contaminating glycine, before use.
Single-channel current recording, processing, kinetic analyses, and modeling
Recording, processing, and idealization of on-cell one-channel NMDA receptor currents are extensively explained and illustrated in previous publications (Kussius et al., 2009; Maki et al., 2014). In brief, we used fire-polished glass pipettes of 12–25-MΩ resistance, when filled with extracellular solution containing (mM): 150 NaCl, 2.5 KCl, 1 EDTA, 10 HEPBS, pH 8.0 (NaOH), 1 glutamate, and several different concentrations of either glycine, l-serine, or 3,3,3-trifluoro-dl-alanine. We recorded inward Na+ current through a cell-attached patch containing one active channel, with an applied pipette potential of 100 mV. Data were amplified at 100× gain, analogue filtered at 10 kHz (Axopatch 200B, four-pole Bessel), and sampled at 40 kHz (NiDAq, PCI-6229, M series card; National Instruments). Currents were recorded and stored using QuB software. Only data from patches with no apparent multiple channel openings were used for analyses.
Data were conservatively and minimally processed before subsequent analysis and modeling to correct for recording artifacts, including low frequency, high amplitude noise spikes and baseline drift. Current spikes (shorter than 0.05–0.15 ms) were eliminated by replacing the sampled amplitudes within the spike with amplitude levels averaged from the immediately adjacent (open or closed) samples. Baseline drift was corrected by forcing the amplitude level for closed events to zero, at nodal points throughout the file, as necessary.
Idealization was done with the segmental k-means (SKM) algorithm on digitally filtered data (12 kHz) with no imposed dead time (Qin, 2004). This procedure assigns each sampled data point to either the open (O; ∼8 pA) or closed (C; 0 pA) class and produces lists of open and closed intervals (i.e., periods containing consecutive O or C class samples), with their associated durations. The list of events thus generated preserves the sequence in which these intervals occurred.
Modeling was done in QuB by fitting user-defined state models to the sequence of idealized intervals generated by SKM idealized intervals. Fitting was done with a maximum interval likelihood (MIL) algorithm using an imposed dead time of 0.15 ms (Qin et al., 1997), and models were ranked according to their calculated maximum value of the log likelihood (LL) function.
Simulations
For each candidate model, macroscopic responses were calculated for 100 channels of 10-pA unitary amplitude, as the accretion of receptors in open states over time, following a variety of stimulation protocols. Glutamate-binding steps were appended to the kinetic state C3 as described previously (Popescu et al., 2004), and glycine-binding steps were appended in turn to C3, C2, or C1 closed states using the rates calculated for each model from global fits across glycine concentrations. Receptors were first equilibrated in 1 mM glutamate or 0.5 or 100 µM glycine, and then stepped into the indicated glycine or glutamate concentrations, as indicated, for a given time interval. The resulting simulated traces were analyzed as described below for experimentally recorded macroscopic currents.
Macroscopic current recordings and analyses
Macroscopic currents were acquired using the whole-cell or outside-out patch configurations with intracellular solutions containing (mM): 135 CsCl, 33 CsOH, 2 MgCl2, 1 CaCl2, 10 HEPES, pH 7.4/CsOH, and 11 EGTA using pipettes of 5–10-MΩ resistance and a holding potential of −70 mV. All extracellular solutions contained (mM): 150 NaCl, 2.5 KCl, 10 HEPBS, pH 8.0/NaOH, 0.5 CaCl2, 0.01 EDTA, and 1 glutamate, 0.1 or 100 µM glycine, or 2.1 mM l-serine. Currents were elicited by switching patches or cells expressing GluN1/GluN2A receptors into extracellular solution containing the above components plus the required coagonist. For experiments assaying responses to glycine and l-serine, currents were elicited by exposing cells or patches to glycine or l-serine in the continuous presence of glutamate. For experiments assaying responses to glutamate, currents were elicited by exposing cells or patches to glutamate in the continuous presence of glycine or l-serine. For outside-out patches, only those records with open-tip response rise times of 0.2–0.4 ms were processed and analyzed. We calculated, in pClamp 10.2 (Molecular Devices), rise time as the 10–90% time to peak, decay kinetics as exponential decline for outside-out patch currents, and Iss/Ipk ratios for whole-cell currents. All dose–response calculations were done by fitting the Hill equation with fixed minimum and maximum response values to experimental or simulated data, as indicated.
RESULTS
Modeling one-channel activity in low glycine concentrations
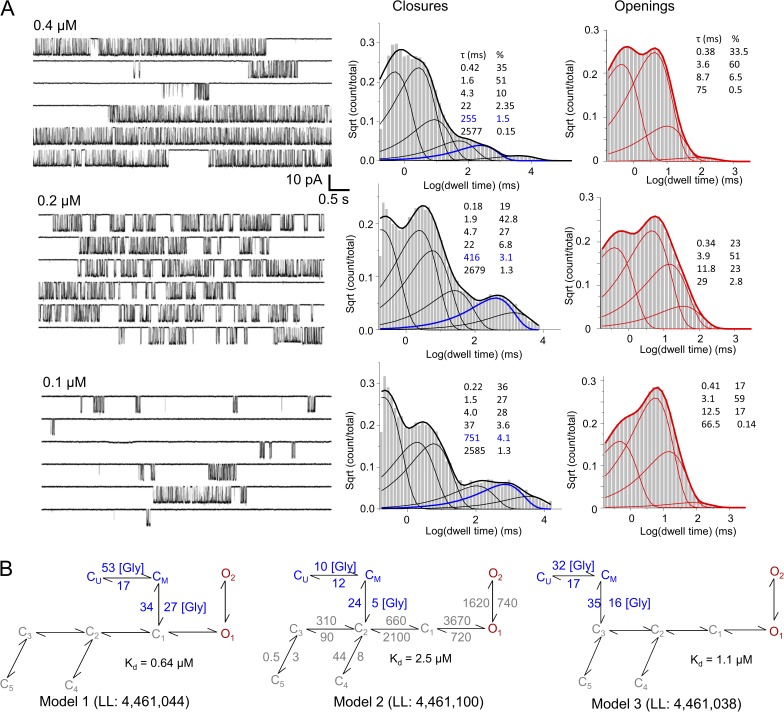
To investigate the microscopic kinetics of glycine binding to NMDA receptors, we took a modeling approach that was previously successful in delineating the sequence of kinetic states populated by NMDA receptors after glutamate exposure (Popescu et al., 2004; Amico-Ruvio and Popescu, 2010; Kussius and Popescu, 2010; Murthy et al., 2012). For this, we recorded steady-state currents from cell-attached patches that contained one GluN1/GluN2A receptor with pipettes containing a high glutamate concentration (1 mM; Kd = 3 µM) and several subsaturating glycine concentrations (0.1, 0.2, and 0.4 µM; EC50 of ∼1 µM) (Patneau and Mayer, 1990; Schorge et al., 2005; Chen et al., 2008) (Fig. 1 A, left). We used these data to evaluate a series of kinetic models for their goodness of fit, with the goal of first establishing the minimum number of closed and open states in the model, and then discerning the most likely arrangement of these states.
Figure 1.
NMDA receptor activity in subsaturating concentrations of glycine and 1 mM glutamate. (A) Representative current traces (left) and histograms of closed (black) and open (red) event durations calculated from data within one full record, in three glycine concentrations, as indicated. The concentration-dependent closed component is highlighted in blue. (B) Three top-ranking models, and their respective LL values, incorporate explicit glycine-binding steps; they were each fitted globally to data obtained with several glycine concentrations (n = 9; >2 × 106 events). Glycine concentration is in micromolars. Rate constants are in s−1, except for the glycine-binding rate constants, which are in µM−1s−1.
To determine the optimum number of states in the model, we started with the simplest possible reaction: 1C1O or C↔O; we constructed models with increasing numbers of states by adding closed or open states one at a time, in no order. Using the MIL algorithm, we fitted each model directly to the sequence of idealized intervals obtained from each single-channel data file and recorded the increase in the maximum value of the LL function with each added state. We note that MIL calculates probability density functions rather than simple probabilities. As a result, LL values increase proportionally with the number of events in the idealized data and with the number of free parameters in the model. Numerous studies have found that, in practice, when evaluating models of increasing complexity against a fixed dataset, a cutoff value of 10 LL units per added state is a useful even if empirical criterion to select a minimally complex but sufficiently detailed kinetic model (Horn, 1987). In our modeling procedure, for traces recorded with subsaturating glycine concentrations, we found that for all files, closed intervals had six kinetic components (Fig. 1 A, right). As in previous reports for data obtained with saturating concentrations of glycine, the files obtained here differed in the number of open states that could be discerned with the 10 LL–unit threshold, with most files requiring three or four open states for optimum fit (Popescu and Auerbach, 2003). Given the present understanding of the NMDA receptor gating mechanisms, we aimed to select a model that would adequately fit all files in our dataset, so that we could ultimately fit it globally to data obtained at several subsaturating concentrations of glycine.
The basic reaction mechanism of fully liganded GluN1/GluN2A receptors consists of transitions on three time scales, referred to as activation, desensitization, and mode shifts (Popescu, 2012). Upon binding glutamate, receptors enter the activation sequence, which can be described as the passage through three consecutive closed states (C3↔C2↔C1) that lead into two coupled open states (O1↔O2) of similar conductance (Popescu and Auerbach, 2003; Popescu et al., 2004; Auerbach and Zhou, 2005). On a slower time scale, receptors can escape this linear 3C2O sequence by branching off from C3 or C2 states into two kinetically distinct desensitized states, D1 or D2, respectively (Kussius et al., 2009). This branched seven-state reaction mechanism, 3C2D2O, can generate three distinct kinetic patterns that differ mainly in the stability of open states, and these three modes interconvert on a seconds-to-minutes time scale (Popescu and Auerbach, 2004). Therefore, a comprehensive statistical description for the activity of a fully liganded one-channel that includes all three kinetic modes must be represented by a 21-state three-tier model, such that each channel cycles extensively through each tier, described by the 3C2D2O scheme, before it slips stochastically into a mode with distinct open lifetimes but similar 3C2D2O sequence. It was proposed previously that glutamate dissociation occurs with observable probability only from the C3 states of either mode, and the association and dissociation kinetics of glutamate are similar for all three tiers (Popescu and Auerbach, 2003). Clearly, after incorporating glutamate-binding steps to each of the three tiers, the resulting 27-state model, although grossly simplified relative to the myriad of conformational transitions that most likely accompany gating, is too complex to be practical for quantitative investigations of steady-state one-channel data. Instead, depending on the experimental conditions and the scope of the analyses, simpler models can adequately fit the purpose. One common simplification is to ignore modal behavior and fit a 3C2D2O model to the entire dataset (Kazi et al., 2013, 2014). If the behavior investigated is independent of gating mode, the analysis remains valid with the understanding that values for opening and closing rate constants represent weighted averages across the modes captured in each file and thus may vary somewhat from one record to another. For this work, we also elected to use a model with two aggregated open states. When compared with results from files obtained with high glycine concentrations (100 µM; n = 14; >3.4 × 106 events) (Table 1), decreasing glycine concentration (0.1 µM, n = 5; 0.2 µM, n = 6; and 0.4 µM, n = 5) had no observable effect on the mean open durations or on the open interval distributions (Fig. 1 A, right). Therefore, aggregating the modal behavior (indicated by the open interval distributions) into an average model with only two open states is unlikely to confound our investigations of glycine binding.
Table 1.
Single-channel parameters for GluN1/GluN2A receptors in several glycine concentrations
| [Glycine] | n | Amplitude | Poa | MOTb | MCTb | Events analyzed |
| µM | pA | ms | ms | |||
| 0.1 | 3 | 9.2 ± 0.2 | 0.08 ± 0.01 | 6.1 ± 0.9 | 57 ± 14c | 3.5 × 105 |
| 0.2 | 3 | 8.9 ± 0.7 | 0.16 ± 0.05 | 6.7 ± 0.8 | 31 ± 4c | 7.3 × 105 |
| 0.4 | 3 | 9.5 ± 0.5 | 0.29 ± 0.03 | 6.2 ± 0.5 | 14 ± 1c | 9.7 × 105 |
| 100 | 14 | 8.9 ± 0.3 | 0.52 ± 0.03 | 7.1 ± 0.6 | 6.5 ± 0.7 | 3.4 × 106 |
All values are mean ± SEM.
Values were calculated with full models in MIL.
Values were calculated with two-state models in SKM.
Significant differences relative to 100 µM (P < 0.00002; Student’s t test).
Having selected a model with six closed and two open states, we assumed that the sequence of states within the core 3C2D2O model remains as reported previously for GluN2A- and GluN2B-containing receptors (Kussius et al., 2009; Amico-Ruvio and Popescu, 2010). For the low concentration dataset, we fit manually the seven models where the C6 state was appended in turn to each of the C1,2,3, D1,2, or O1,2 states. This analysis allowed us to calculate average parameters (amplitude, MOT, MCT) for each file (n = 16). In Table 1, we report means ± SEM for a subset of these files (n = 9; see below data selection for global fits). We compared these parameters with those obtained by reanalyzing, with the methodology described here, a combined dataset obtained for previous studies of GluN2A-containing receptors (n = 14) (Kussius et al., 2009; Borschel et al., 2011). For the low glycine dataset, models that had the C6 state appended to D1, D2, or O2 did not converge to an acceptable fit for any of the 16 records tested. Therefore, we did not include these models in our subsequent analyses. However, the remaining four models estimated rates for transitions within the core model that did not change significantly when moving the C6 state. Moreover, each of the 12 rates was within the range of values (means ± SEM) we calculated for the saturation dataset (n = 14) with the core model, 3C2D2O. Of the four models tested, the model that postulated glycine dissociation from the O1 state produced consistently the poorest fits, in line with the observation that neither open durations nor open distributions were affected by changing glycine concentrations. Therefore, we proceeded to evaluate in more detail the remaining three models that had the extra closed state (C6) connected to C3, C2, or C1.
To estimate the microscopic binding and dissociation rate constants predicted by each of these three remaining models, we replaced the concentration-dependent step (C6↔C3,2,1) with two sequential transitions (CU↔CM↔C3,2,1), whose rates were scaled to represent the assumption that the glycine-binding steps are identical and independent. Therefore, with this substitution, the number of free parameters (and thus the LL values) in our models remained unchanged. Next, we aimed to fit these models globally across data obtained at several glycine concentrations. Because of the computational cost of fitting globally to a large dataset, we selected for this analysis three representative files obtained at each 0.1-, 0.2-, and 0.4-µM glycine concentration. Except for scaling the binding steps, all rate constants were allowed to fluctuate freely during the fitting procedure. Because the threshold we used for selecting the minimum number of states was 10 LL units, we would expect that keeping the same number of states and changing only the arrangement of states will likely improve the fit by less than 10 LL units. For this reason, relative to the large value of the maximum LL function for each file, the differences in LL produced by rearranging states were small; however, this analysis provided a first-pass method for ranking goodness of fit for models with the same number of states but different topologies.
Based on LL values, Model 2, which allows glycine dissociation from C2, was the top candidate: Model 1, 4,461,044; Model 2, 4,461,100; Model 3, 4,461,038 (Fig. 1 B). As we observed above with fits to individual files, the rate constants for transitions within the core 3C2D2O model obtained from this global fit (n = 9) were similar to those obtained at saturation (n = 14). Therefore, in Fig. 1 B, we report rounded values for these transitions calculated from the saturation dataset (in gray), and the actual results of the global fit (in blue) for the binding transitions. Model 2 estimated glycine association and dissociation rate constants, kon = 5 × 106 M−1s−1 and koff = 12 s−1, and a calculated dissociation constant, Kd = 2.4 µM, that is well within the range of literature values (Johnson and Ascher, 1987; Kleckner and Dingledine, 1988; Benveniste et al., 1990). The glycine dissociation rate constant estimated here is approximately fivefold slower than the glutamate-dissociating constant estimated previously with the same method (60 s−1) (Popescu et al., 2004; Kussius et al., 2009), and directly reflects the much lower frequency with which glycine dissociation events occurred. In the closed event distribution, the kinetic component whose duration varied with glycine concentration represented <4% of the total (>2 × 106) closed events. In an attempt to increase the resolution for the dissociation step, we repeated these measurements and analyses using two glycine-site agonists with lower affinity, which may dissociate more frequently.
Modeling one-channel activity with partial and supra-agonists
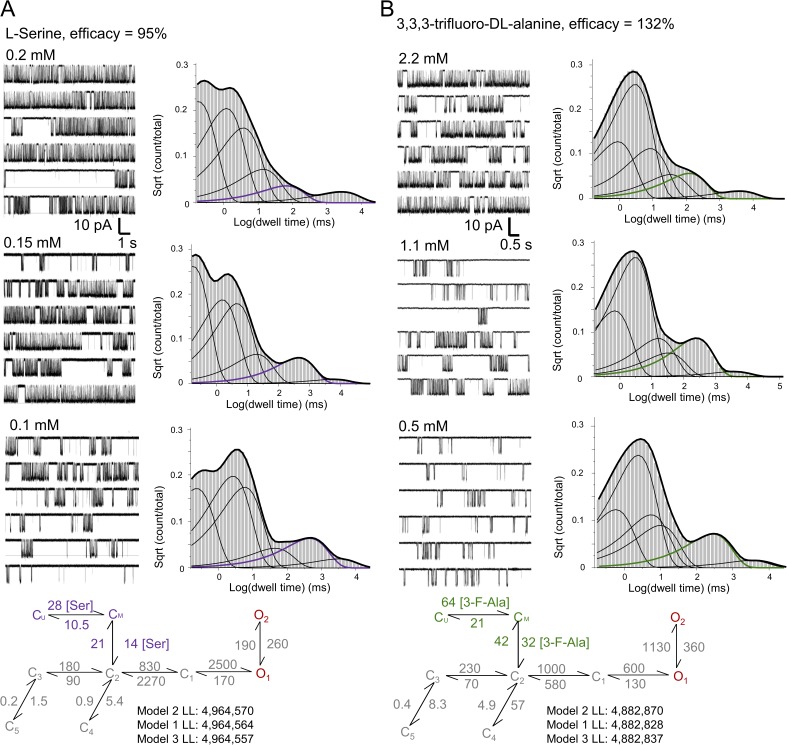
The glycine-binding site resides between two flexible lobes in the ligand-binding domain of the GluN1 subunit. Crystallographic studies revealed that several glycine-site agonists, partial agonists, and antagonists bind at similar locations, even if in slightly different orientations (Furukawa and Gouaux, 2003; Jespersen et al., 2014). In addition, functional studies showed that, when activated with partial agonists at either the glycine or the glutamate site, GluN1/GluN2A receptors preserve the same basic 3C2D2O mechanism (and modes), with only a set of rate constants changed, which account for differences in efficacy (Kussius et al., 2009; Kussius and Popescu, 2010). Based on this knowledge, we reasoned that glycine-site agonists would dissociate from fully liganded receptors from the same closed state as glycine, but may have different association and/or dissociation kinetics to account for their different affinities. When evaluated with macroscopic equilibrium measurements, l-serine, the stereoisomer of the physiologically relevant d-serine, was found to have a higher EC50 value (200 µM) but only slightly lower efficacy (∼95%) relative to glycine (Chen et al., 2008). Similarly, 3,3,3-trifluoro-dl-alanine had much higher EC50 (2.2 mM), but higher (132%) efficacy. Based on these values, we reasoned that both l-serine and 3,3,3-trifluoro-dl-alanine may dissociate more frequently from actively gating receptors, and thus would provide increased resolution in evaluating the model topology and kinetics of glycine binding.
We recorded on-cell one-channel records with several concentrations of l-serine (Fig. 2 A) or 3,3,3-trifluoro-dl-alanine (Fig. 2 B) (and 1 mM glutamate), and we processed and analyzed these data in a manner identical to that described for glycine. As with glycine, neither the mean open durations nor the distributions of open durations changed with changing agonist concentrations. In addition, the closed component distributions displayed the additional concentration-dependent component, as expected. We used the two new datasets to rank a series of models according to their goodness of fit to individual files. As with glycine, we found that for both agonists tested, Model 2 returned the highest LL values (Fig. 2). For global fits with models that include two glycine-binding steps, LL values were for l-serine: 4,964,564 with Model 1, 4,964,570 with Model 2, and 4,964,557 with Model 3; and for 3,3,3-trifluoro-dl-alanine, 4,882,828 with Model 1, 4,882,870 with Model 2, and 4,882,837 with Model 3. As noted, LL values provide only a first-pass measure for model selection; therefore, additional tests will be necessary to validate this arrangement.
Figure 2.
NMDA activity in subsaturating concentrations of (A) l-serine (B) and 3,3,3-trifluoro-dl-alanine. Each panel illustrates representative one-channel traces for the indicated agonist concentrations, closed-interval distributions calculated from one file, and the best-fitting models, with associated LL values. The concentration-dependent closed component is highlighted in color.
The topology of Model 2 implies that receptor conformations aggregated in the kinetic state C3 have low affinity for glutamate and high affinity for glycine and that those in state C2 have high affinity for glutamate and low affinity for glycine, whereas all other conformations (in states C1, O, D1, and D2) have high affinity for both glutamate and glycine. Therefore, Model 2 postulates that a conformation change of measurable kinetics (C3↔C2) intervenes between the dissociation steps for glutamate and glycine, and places the dissociation transitions for these agonists at different kinetic distances from the open states. Therefore, this model predicts that changes in glutamate and glycine concentrations will have discrete effects on the relaxation of macroscopic NMDA receptor responses. These differential predictions represent an opportunity to test the model by comparing traces predicted by the three models with responses we can measure experimentally.
Macroscopic testing of the glycine activation schemes deduced from one-channel data
If a scheme with glycine dissociating from C2 represents a reality with higher probability than the alternatives considered, then this scheme, even if deduced from steady-state data, should also account better for all other glycine-dependent behaviors, including dose dependence, rise and deactivation time course, and desensitization kinetics. We proceeded to test the microscopic Model 2 against these macroscopic standards.
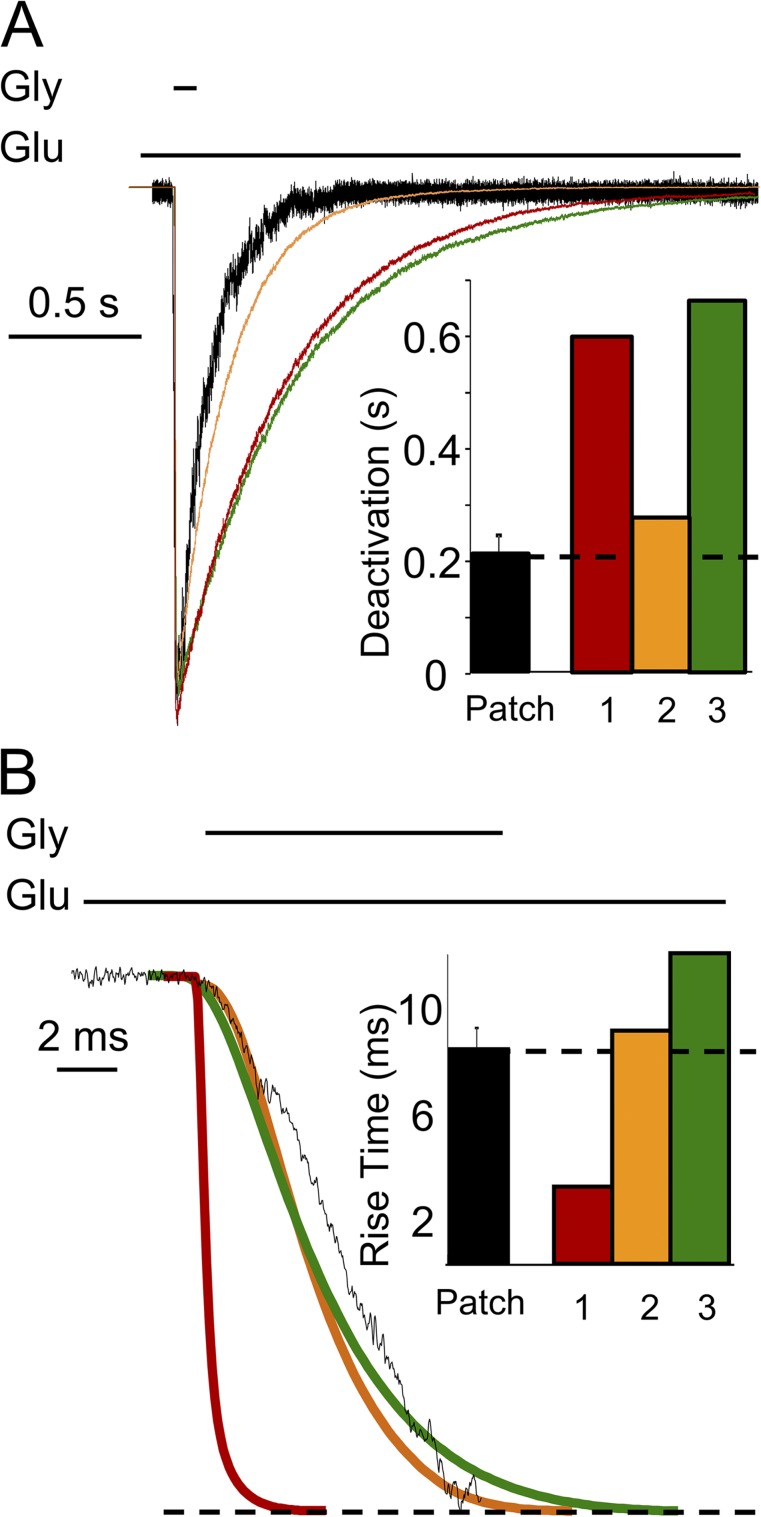
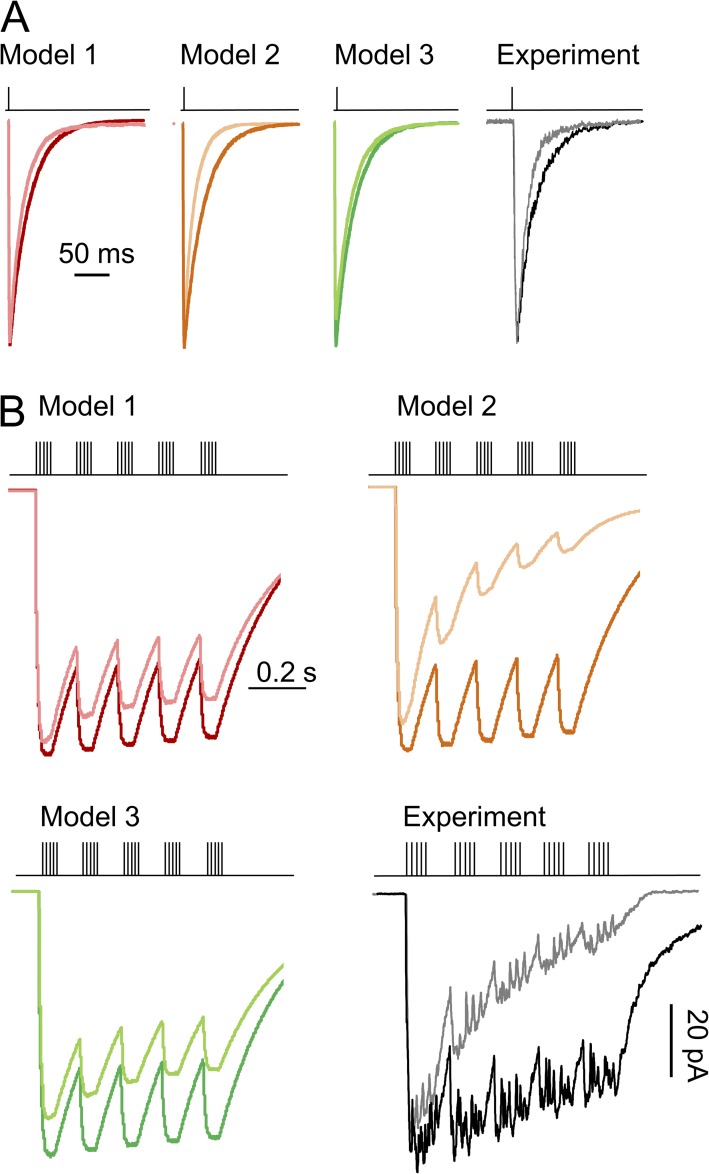
We first examined the rise and deactivation time course predicted by the three models. We simulated macroscopic currents (100 channels, 10 pA/each) with each model, elicited with a brief (10-ms) pulse of glycine (0.1 mM). For the predicted traces, we measured rise and deactivation time courses to compare these to experimentally measured values (Fig. 3). Results show that Models 1 and 3 predicted similarly slow deactivation (0.61 and 0.66 s, respectively), and Model 2 predicted much faster deactivation (0.28 s) (Fig. 3 A). Further, Models 3 and 1 produced responses with the slowest (12.1-ms) and fastest (2.3-ms) rise time, respectively, whereas Model 2 predicted an intermediate rise time (8.6 ms) (Fig. 3 B). Using a piezo-driven perfusion system, we recorded responses from excised outside-out patches exposed briefly to glycine (10 ms; 0.1 mM), with 1 mM glutamate continuously present. For these experimental currents (n = 4), the rise time was 8.1 ± 0.8 ms and the deactivation time was 0.21 ± 0.03 s. These results also support Model 2 as the best representation of the true NMDA receptor activation mechanism.
Figure 3.
Macroscopic response to brief glycine exposure. Currents were recorded from multichannel outside-out patches exposed to 0.1 mM glycine (10 ms) and with 1 mM glutamate present (black; error bars are ± SEM), or were simulated in similar conditions with Models 1 (red), 2 (orange), and 3 (green) illustrated in Fig. 1. (A) Deactivation and (B) rise times were quantified for experimental and simulated traces.
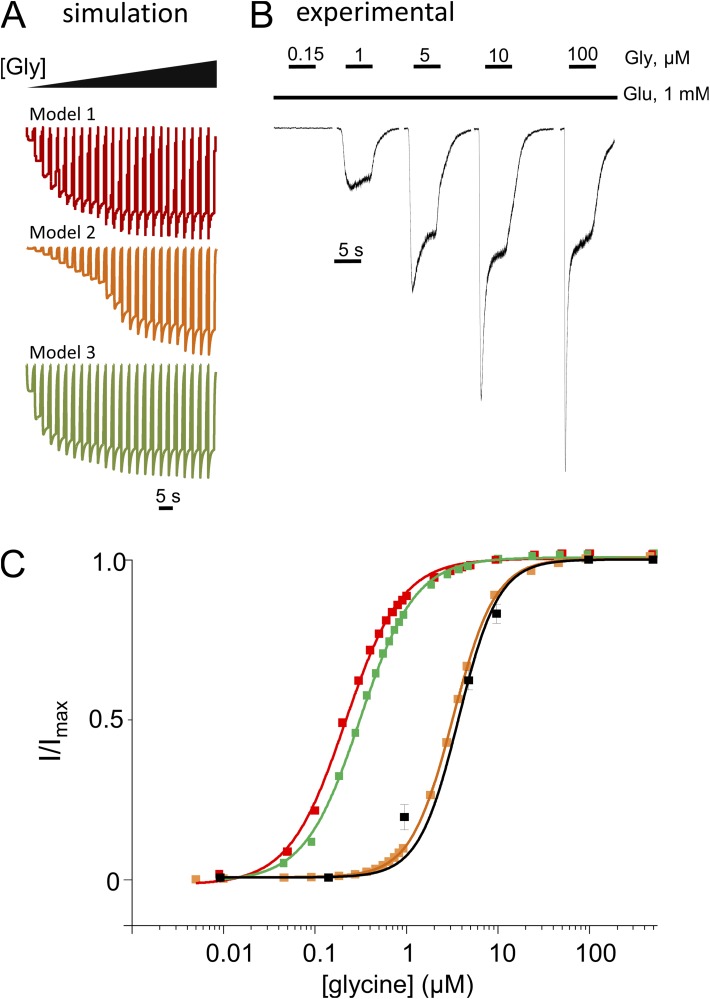
Next, we simulated macroscopic traces in response to several glycine concentrations using the three models (Fig. 4 A), and we recorded whole-cell currents in response to several concentrations of glycine, with 1 mM glutamate continuously present (Fig. 4 B). As noted in previous reports, the microscopic rate constants derived from cell-attached recordings predicted more modest desensitization at equilibrium (larger Iss/Ipk) than was measured from receptors residing in excised patches (Zhang et al., 2008) or whole-cell (Kussius et al., 2009) recordings. This increase in intrinsic desensitization reflects genuine changes in microscopic desensitization rates (C3↔C5 and C2↔C4) and has been attributed to differences in the intracellular milieu of receptors in the three recording configurations (Zorumski et al., 1989; Lester and Jahr, 1992; Lester et al., 1993; Krupp et al., 2002). To overcome this potential obstacle, we constructed dose–response plots using the predicted and measured peak current values, which appear to be less affected by intracellular factors. With this analysis, the predicted EC50 values were: 0.2, 3.6, and 0.3 µM for Models 1, 2, and 3, respectively (Fig. 4 C). The measured glycine EC50 = 3.9 µM was closest to the value predicted by Model 2. Therefore, these results also lend support to Model 2.
Figure 4.
NMDA receptor glycine dose–response. (A) Simulated traces (100 channels, 10 pA/each) with Models 1, 2, and 3 (from Fig. 1), and 5-s pulses of glycine of increasing concentrations (5 nM–500 µM). (B) Recorded whole-cell currents in response to 5-s pulses of glycine (as indicated) and 1 mM Glu. (C) Dose–response curves were calculated by fitting the Hill equation to peak current amplitudes measured experimentally (black) or obtained from simulations (color). EC50 = 3.9 µM, for the experimental traces (error bars are ± SEM) and 0.2, 3.6, and 0.3 µM, for Models 1 (red), 2 (orange), and 3 (green), respectively.
Glycine dependence of macroscopic current desensitization
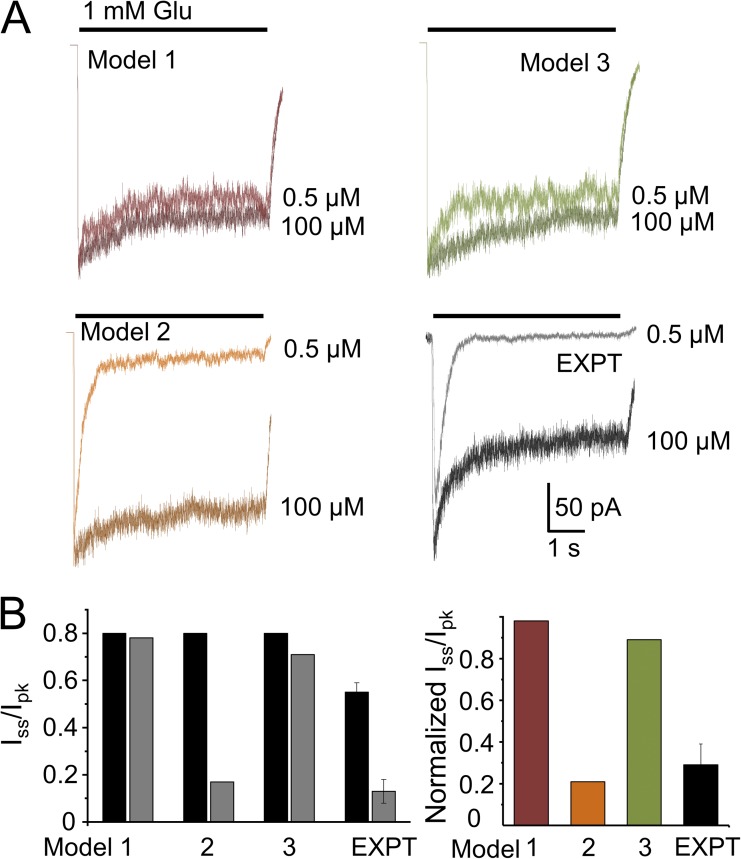
Next, we simulated macroscopic currents after long (5-s) applications of glutamate (1 mM) in the continuous presence of high (100 µM) or low (0.5 µM) glycine concentrations and examined the extent (Iss/Ipk) and time course of macroscopic desensitization (Fig. 5 A). Models 1 and 3 predicted currents with similar shapes regardless of glycine concentration: Iss/Ipk, 0.8 versus 0.78 and 0.79 versus 0.73 for Models 1 and 3, respectively (Fig. 5 B). However, Model 2, which also predicted a modest desensitization level in high glycine (Iss/Ipk, 0.8), produced current traces that desensitized more (Iss/Ipk = 0.17) and faster in low glycine. As illustrated in Fig. 5 A, and noted above, experimentally recorded whole-cell currents elicited by glutamate in high glycine desensitize deeper (smaller Iss/Ipk ratio) than any of our single-channel–derived models predict. However, when comparing the effects of glycine on the current desensitization level, only Model 2 predicted the dramatic change observed experimentally. Remarkably, this result shows that a kinetic model developed purely on statistical analyses of stationary single-channel data, and which does not assume a glutamate-dependent change in glycine affinity, reproduced the well-documented “glycine-dependent desensitization” of macroscopic NMDA receptor responses (Fig. 5). This result represents strong evidence for our hypothesis that Model 2 most closely approximates the channel activation mechanism, and brings into question the hypothesis of negative cooperativity between the glycine- and glutamate-binding sites in NMDA receptors.
Figure 5.
Glycine-dependent desensitization. (A) Whole-cell currents were elicited with long glutamate pulses (5 s, 1 mM) in the continuous presence of two glycine concentrations, as indicated (black and gray) and compared with traces simulated from Models 1, 2, and 3 (dark and light). (B) Summary data for extent of desensitization (Iss/Ipk) and the glycine dose dependence of desensitization extent (Iss/Ipk in low glycine relative to Iss/Ipk in high glycine). Error bars are ± SEM.
Macroscopic testing of the l-serine activation schemes deduced from one-channel data
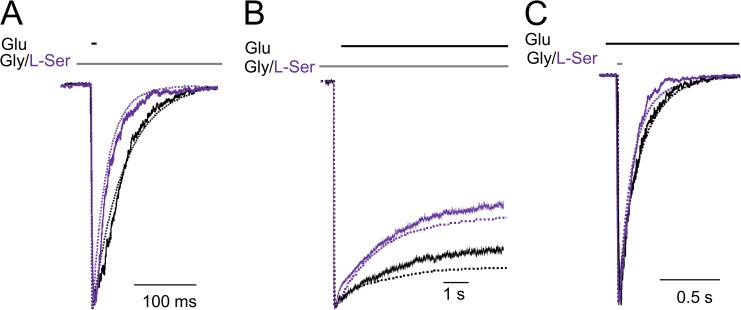
Having derived an activation mechanism for l-serine, we took the opportunity to further test Model 2 for its ability to reproduce macroscopic currents with this coagonist. We decided to measure three characteristic macroscopic parameters: deactivation kinetics upon removal of glutamate in the presence of glycine or l-serine (Fig. 6 A), desensitization kinetics upon glutamate application in a background of glycine or l-serine (Fig. 6 B), and deactivation kinetics upon removal of the glycine-site agonist (glycine or l-serine) (Fig. 6 C). First, we found that, as for glycine (solid black traces), currents recorded with l-serine (solid purple traces) matched well simulations (dotted traces) with the rates and topology illustrated for Model 2 in Fig. 2 A. Currents elicited with brief pulses of 1 mM glutamate (10 ms) deactivated faster in the presence of l-serine (τdeact = 51 ± 4 ms) relative to those elicited in the presence of glycine (τdeact = 68 ± 5 ms). This differs from previous reports that found similar deactivation time courses for NMDA receptor–mediated excitatory postsynaptic currents with ambient glycine or l-serine (Lester et al., 1993). This discrepancy is likely caused by differences in the agonist concentrations, recording configurations, and in receptor subunit composition used in the two experiments. Second, we found that currents elicited with long pulses of 1 mM glutamate (7 s) desensitized deeper and faster with l-serine (Iss/Ipk = 0.5 ± 0.03; τD = 1.6 ± 0.2 s) relative to those elicited in the presence of glycine (Iss/Ipk = 0.65 ± 0.04; τD = 2.2 ± 0.1 s) (Fig. 3 B). Last, we found that deactivation, upon removal of l-serine, was faster (τdeact = 84 ± 10 ms) than upon removal of glycine (τdeact = 209 ± 8 ms), similar to previous findings in response to d,l-homoserine and glycine (Benveniste et al., 1990). Importantly, the faster deactivation was not caused by faster dissociation rates as previously hypothesized, but rather because in the presence of l-serine mean open durations were shorter (5.1 ± 0.4 ms). In all six cases illustrated in Fig. 6, traces simulated with Model 2 (dotted lines) superimposed well with experimental traces (solid lines).
Figure 6.
Macroscopic NMDA receptors produced with glutamate and glycine or l-serine. The stimulus was: (A) 10 ms glutamate, (B) 7 s glutamate, and (C) 1 ms glycine or l-serine. Experimental (solid lines) and simulated traces (dotted lines) are overlaid; currents were recorded from outside-out patches (A and C) and whole cells (B). Simulations were done with Model 2 illustrated in Fig. 1 B for 0.1 mM glycine (black) and in Fig. 2 A for 2.1 mM l-serine (purple).
Collectively, these results offer strong support for the hypothesis that unlike glutamate, which dissociates primarily from C3, glycine and other glycine-site agonists dissociate with highest probability from the kinetic state C2. Our model predicts that, when activated with long pulses of glutamate in low concentrations of glycine, glycine dissociation will trap receptors in glutamate-bound closed states that are functionally but not structurally similar to desensitized states, thus explaining the observed “glycine-dependent” decay of the macroscopic current. Importantly, this effect occurs without invoking binding cooperativity between glycine and glutamate and in the absence of changes in microscopic desensitization.
Implications for in situ NMDA receptor signals
With a new model in hand, we began to explore its predictions for responses to physiological patterns of synaptic stimulation. For this, we asked whether and how the glutamate-elicited response would be affected by changing levels of ambient glycine, and whether the model we propose confers unique features on the response time course. For this, we used the three models in Fig. 1 to simulate currents from receptors (100 receptors, 10 pA/each) that were pre-equilibrated with low (0.5 µM) or high (100 µM) glycine and then exposed these to glutamate (1 mM) (Fig. 7) under two experimental paradigms: low and high frequency stimulation patterns.
Figure 7.
NMDA receptor responses in low and high glycine concentrations. Response to brief (1-ms) application of 1 mM glutamate (A) and responses to theta-like bursts (5 × 1-ms pulses at 100 Hz, repeated at 10 Hz) (B) were simulated with the three models in Fig. 1 B equilibrated in low (0.5 µM; lighter color) or high (0.1 mM; darker color) glycine concentration; representative currents recorded experimentally from outside-out patches in low (gray) and high (black) glycine.
With low frequency synaptic-like glutamate applications (1-ms pulse of 1 mM glutamate), when using Models 1 and 3, predictions were similar for both peak current amplitude and decay kinetics between low (0.5 µM) and high (100 µM) glycine conditions: Model 1, 75 versus 77 ms; Model 3, 82 versus 89 ms (Fig. 7 A). However, currents simulated with Model 2 decayed appreciably faster in low versus high glycine (60 vs. 73 ms). To test this prediction, we measured responses from excised patches with a similar (low vs. high glycine) protocol and found that deactivation times of recorded currents were in strong agreement with those predicted by Model 2: 58 ± 7 versus 71 ± 5 ms. Therefore, Model 2 recapitulates the strong effect of ambient glycine concentration on current deactivation, as reported previously (Mayer et al., 1989); importantly, this effect was not predicated on changing desensitization or binding rates; rather, the model explains it as a natural consequence of changing the ratio between the glycine concentration–dependent association rate and its concentration-independent dissociation rate.
With high frequency theta-like glutamate stimuli (five pulses of 1-ms duration applied at 100 Hz, repeated at 10 Hz), all three models predicted a reduction in total charge transfer upon lowering glycine concentration (Fig. 7 B), and Model 2 indicated the strongest sensitivity to fluctuations in glycine concentration: Model 1, 9%; Model 2, 42%; Model 3, 15%. We measured experimentally current traces in response to a theta burst–like application of glutamate onto outside-out patches in the continuous presence of 0.5 or 100 µM glycine and found that the response amplitude was strongly dependent on glycine concentration, with 41 ± 6% (n = 3) reduction in charge transfer upon lowering glycine concentration, similar to traces simulated with Model 2. Importantly, for the conditions used here, we found that increasing glycine concentration above 0.5 µM boosted NMDA receptor currents up to approximately twofold, which represents a substantial range of potentiation. Therefore, these observations strongly support a role of glycine concentrations in modulating NMDA receptor responses to tonic as well as high frequency stimulation.
Because Model 2, but not Models 1 and 3, produced responses that were clearly distinct in low versus high glycine conditions, with both high and low frequency stimulation patterns, we suggest that the precise arrangement of glutamate and glycine dissociation steps along the NMDA receptor reaction mechanism may have direct implications for the physiology of synaptic responses. The kinetics of NMDA receptor deactivation sets the window over which signals are declared coincident across a dendritic tree and, thus, has important consequences for synaptic integration and plasticity. In addition, given their high Ca2+ permeability, the total charge transferred by NMDA receptors during high frequency or tonic stimulation has critical consequences for cellular physiology, as it sets a threshold between plasticity and cellular death. The model we propose may represent a valuable instrument to explore how dynamic fluctuations in the ambient glycine level control the NMDA receptor deactivation kinetics and its capacity for charge transfer.
DISCUSSION
We used kinetic analyses and modeling of single-molecule currents and developed a new kinetic model for NMDA receptor activation that includes microscopic glycine association/dissociation steps. This model postulates that during receptor deactivation, glycine and glutamate dissociate most likely from closed receptors that differ in their stability and in their position along the deactivation sequence. The model accounts for a wide range of microscopic and macroscopic behaviors of NMDA receptors, and therefore it can be used to predict receptor responses under a variety of stimulation protocols that mimic physiological and pathological conditions.
Glutamate and glycine are obligatory coagonists of NMDA receptors, and they control receptor activity in a concentration-dependent manner. At excitatory synapses in brain and spinal cord, glutamate is released in brief transient pulses from presynaptic terminals, whereas glycine originates from several sources, predominantly from glia (Harsing and Matyus, 2013). The activity of glycine transporters sets the resting levels of ambient glycine. Synaptic concentrations of glycine are difficult to ascertain; microdialysis methods indicate a range from nanomolar to low micromolar (Semba and Patsalos, 1993; Vandenberg and Aubrey, 2001; Papp et al., 2008) and may, therefore, be below those necessary to saturate NMDA receptors, as determined in this work and previously (Bergeron et al., 1998; Chen et al., 2008). However, synaptic activity can increase extracellular glycine concentrations close to 1 mM (Vandenberg and Aubrey, 2001). Notably, glycine homeostasis is perturbed in several neurological disorders, including schizophrenia and autism, and modulating the endogenous concentration of glycine-site agonists ameliorates symptoms, and thus, is considered a viable therapeutic approach (Javitt et al., 1994; Coyle and Tsai, 2004; Heresco-Levy and Javitt, 2004; Hons et al., 2010; Ghanizadeh, 2011). Therefore, how ambient glycine levels control NMDA receptor responses under a variety of stimulation patterns is highly relevant to both physiological and pathological situations.
High resolution crystal structures illustrate the atomic details of glycine and glutamate binding on the GluN1 and GluN2B subunits (Karakas and Furukawa, 2014; Lee et al., 2014). However, similarly detailed kinetic information is still lacking, as is the knowledge of how to ascertain the functional consequences of glycine-site agonists on the NMDA receptor signals. We report here the first kinetic model for NMDA receptor activation that includes microscopic glycine-binding steps. It accounts for a series of microscopic and macroscopic behaviors of NMDA receptors and predicts substantial changes in the synaptic NMDA receptor responses by fluctuations of glycine concentrations in the physiological range.
A distinctive feature of this new model is its postulate that, during active gating of fully liganded receptors, which is described with a linear sequence of closed and open states, glutamate and glycine dissociate with maximum probability from separate kinetic states: glutamate from C3 and glycine from C2. The idea that glutamate dissociates most likely from C3 is largely predicated on three pieces of previously reported evidence. First, a model where glutamate dissociates from C3 best represents statistically the pattern of single-channel traces obtained in low concentrations of glutamate (Popescu et al., 2004); second, only models with at least two slow steps (C3↔C2 and C2↔C1) interposed between glutamate binding and channel opening predict the observed slow rise (∼10 ms) of the macroscopic current (Auerbach and Zhou, 2005); and last, only this model predicts that the peak amplitude of the macroscopic response is dependent on stimulation frequency, an unexpected property that was subsequently verified experimentally (Popescu et al., 2004). In the work we describe here, we present three lines of evidence that glycine dissociates most likely from C2. First, probability density functions calculated with Model 2 enveloped with the highest probability the single-channel data obtained in low concentrations of glycine (Fig. 1 B); second, the same model returned the highest fitting scores for two additional sets of single-channel data obtained with low concentrations of the glycine-site agonists l-serine or 3,3,3-trifluoro-dl-alanine (Fig. 2, A and B); and last, Model 2, but not Models 1 or 3, predicted accurately several glycine-dependent macroscopic behaviors such as deactivation after brief (10-ms) pulses of 100 µM glycine (Fig. 3), glycine dose dependence of peak currents (Fig. 4), and glycine concentration dependence of the macroscopic current desensitization during long glutamate exposure (Fig. 5).
Notably, the model predicts correctly glycine-dependent desensitization without assuming a decrease in glycine affinity upon binding glutamate (Figs. 5 and 1 B), as previously postulated by the negative cooperativity hypothesis. Instead, the model developed and validated here incorporates high and low affinity states for both glutamate and glycine, as in previous work (Benveniste et al., 1990), but proposes that the low affinity states for each agonist, those from which the agonist dissociates with highest probability, are kinetically and topologically distinct for glutamate (C3) and glycine (C2). This arrangement is consistent with the hypothesis that the C3↔C2 and C2↔C1 transitions may represent separate conformational changes that can be traced to the activation of the GluN2 and GluN1 subunits, respectively (Banke and Traynelis, 2003; Murthy et al., 2012). Such an arrangement is only functionally discernible and thus, physiologically relevant, for receptors with complex kinetic mechanisms, where several closed states with separable stabilities precede channel opening. Similar kinetic resolution has not been achieved for other neurotransmitter receptors, perhaps because of their much faster kinetics. Therefore, the slow kinetics of NMDA receptors may be an adaptation that makes possible the modulation of its physiologically relevant parameters by endogenous modulators, including fluctuating concentrations of ambient glycine. This may explain why, of all neurotransmitter receptors, NMDA receptors alone require for activation a coagonist, an issue that has not yet been addressed.
Initially, kinetic models of NMDA receptor activation assumed a simple two-state mechanism in which the entire gating reaction was represented by a global C↔O conformational change. Because the NMDA receptor mean open durations are independent of glycine concentration, glycine dissociation occurs most likely from closed states. To explain how macroscopic NMDA receptor response depended on glycine concentration, Benveniste et al. (1990) postulated a concerted change in receptor conformation upon binding glutamate that would result in a lower affinity for glycine, but did not explicitly incorporate this conformational change in their model, and appended both the glutamate and glycine dissociation transitions to the same aggregated preopen state. The assumption that glutamate and glycine dissociate from the same preopen state has been perpetuated without evidence to subsequent multistate gating models, which were necessarily developed to explain the multiplicity of states observed in single-channel records (Benveniste et al., 1990; Nahum-Levy et al., 2001; Schorge et al., 2005). As in other previously reported multistate models, the new mechanism we developed here also incorporates a series of fully liganded preopen states, which represent receptor isomerizations that precede channel opening (C3↔C2↔C1); however, it is unique in that it proposes that glutamate and glycine dissociate with highest probability from distinct kinetic states, C3 and C2, respectively. This more granular description of the glutamate and glycine activation reactions will help in investigating and better understanding the structural changes that underlie NMDA receptor currents, and may be instrumental in understanding the physiological and pathological consequences of glycine-dependent modulation of NMDA receptor responses.
The model we propose here recapitulates well macroscopic behaviors observed in experimental recordings; however, some discrepancy exists in the level of desensitization predicted from rates measured in cell-attached and those measured in whole-cell and excised-patch preparations, as reported previously (Zhang et al., 2008). We attribute this to real differences in receptor kinetics in intact versus ruptured cells, which may reflect Ca2+-dependent changes in desensitization kinetics (Medina et al., 1995; Ehlers et al., 1996).
Structural models of NMDA receptors illustrate extensive contacts between the ligand-binding domains on GluN1 and GluN2 subunits, and it was speculated that these may represent the basis of negative cooperativity, understood strictly as a decrease in affinity for one agonist upon coagonist binding; our results do not exclude allosteric communication between ligand-binding domains as a result of binding glutamate and/or glycine. We also note that the glycine association and dissociation rate constants we estimated here are only valid for glutamate-bound receptors. Glycine cannot elicit currents from glutamate-free receptors and, thus, the dissociation constant from glutamate-free receptors is not accessible with our approach. The rate constants we estimate for glycine and for the glycine-site agonists l-serine and 3,3,3-trifluoro-dl-alanine are much slower than diffusion constants. Therefore, these values most likely incorporate movements in the ligand-binding domain that accompany agonist binding and dissociation (Abele et al., 2000; Cheng et al., 2005). In all likelihood, what we call here microscopic rate constants can be further separated into the nanoscopic steps of docking, cleft-closing, and cleft-locking (Sasmal and Lu, 2014).
The model we present here is an advancement over previous kinetic models of the NMDA receptor activation sequence. Early models approximated the gating process with a two-state C↔O reaction, which could be accessed either from resting conformations by binding glutamate or from desensitized conformations by resensitization transitions (Lester and Jahr, 1992). This model reproduced several fundamental features of the macroscopic NMDA receptor response, but it did not consider explicitly glycine binding. Since then, several reports expanded this basic model by including glycine association and dissociation steps (Benveniste and Mayer, 1991; Clements and Westbrook, 1991; Lester et al., 1993; Nahum-Levy et al., 2001). A limitation of these studies is that they relied exclusively on macroscopic measurements, which aggregate gating, binding, and desensitization events and thus lack the necessary detail to resolve microscopic events. More recently, single-channel analyses were leveraged to produce a more detailed model, which includes a multistate gating sequence as well as microscopic steps for glutamate and glycine binding (Schorge et al., 2005). However, in this work, glutamate and glycine were assumed to dissociate from the same closed kinetic state without attempts to validate this arrangement.
The advance we present here was made possible by several precautions that allowed us to focus on the glycine-binding reaction. First, we observed activations from the same channel continuously over long periods. With this approach, the long closed periods between channel activations can be assigned with reasonable confidence to unliganded or desensitized receptors, thus reducing errors from cluster selection. Second, we examined records obtained with glycine-site agonists that differed in their binding kinetics to increase the resolution of the modeling results at this kinetic step. Last, the channels we observed were free of additional modulatory perturbations; the extracellular milieu was virtually free of inhibitory or blocking external cations (H+, Mg2+, Zn2+, Ca2+) and resided in intact membranes, such that the intracellular interactions remained undisturbed.
The model we propose here predicts synaptic responses that are clearly distinct in low versus high glycine conditions and indicates that the precise arrangement of glutamate and glycine dissociation steps along the NMDA receptor activation reaction may have direct implications for synaptic physiology. The kinetics of NMDA receptor deactivation sets the window over which signals are perceived coincident, and thus, has important consequences for synaptic integration and plasticity. In addition, given their high Ca2+ permeability, the total charge transferred by NMDA receptors during tonic or high frequency stimulation has critical consequences for cellular physiology, as it sets a threshold between plasticity and cellular death. Therefore, the more granular model we propose here may represent a valuable instrument to further explore how dynamic fluctuations in the ambient glycine level control the NMDA receptor deactivation kinetics and its capacity for charge transfer during repetitive firing.
Acknowledgments
We thank E.M. Kasperek for technical assistance with tissue culture.
Funding was from the National Institutes of Health (RO1NS052669 to G.K. Popescu and F31NS086765 to K.A. Cummings) and American Heart Association (EIA9100012 to G.K. Popescu).
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- CNS
- central nervous system
- LL
- log likelihood
- MIL
- maximum interval likelihood
- SKM
- segmental k-means
References
- Abele R., Keinanen K., and Madden D.R.. 2000. Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J. Biol. Chem. 275:21355–21363. 10.1074/jbc.M909883199 [DOI] [PubMed] [Google Scholar]
- Amico-Ruvio S.A., and Popescu G.K.. 2010. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys. J. 98:1160–1169. 10.1016/j.bpj.2009.12.4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A., and Zhou Y.. 2005. Gating reaction mechanisms for NMDA receptor channels. J. Neurosci. 25:7914–7923. 10.1523/JNEUROSCI.1471-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banke T.G., and Traynelis S.F.. 2003. Activation of NR1/NR2B NMDA receptors. Nat. Neurosci. 6:144–152. 10.1038/nn1000 [DOI] [PubMed] [Google Scholar]
- Benveniste M., and Mayer M.L.. 1991. Kinetic analysis of antagonist action at N-methyl-d-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys. J. 59:560–573. 10.1016/S0006-3495(91)82272-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M., Clements J., Vyklický L. Jr, and Mayer M.L.. 1990. A kinetic analysis of the modulation of N-methyl-d-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J. Physiol. 428:333–357. 10.1113/jphysiol.1990.sp018215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron R., Meyer T.M., Coyle J.T., and Greene R.W.. 1998. Modulation of N-methyl-d-aspartate receptor function by glycine transport. Proc. Natl. Acad. Sci. USA. 95:15730–15734. 10.1073/pnas.95.26.15730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel W.F., Murthy S.E., Kasperek E.M., and Popescu G.K.. 2011. NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat. Commun. 2:498 10.1038/ncomms1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., and Okayama H.. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.E., Geballe M.T., Katz E., Erreger K., Livesey M.R., O’Toole K.K., Le P., Lee C.J., Snyder J.P., Traynelis S.F., and Wyllie D.J.. 2008. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J. Physiol. 586:227–245. 10.1113/jphysiol.2007.143172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Du M., Ramanoudjame G., and Jayaraman V.. 2005. Evolution of glutamate interactions during binding to a glutamate receptor. Nat. Chem. Biol. 1:329–332. 10.1038/nchembio738 [DOI] [PubMed] [Google Scholar]
- Clements J.D., and Westbrook G.L.. 1991. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-d-aspartate receptor. Neuron. 7:605–613. 10.1016/0896-6273(91)90373-8 [DOI] [PubMed] [Google Scholar]
- Coyle J.T., and Tsai G.. 2004. The NMDA receptor glycine modulatory site: a therapeutic target for improving cognition and reducing negative symptoms in schizophrenia. Psychopharmacology (Berl.). 174:32–38. 10.1007/s00213-003-1709-2 [DOI] [PubMed] [Google Scholar]
- Curtis D.R., Hösli L., and Johnston G.A.. 1967. Inhibition of spinal neurons by glycine. Nature. 215:1502–1503. 10.1038/2151502a0 [DOI] [PubMed] [Google Scholar]
- Ehlers M.D., Zhang S., Bernhadt J.P., and Huganir R.L.. 1996. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 84:745–755. 10.1016/S0092-8674(00)81052-1 [DOI] [PubMed] [Google Scholar]
- Furukawa H., and Gouaux E.. 2003. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 22:2873–2885. 10.1093/emboj/cdg303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H., Singh S.K., Mancusso R., and Gouaux E.. 2005. Subunit arrangement and function in NMDA receptors. Nature. 438:185–192. 10.1038/nature04089 [DOI] [PubMed] [Google Scholar]
- Ghanizadeh A. 2011. Targeting of glycine site on NMDA receptor as a possible new strategy for autism treatment. Neurochem. Res. 36:922–923. 10.1007/s11064-010-0381-2 [DOI] [PubMed] [Google Scholar]
- Hansen K.B., Ogden K.K., and Traynelis S.F.. 2012. Subunit-selective allosteric inhibition of glycine binding to NMDA receptors. J. Neurosci. 32:6197–6208. 10.1523/JNEUROSCI.5757-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsing L.G. Jr, and Matyus P.. 2013. Mechanisms of glycine release, which build up synaptic and extrasynaptic glycine levels: The role of synaptic and non-synaptic glycine transporters. Brain Res. Bull. 93:110–119. 10.1016/j.brainresbull.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Heresco-Levy U., and Javitt D.C.. 2004. Comparative effects of glycine and d-cycloserine on persistent negative symptoms in schizophrenia: a retrospective analysis. Schizophr. Res. 66:89–96. 10.1016/S0920-9964(03)00129-4 [DOI] [PubMed] [Google Scholar]
- Hirai H., Kirsch J., Laube B., Betz H., and Kuhse J.. 1996. The glycine binding site of the N-methyl-d-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3-M4 loop region. Proc. Natl. Acad. Sci. USA. 93:6031–6036. 10.1073/pnas.93.12.6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hons J., Zirko R., Ulrychova M., Cermakova E., Doubek P., and Libiger J.. 2010. Glycine serum level in schizophrenia: Relation to negative symptoms. Psychiatry Res. 176:103–108. 10.1016/j.psychres.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Horn R. 1987. Statistical methods for model discrimination. Applications to gating kinetics and permeation of the acetylcholine receptor channel. Biophys. J. 51:255–263. 10.1016/S0006-3495(87)83331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt D.C., Zylberman I., Zukin S.R., Heresco-Levy U., and Lindenmayer J.P.. 1994. Amelioration of negative symptoms in schizophrenia by glycine. Am. J. Psychiatry. 151:1234–1236. 10.1176/ajp.151.8.1234 [DOI] [PubMed] [Google Scholar]
- Jespersen A., Tajima N., Fernandez-Cuervo G., Garnier-Amblard E.C., and Furukawa H.. 2014. Structural insights into competitive antagonism in NMDA receptors. Neuron. 81:366–378. 10.1016/j.neuron.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.W., and Ascher P.. 1987. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 325:529–531. 10.1038/325529a0 [DOI] [PubMed] [Google Scholar]
- Johnson J.W., and Ascher P.. 1992. Equilibrium and kinetic study of glycine action on the N-methyl-d-aspartate receptor in cultured mouse brain neurons. J. Physiol. 455:339–365. 10.1113/jphysiol.1992.sp019305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E., and Furukawa H.. 2014. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 344:992–997. 10.1126/science.1251915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R., Gan Q., Talukder I., Markowitz M., Salussolia C.L., and Wollmuth L.P.. 2013. Asynchronous movements prior to pore opening in NMDA receptors. J. Neurosci. 33:12052–12066. 10.1523/JNEUROSCI.5780-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R., Dai J., Sweeney C., Zhou H.-X., and Wollmuth L.P.. 2014. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 17:914–922. 10.1038/nn.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N.W., and Dingledine R.. 1988. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 241:835–837. 10.1126/science.2841759 [DOI] [PubMed] [Google Scholar]
- Krupp J.J., Vissel B., Thomas C.G., Heinemann S.F., and Westbrook G.L.. 2002. Calcineurin acts via the C-terminus of NR2A to modulate desensitization of NMDA receptors. Neuropharmacology. 42:593–602. 10.1016/S0028-3908(02)00031-X [DOI] [PubMed] [Google Scholar]
- Kussius C.L., and Popescu G.K.. 2010. NMDA receptors with locked glutamate-binding clefts open with high efficacy. J. Neurosci. 30:12474–12479. 10.1523/JNEUROSCI.3337-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius C.L., Kaur N., and Popescu G.K.. 2009. Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J. Neurosci. 29:6819–6827. 10.1523/JNEUROSCI.0281-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube B., Kuhse J., and Betz H.. 1998. Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 18:2954–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Lü W., Michel J.C., Goehring A., Du J., Song X., and Gouaux E.. 2014. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 511:191–197. 10.1038/nature13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J., Zukin R.S., and Bennett M.V.. 1990. Glycine decreases desensitization of N-methyl-d-aspartate (NMDA) receptors expressed in Xenopus oocytes and is required for NMDA responses. Proc. Natl. Acad. Sci. USA. 87:2354–2358. 10.1073/pnas.87.6.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R.A., and Jahr C.E.. 1992. NMDA channel behavior depends on agonist affinity. J. Neurosci. 12:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R.A., Tong G., and Jahr C.E.. 1993. Interactions between the glycine and glutamate binding sites of the NMDA receptor. J. Neurosci. 13:1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki B.A., Cummings K.A., Paganelli M.A., Murthy S.E., and Popescu G.K.. 2014. One-channel cell-attached patch-clamp recording. J. Vis. Exp. 88:e51629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L., Vyklicky L. Jr, and Clements J.. 1989. Regulation of NMDA receptor desensitization in mouse hippocampal neurons by glycine. Nature. 338:425–427. 10.1038/338425a0 [DOI] [PubMed] [Google Scholar]
- Medina I., Filippova N., Charton G., Rougeole S., Ben-Ari Y., Khrestchatisky M., and Bregestovski P.. 1995. Calcium-dependent inactivation of heteromeric NMDA receptor-channels expressed in human embryonic kidney cells. J. Physiol. 482:567–573. 10.1113/jphysiol.1995.sp020540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S.E., Shogan T., Page J.C., Kasperek E.M., and Popescu G.K.. 2012. Probing the activation sequence of NMDA receptors with lurcher mutations. J. Gen. Physiol. 140:267–277. 10.1085/jgp.201210786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahum-Levy R., Lipinski D., Shavit S., and Benveniste M.. 2001. Desensitization of NMDA receptor channels is modulated by glutamate agonists. Biophys. J. 80:2152–2166. 10.1016/S0006-3495(01)76188-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahum-Levy R., Tam E., Shavit S., and Benveniste M.. 2002. Glutamate but not glycine agonist affinity for NMDA receptors is influenced by small cations. J. Neurosci. 22:2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P., Bellone C., and Zhou Q.. 2013. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14:383–400. 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- Papp A., Juranyi Z., Nagymajtenyi L., Matyus P., and Harsing L.G. Jr. 2008. The synaptic and nonsynaptic glycine transporter type-1 inhibitors Org-24461 and NFPS alter single neuron firing rate in the rat dorsal raphe nucleus. Further evidence for a glutamatergic-serotonergic interaction and its role in antipsychotic action. Neurochem. Int. 52:130–134. 10.1016/j.neuint.2007.06.030 [DOI] [PubMed] [Google Scholar]
- Patneau D.K., and Mayer M.L.. 1990. Structure-activity relationships for amino acid transmitter candidates acting at N-methyl-d-aspartate and quisqualate receptors. J. Neurosci. 10:2385–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G.K. 2012. Modes of glutamate receptor gating. J. Physiol. 590:73–91. 10.1113/jphysiol.2011.223750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G., and Auerbach A.. 2003. Modal gating of NMDA receptors and the shape of their synaptic response. Nat. Neurosci. 6:476–483. [DOI] [PubMed] [Google Scholar]
- Popescu G., and Auerbach A.. 2004. The NMDA receptor gating machine: Lessons from single channels. Neuroscientist. 10:192–198. 10.1177/1073858404263483 [DOI] [PubMed] [Google Scholar]
- Popescu G., Robert A., Howe J.R., and Auerbach A.. 2004. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 430:790–793. 10.1038/nature02775 [DOI] [PubMed] [Google Scholar]
- Qin F. 2004. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys. J. 86:1488–1501. 10.1016/S0006-3495(04)74217-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Auerbach A., and Sachs F.. 1997. Maximum likelihood estimation of aggregated Markov processes. Proc. Biol. Sci. 264:375–383. 10.1098/rspb.1997.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmal D.K., and Lu H.P.. 2014. Single-molecule patch-clamp FRET microscopy studies of NMDA receptor ion channel dynamics in living cells: Revealing the multiple conformational states associated with a channel at its electrical off state. J. Am. Chem. Soc. 136:12998–13005. 10.1021/ja506231j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge S., Elenes S., and Colquhoun D.. 2005. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J. Physiol. 569:395–418. 10.1113/jphysiol.2005.095349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semba J., and Patsalos P.N.. 1993. Milacemide effects on the temporal inter-relationship of amino acids and monoamine metabolites in rat cerebrospinal fluid. Eur. J. Pharmacol. 230:321–326. 10.1016/0014-2999(93)90568-3 [DOI] [PubMed] [Google Scholar]
- Snyder S.H., and Ferris C.D.. 2000. Novel neurotransmitters and their neuropsychiatric relevance. Am. J. Psychiatry. 157:1738–1751. 10.1176/appi.ajp.157.11.1738 [DOI] [PubMed] [Google Scholar]
- Vandenberg R.J., and Aubrey K.R.. 2001. Glycine transport inhibitors as potential antipsychotic drugs. Expert Opin. Ther. Targets. 5:507–518. 10.1517/14728222.5.4.507 [DOI] [PubMed] [Google Scholar]
- Vyklický L. Jr, Benveniste M., and Mayer M.L.. 1990. Modulation of N-methyl-d-aspartic acid receptor desensitization by glycine in mouse cultured hippocampal neurones. J. Physiol. 428:313–331. 10.1113/jphysiol.1990.sp018214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werman R., Davidoff R.A., and Aprison M.H.. 1967. Inhibition of motoneurones by iontophoresis of glycine. Nature. 214:681–683. 10.1038/214681a0 [DOI] [PubMed] [Google Scholar]
- Zafra F., Aragón C., Olivares L., Danbolt N.C., Giménez C., and Storm-Mathisen J.. 1995. Glycine transporters are differentially expressed among CNS cells. J. Neurosci. 15:3952–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Howe J.R., and Popescu G.K.. 2008. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat. Neurosci. 11:1373–1375. 10.1038/nn.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorumski C.F., Yang J., and Fischbach G.D.. 1989. Calcium-dependent, slow desensitization distinguishes different types of glutamate receptors. Cell. Mol. Neurobiol. 9:95–104. 10.1007/BF00711446 [DOI] [PMC free article] [PubMed] [Google Scholar]