Abstract
A positive effect of all-trans retinoic acid (ATRA) on white adipose tissue (WAT) oxidative and thermogenic capacity has been described and linked to an in vivo fat-lowering effect of ATRA in mice. However, little is known about the effects of ATRA on mitochondria in white fat. Our objective has been to characterize the effect of ATRA on mitochondria biogenesis and oxidative phosphorylation (OXPHOS) capacity in mature white adipocytes. Transcriptome analysis, oxygraphy, analysis of mitochondrial DNA (mtDNA), and flow cytometry-based analysis of mitochondria density were performed in mature 3T3-L1 adipocytes after 24 h incubation with ATRA (2 µM) or vehicle. Selected genes linked to mitochondria biogenesis and function and mitochondria immunostaining were analyzed in WAT tissues of ATRA-treated as compared with vehicle-treated mice. ATRA upregulated the expression of a large set of genes linked to mtDNA replication and transcription, mitochondrial biogenesis, and OXPHOS in adipocytes, as indicated by transcriptome analysis. Oxygen consumption rate, mtDNA content, and staining of mitochondria were increased in the ATRA-treated adipocytes. Similar results were obtained in WAT depots of ATRA-treated mice. We conclude that ATRA impacts mitochondria in adipocytes, leading to increased OXPHOS capacity and mitochondrial content in these cells.
Keywords: vitamin A, adipose tissue, browning
Typical white adipocytes are poor in mitochondria and have a low oxidative capacity. Because of this, the contribution of white adipose tissue (WAT) to whole body energy expenditure is considered relatively small. However, there are studies in both humans and rodents documenting a negative association between mitochondrial content in WAT and obesity, as well as examples of nutritional and pharmacological interventions in animals resulting in obesity resistance that associate with increased oxidative capacity in WAT [reviewed in (1)]. Stimulation of mitochondrial biogenesis and oxidative capacity in white adipocytes, when linked to increased energy expenditure in these cells through increased energy uncoupling and/or waste (e.g., futile cycles), emerges therefore as a potential novel target in the control of obesity and its related medical complications (1).
Vitamin A metabolites (i.e., retinoids) modulate the growth and differentiation of a wide range of cells and tissues. Dietary vitamin A and pro-vitamin A are stored as retinyl esters or intracellularly metabolized to retinoic acid (RA), the main active form of vitamin A (2). There are two isoforms of RA, all-trans-RA (ATRA) and 9-cis-RA, which exert their effects on cell processes through both genomic and nongenomic mechanisms (3). After the liver, adipose tissue is a major site of vitamin A storage and metabolism, as well as a main target of ATRA action (4, 5).
Previous studies have shown that in mice treatment with ATRA reduces body weight by decreasing adiposity independently of changes in food intake and improves glucose tolerance and insulin sensitivity in lean and obese animals (6–12). ATRA-induced body fat loss is unaccompanied by increased circulating nonesterified fatty acids (11), suggesting that fatty acids mobilized from fat stores are efficiently oxidized in cells. The latter may include the white adipocytes themselves. In fact, ATRA treatment has been shown to increase the expression of genes related to oxidative metabolism, fatty acid oxidation, and thermogenesis in WAT depots (11, 12), besides activating brown adipose tissue (BAT) (6, 7) and increasing lipid oxidation capacity in additional tissues such as skeletal muscle (12, 13) and the liver (14). Moreover, cell-autonomous effects of ATRA in mature adipocytes reducing intracellular lipids, enhancing fatty acid oxidation, and triggering gene expression changes consistent with increased lipid turnover and oxidation have been described (15).
Whether the above effects are linked to an ATRA-induced increase in mitochondria in white adipocytes has, however, not been addressed. Likewise, a systematic analysis of the impact of ATRA on oxidative phosphorylation (OXPHOS) capacities in mature adipocytes was lacking. The objective of the present work has been to characterize the effect of ATRA on mitochondria biogenesis and function, particularly OXPHOS capacity, in mature white adipocytes.
MATERIALS AND METHODS
Cell culture
3T3-L1 preadipocytes (ATCC, Manassas, VA) were seeded in 3.5 cm diameter dishes at a density of 15 × 104 cells per well. Cells were grown in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 humidified atmosphere, as previously reported (16, 17). To induce differentiation, 2-day postconfluent 3T3-L1 preadipocytes (day 0) were stimulated for 48 h with 0.5 mM isobutylmethylxanthine, 0.25 µM dexamethasone and 1 µg/ml insulin in DMEM supplemented with 10% FBS. Cells were then maintained in DMEM supplemented with 10% FBS and 1 µg/ml insulin. To examine the effect of ATRA on gene expression, 3T3-L1 adipocytes were incubated with 2 µM of this molecule for 24 h, as previously reported (18). To study the impact of retinoid acid receptor (RAR) and PPARδ antagonists, adipocytes were preincubated with RAR antagonist (AGN 193109; 10 µM) or PPARδ antagonist (GSK0660; 10 µM) for 1 h and exposed to 2 µM of ATRA for 24 h. The data are the mean of three independent experiments each performed in triplicate.
Animal experiments
The care and use of mice were in accordance with the guidelines of the University of the Balearic Islands (UIB), and the protocols were submitted to, and approved by, the UIB institutional review board. Twelve adult Naval Medical Research Institute (NMRI) male mice (CRIFFA, Barcelona, Spain) fed ad libitum regular laboratory chow (Panlab, Barcelona, Spain; 73.4% carbohydrate-, 18.7% protein-, and 7.9% lipid-derived energy; 5 IU vitamin A/kcal) and kept at 22°C under 12 h light/12 h dark cycles (lights on at 08:00) were used. Six animals received one daily subcutaneous injection of ATRA at a dose of 50 mg/kg body weight during the 4 days before they were euthanized, as previously described (11, 13, 14). The other six animals were used as controls and injected the vehicle (100 μl olive oil). The animals were euthanized by decapitation at the start of the light cycle. Inguinal, retroperitoneal, and epididymal WAT depots were excised in their entirety, weighed, snap-frozen in liquid nitrogen, and stored at −80°C until analysis. A fragment of each WAT depot was fixed for morphological and immunohistochemical analysis (see below).
RNA isolation and quantitative PCR
Total cellular RNA was extracted from 3T3-L1 cells and mice fat pads using TRIzol reagent according to the manufacturer’s instructions. The cDNA was synthesized from 1 µg of total RNA in 20 µl using random primers and Moloney murine leukemia virus reverse transcriptase. Real-time quantitative RT-PCR analyses for the genes were performed using the Mx3005P Real-Time PCR System (Stratagene, La Jolla, CA) as previously described (19). For each sample, expression was quantified in duplicate, and 18S rRNA was used as the endogenous control in the comparative cycle threshold (CT) method.
Mitochondrial DNA quantification
Total DNA was extracted from cells using DNAzol (Euromedex, France). The content of mitochondrial DNA (mtDNA) was calculated using real-time quantitative PCR by measuring the threshold cycle ratio (ΔCt) of a mitochondrial-encoded gene Cox1 versus a nuclear-encoded gene cyclophilin A.
Hybridization arrays and microarray data analysis
RNA quality control was performed on an Agilent 2100 Bioanalyzer (Massy, France) with 6000 Nano Chips, according to the manufacturer’s instructions and as previously reported (20). RNA from three independent experiments was hybridized to Agilent Whole Mouse Genome arrays (4 × 44k; Massy). All labeling, hybridization, washing, and scanning were performed as described in the manufacturer’s protocol. Arrays were scanned with an Agilent Scanner (Massy). Data were extracted with Agilent Feature Extraction v9.5.3 and analyzed with Agilent GeneSpring GX v11 (Massy). Data were normalized according to the Lowess method, and multiple correction test false discovery rate was applied, then data were filtered on the P value (P < 0.05) for further analyses. Pathway analyses were performed with Metacore (http://www.genego.com/metacore.php), and gene set enrichment analyses (GSEAs) were performed with GSEA software (http://www.broadinstitute.org/gsea) as previously described (21, 22).
Mitochondria labeling
3T3-L1 adipocytes (after 8 days of differentiation, in 12-well plates) treated or not with ATRA (2 µM for 24 h) were incubated with 50 nM MitoTracker® Green FM (Life Technologies) in complete adipose medium (DMEM 4.5 g/l glucose, 17 µM biotin, 33 µM panthonetate, 170 nM insulin) for 30 min at 37°C. Cells were then washed with PBS, trypsinized, and resuspended in 400 µl PBS + 2% FBS. MitoTracker® Green FM fluorescent intensity was measured in 10,000 cells using a C6 Accuri flow cytometer. Subsequently, the scatter signals, fluorescence intensity, and X-mode were analyzed using Kaluza Flow analysis software (Beckman Coulter).
Oxygen consumption
Oxygen consumption of 3T3-L1 adipocytes (after 8 days of differentiation, in 6-well plates) treated or not with ATRA (2 µM for 24 h) was measured by using a Clark-type oxygen electrode (Hansatech Instruments, Cergy St. Christophe, France). Each sample was analyzed by incubating cells (contained in a well of the 6-well plate) with Krebs Ringer solution buffered with HEPES, 0.5% BSA, and digitonin in a magnetically stirred chamber thermostated at 37°C. After the addition of the cells, the chamber was closed, and basal respiration was measured for 20 min. Oxygen consumption (expressed in nmol of O2/ml/min) was normalized to the quantity of protein of the samples, measured by BCA protein assay kit (Thermo Fisher Scientific, Courtaboeuf, France).
Histology and immunohistochemistry
Tissue specimens of inguinal, retroperitoneal, and epididymal WAT were fixed by immersion in 4% paraformaldehyde in 0.1 M sodium phosphate buffer (PB), pH 7.4. After washing in PB overnight, the samples were dehydrated in a graded series of ethanol and embedded in paraffin blocks for light microscopy and immunohistochemistry. Sections (5 µm) were immunostained for CoxIV by means of the avidin-biotin technique (23). Briefly, sections were incubated first with primary polyclonal anti-CoxIV antibody raised in rabbits (Cell Signaling Technology, Danvers, MA) diluted 1:100 dilution in PBS, then with the corresponding biotinylated anti-rabbit IgG secondary antibody, raised in goat, diluted 1:200 (Vector Laboratories, Burlingame, CA), and finally, with ABC complex (Vectastain ABC kit; Vector Laboratories). Peroxidase activity was revealed using 0.075% 3,3′-diaminobenzidine hydrochloride as chromogen (Sigma, St. Louis, MO) in Tris buffer 0.05 M, pH 7.6. Sections were counterstained with hematoxylin and mounted in Eukitt (Kindler, Freiburg, Germany). Sections were observed with Zeiss Axioskop 2 microscope equipped with AxioCam Icc3 digital camera (Carl Zeiss, S.A., Barcelona, Spain). Appropriate positive controls were used to check antibody specificity.
Statistical analysis
Data are expressed as means ± SEM. Significant differences between control and treated cells/groups were determined by ANOVA following by Tukey Kramer post hoc test using Statview software (SAS Institute, Cary, NC). Values of P < 0.05 were considered significant.
RESULTS
ATRA upregulates genes linked to mitochondria and OXPHOS and increases oxygen consumption in adipocytes
To study in detail the impact of ATRA, in terms of gene expression, on adipocytes, and notably its putative impact on mitochondrial function, we performed microarray experiments. Mature 3T3-L1 adipocytes were incubated with ATRA (2 µM for 24 h). The treatment affected as expected the expression of genes previously shown to be modulated by ATRA in adipocytes and/or adipose tissue (Table 1), which validated our microarray experiments. Thus, changes brought about by ATRA treatment included the induction of Rarβ, Rarres1, Ucp1 and 2, Ppargc1α, Pparα, or Cyp26b1 and the repression of the leptin, resistin, Cebpα, adiponectin, and Rxrα genes (4, 9, 15, 18, 24–27).
TABLE 1.
Known ATRA target genes found changed by microarray analysis in 3T3-L1 adipocytes exposed to 2 μM ATRA
| Probe Number | Fold Change | Refseq Number | Gene Name | P |
| A_51_P202440 | 1.35 | NM_011243 | Rarb | 0.002 |
| A_51_P401184 | 2.43 | XM_130987 | Rarres1 | 1.08 E-05 |
| A_51_P501844 | 46.29 | NM_175475 | Cyp26b1 | 6.31 E-12 |
| A_52_P374882 | −24.74 | NM_008493 | Lep | 9.75 E-14 |
| A_51_P233597 | −1.87 | NM_022984 | Retn | 4.16 E-04 |
| A_52_P168567 | −3.00 | NM_007678 | Cebpα | 1.41 E-07 |
| A_51_P458451 | −1.42 | NM_009605 | Adipoq | 0.008 |
| A_52_P188139 | −1.66 | NM_011305 | Rxrα | 3.97 E-04 |
| A_52_P499675 | 3.33 | NM_011671 | Ucp2 | 0.04 |
| A_51_P426353 | 32.54 | NM_009463 | Ucp1 | 2.66 E-07 |
| A_52_P5945 | 1.32 | AK032149 | Ppargc1α | 4.21 E-04 |
| A_51_P348334 | 3.56 | NM_011144 | Pparα | 3.81 E-06 |
Then, transcriptomic data were analyzed by GSEA. Among the top 20 gene sets positively regulated by ATRA, 9 genes sets contained the term “mitochondria” in their title (Table 2). Notably, the gene set called “Mitochondrion,” which regroups 343 genes related to mitochondria was significantly enriched (NES = 1.93, P value = 0.002, FDR q value = 0.289). In addition, several others gene sets related to organelle membranes or transporters were also indirectly linked to mitochondria, suggesting a global positive impact of ATRA treatment on mitochondria function. To complete this analysis, we conducted a Metacore pathway analysis (data not shown). Interestingly, of the pathways identified, the one called “oxidative phosphorylation” (Metacore nomenclature) was deeply affected: of the 105 genes involved in this pathway, 62 were upregulated by ATRA (P < 2.73e-10). These genes code for subunits of the complexes I through IV, involved in the electron transport chain and found in the inner membrane of the mitochondrion (supplementary Table 1). Among regulated genes, Cycs, encoding cytochrome c, appeared as one of the most upregulated (3.6-fold induction). Together these data support that ATRA strongly impacted mitochondria function, notably through an induction of OXPHOS capacity.
TABLE 2.
Top 20 gene sets impacted by (2 μM) ATRA incubation in 3T3-L1 adipocytes as indicated by GSEA of microarray data
| Gene Set | NES | NOM P | FDR q |
| GLUCOSE_METABOLIC_PROCESS | 2.54 | 0.004 | 0.384 |
| LIPID_TRANSPORTER_ACTIVITY | 2.45 | 0.004 | 0.348 |
| MITOCHONDRIAL_PART | 2.33 | 0.002 | 0.403 |
| MITOCHONDRIAL_MEMBRANE_PART | 2.22 | 0.004 | 0.431 |
| CATION_TRANSPORT | 2.21 | 0.011 | 0.355 |
| MONOVALENT_INORGANIC_CATION_TRANSPORT | 2.20 | 0.020 | 0.302 |
| MITOCHONDRIAL_INNER_MEMBRANE | 2.16 | 0.007 | 0.289 |
| ANION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 2.10 | 0.007 | 0.303 |
| MITOCHONDRIAL_MATRIX | 2.06 | 0.018 | 0.309 |
| MITOCHONDRIAL_LUMEN | 2.05 | 0.011 | 0.282 |
| MITOCHONDRIAL_ENVELOPE | 2.02 | 0.002 | 0.281 |
| ORGANELLE_INNER_MEMBRANE | 1.95 | 0.022 | 0.313 |
| SUBSTRATE_SPECIFIC_TRANSPORTER_ACTIVITY | 1.94 | 0.004 | 0.299 |
| MITOCHONDRION | 1.93 | 0.002 | 0.289 |
| TRANSCRIPTION_REPRESSOR_ACTIVITY | 1.88 | 0.017 | 0.316 |
| MITOCHONDRIAL_MEMBRANE | 1.86 | 0.014 | 0.317 |
| MITOCHONDRIAL_RESPIRATORY_CHAIN | 1.86 | 0.023 | 0.302 |
| VOLTAGE_GATED_CHANNEL_ACTIVITY | 1.85 | 0.041 | 0.290 |
| ION_TRANSPORT | 1.81 | 0.023 | 0.317 |
| ION_CHANNEL_ACTIVITY | 1.79 | 0.027 | 0.323 |
FDR, false discovery rate; NES, normalized enrichment score; NOM P, nominal P value.
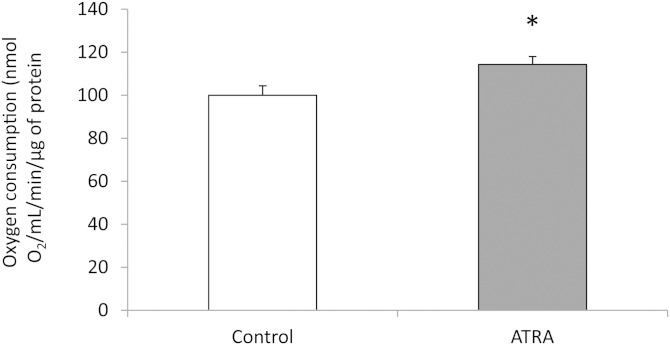
To determine whether these gene expression modifications were accompanied by functional changes in cellular metabolism, the rate of oxygen consumption by cells treated or not with ATRA was compared. As shown on Fig. 1, ATRA slightly but significantly induced oxygen consumption by 15%.
Fig. 1.
Oxygen consumption in 3T3-L1 adipocytes exposed to 2 µM of ATRA was measured as described in Materials and Methods using Clarke’s electrode. Control refers to control cells, which received the vehicle (DMSO). Data are the mean ± SEM of three independent cultures per treatment condition.
ATRA upregulates several transcription factors and coactivators linked to mtDNA transcription, biogenesis, and function in adipocytes
To go into further detail regarding the analysis of genes regulated by ATRA in adipocytes, a deep analysis of genes present within the enriched genes sets was conducted. We identified several genes coding for transcription factors strongly involved in mitochondrial biogenesis and function. Gene expression of classical transcription factors controlling the transcription as well as the replication of mitochondrial DNA (i.e., Tfam, Mterf, and Tfb2m) was significantly upregulated (Table 3). In addition, genes encoding transcription factors governing respiratory gene expression (i.e., Nrf2, Errα, Yy1, or Pparα) were also induced by ATRA in adipocytes, as well as genes for nuclear coactivators of mitochondrial biogenesis Ppargc1α and Ppargc1β (encoding peroxisome proliferator-activated receptor gamma coactivator (PGC)1α and PGC1β, respectively), the latter being strongly upregulated (×2.5). Finally, gene coding for proteins involved in mitochondria fusion (Opa1, Mfn1, and Mfn2) and fission (Dnm1L) were slightly but significantly upregulated by ATRA (Table 3).
TABLE 3.
Transcription factors regulated by ATRA in 3T3-L1 adipocytes and involved in mitochondria biogenesis and genes involved in mitochondria fusion and fission
| Probe Number | Fold Change | Refseq Number | Gene Name | P |
| A_52_P45724 | 1.52 | NM_172135 | Mterf | 2.76 E-07 |
| A_51_P493720 | 1.22 | NM_009360 | Tfam | 2.79 E-06 |
| A_51_P374499 | 1.55 | NM_008249 | Tfb2m | 7.43 E-05 |
| A_52_P526724 | 2.54 | NM_133249 | Ppargc1β | 4.65 E-07 |
| A_51_P279038 | 1.21 | NM_008904 | Ppargc1α | 0.016 |
| A_52_P396436 | 1.63 | NM_010902 | Nrf2 | 2.91 E-09 |
| A_51_P456320 | 1.31 | NM_009537 | Yy1 | 0.001 |
| A_51_P248580 | 1.35 | NM_007953 | Errα | 6.51 E-05 |
| A_51_P348334 | 3.56 | NM_011144 | Pparα | 3.81 E-06 |
| A_52_P547795 | 1.48 | NM_133752 | Opa1 | 1.61 E-04 |
| A_52_P2732 | 1.35 | NM_024200 | Mfn1 | 5.00 E-04 |
| A_51_P266774 | 1.17 | NM_133201 | Mfn2 | 0.002 |
| A_51_P185701 | 1.33 | NM_152816 | Dnm1L | 1.19 E-04 |
In order to identify nuclear receptors putatively involved in this regulation, we used RAR and PPARδ antagonists. Results strongly suggested that ATRA-mediated regulation of mitochondria-related genes was at least in part driven by RAR because the induction of Ppargc1β, Nrf2, Ucp1, and Aco was partly blunted, together with that of Cyp26b1, when the cells were preincubated with an RAR antagonist (AGN 193109; supplementary Table 2), whereas a PPARδ antagonist (GSK0660) was ineffective (supplementary Table 3).
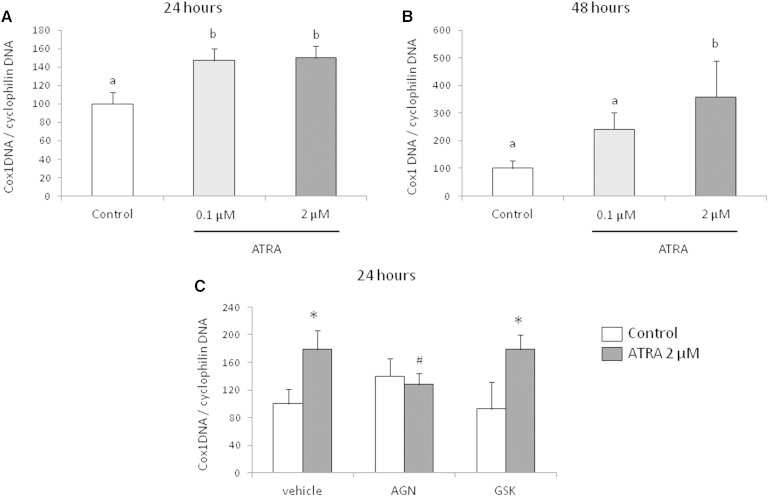
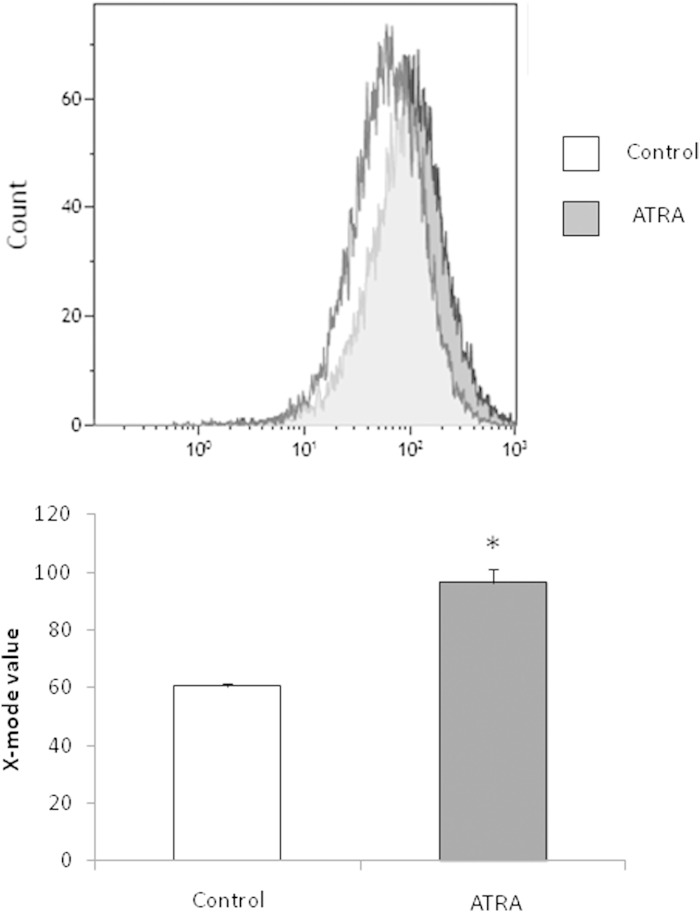
Importantly, results on gene expression were extended by quantification of mtDNA and mitochondria in adipocytes. In parallel with the induction of transcription factors linked to mtDNA transcription and replication, there was a significant increase in mtDNA quantified by the ratio Cox1 DNA/cyclophilin DNA in the ATRA-treated adipocytes (Fig. 2). This increase was significant 24 h after the beginning of incubation of cells with ATRA (Fig. 2A) and was maintained after 48 h for the 2 µM condition (Fig. 2B). Interestingly, as reported for gene expression, addition of an RAR antagonist blunted the mtDNA induction mediated by ATRA, whereas PPARδ antagonist was without effect (Fig. 2C). In addition, consistently with the upregulation of transcription factors for mitochondria biogenesis, the labeling of mitochondria, determined by cytometry after MitoTracker® incubation, was increased by ATRA treatment (Fig. 3).
Fig. 2.
mtDNA to nuclear DNA ratio in 3T3-L1 adipocytes exposed to the indicated ATRA doses for 24 h (A) and 48 h (B). Control refers to control cells, which received the vehicle (DMSO). Data are the mean ± SEM of three independent cultures per treatment condition. mtDNA to nuclear DNA ratio in 3T3-L1 adipocytes preincubated with RAR antagonist (AGN 193109; 10 µM) or PPARδ antagonist (GSK0660; 10 µM) for 1 h and exposed to 2 µM of ATRA for 24 h (C). Control refers to control cells, which received the vehicles (ethanol and DMSO). Data are the mean ± SEM of two independent cultures per treatment condition.
Fig. 3.
MitoTracker® Green FM staining of 3T3-L1 adipocytes after treatment for 24 h with vehicle (control) or 2 µM ATRA. Incubation with the dye and flow cytometry sorting of the stained cells were conducted as described in Materials and Methods.
ATRA treatment in mice induces mitochondria-related genes and increases the number of mitochondria in WAT
To investigate the impact of ATRA in vivo, NMRI mice received ATRA (50 mg/kg of body weight/day) subcutaneously for 4 days. Gene expression analysis of inguinal, retroperitoneal, and epididymal fat pads was conducted by quantitative PCR. The effectiveness of the treatment was confirmed by the upregulation of classical RAR target genes such as Cyp26a1 and Ucp1, which were strongly induced in the three WAT depots (Table 4). Because Ppargc1α, Ppargc1β, Nrf2, Tfam, Yy1, Tfb2m, Cox8b, and Atp5f1 were among the most relevant mitochondria-related genes upregulated by ATRA in cultured adipocytes, we quantified the expression of these mRNAs in vivo. As expected, all mRNAs were significantly upregulated, with some variations between adipose fat pads (Table 4). In some depots, the induction did not reach statistical significance, but still a clear tendency toward increased expression was observed. In addition, we quantified in the three WAT depots several genes related to beige/BRITE phenotypes [Slc27a1, Cd137, Tmem26, Hoxc9, and Tbx1 (28)]. The results showed an upregulation of Cd137 in all WAT depots of ATRA-treated mice examined and an upregulation of Slc27a1, Hoxc9, and Tbx1 in some of them (mainly the visceral depots). To the contrary, we found the adipose expression of Tmem26 to be downregulated by ATRA-treatment.
TABLE 4.
Expression of mitochondria- and beige/BRITE-related genes in WAT depots of ATRA-treated mice
| Gene Name | Inguinal AT | Retroperitoneal AT | Epididymal AT |
| Ppargc1α | 1.24 ± 0.24 | 2.35 ± 0.31a | 1.52 ± 0.19a |
| Ppargc1β | 2.87 ± 0.48a | 2.70 ± 0.52a | 2.67 ± 0.57a |
| Nrf2 | 1.76 ± 0.32 | 1.13 ± 0.14 | 1.63 ± 0.27a |
| Tfam | 2.51 ± 0.58a | 1.29 ± 0.18 | 1.29 ± 0.18 |
| Yy1 | 0.98 ± 0.14 | 1.27 ± 0.08a | 1.23 ± 0.10a |
| Tfb2m | 2.61 ± 0.85a | 1.44 ± 0.23a | 1.78 ± 0.11a |
| Pparα | 3.10 ± 0.69a | 5.48 ± 1.04a | 2.92 ± 0.6a |
| Aco | 0.96 ± 0.20a | 1.99 ± 0.32a | 1.51 ± 0.34 |
| Cox8b | 0.80 ± 0.17 | 3.40 ± 1.04a | 1.87 ± 0.39a |
| Atp5f1 | 0.98 ± 0.11 | 1.53 ± 0.15a | 1.28 ± 0.10a |
| Ucp1 | 3.35 ± 0.81a | 3.89 ± 0.74a | 2.14 ± 0.43a |
| Slc27a1 | 1.67 ± 0.42 | 2.47 ± 0.34a | 3.23 ± 0.62a |
| Cd137 | 3.05 ± 1.01a | 3.34 ± 0.97a | 4.56 ± 1.3a |
| Tmem26 | 0.42 ± 0.07a | 0.51 ± 0.13a | 0.60 ± 0.08a |
| Hoxc9 | 0.88 ± 0.23 | 1.40 ± 0.15a | 1.08 ± 0.10 |
| Tbx1 | 1.28 ± 0.28 | 1.20 ± 0.30 | 2.22 ± 0.36a |
| Cyp26b1 | 47.5 ± 9.0a | 62.6 ± 9.3a | 43.7 ± 7.3a |
AT, adipose tissue. Gene expression values in ATRA-treated mice were normalized to those in control mice (equal to 1). Expression values between control and ATRA-treated mice were compared by Student’s t-test.
P values <0.05 were considered significant.
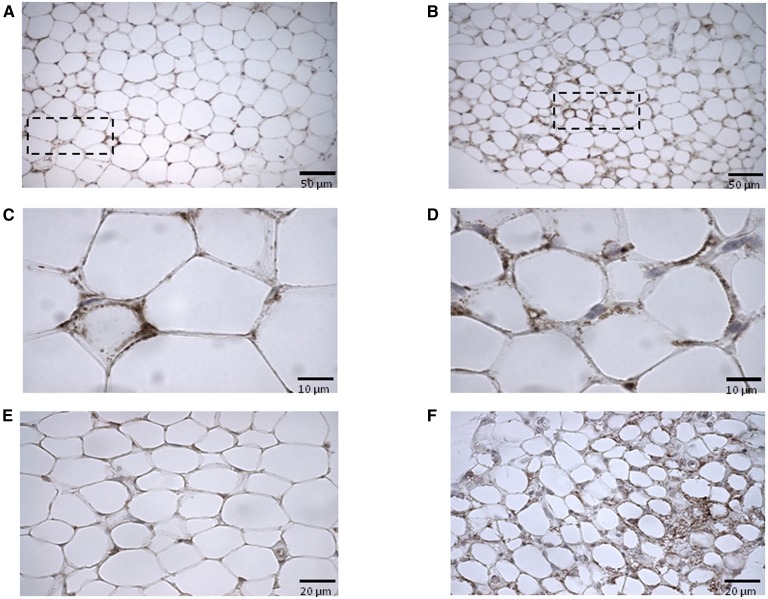
To gain further insight into the impact of ATRA on adipose tissue mitochondria, immunohistochemical staining of WAT with COXIV antibodies was performed. For all WAT depots studied, this analysis revealed increased COXIV positivity in the ATRA-treated mice as compared with control mice. Representative micrographs for the inguinal and retroperitoneal depots are shown in Fig. 4. In the visceral depots (retroperitoneal and epididymal), ATRA treatment led to increased COXIV positivity in the peripheral cytoplasm of unilocular adipocytes without inducing the appearance of multilocular cells (Fig. 4B, D and data not shown). In the subcutaneous (inguinal) WAT, both effects were apparent (Fig. 4F): in concordance with a previous report (11), multilocular adipocytes emerged in this depot in (three out of six) ATRA-treated mice, and these multilocular adipocytes were highly positive for COXIV. In all WAT depots studied, adipocytes of ATRA-treated mice were smaller than that of control mice (Fig. 4 and data not shown).
Fig. 4.
Representative light microscopy micrographs of CoxIV immunostained sections of retroperitoneal (A–D) and inguinal (E, F) white adipose depots of control (A, C, E) and ATRA-treated (B, D, F) mice. Cells from control animals present sparse COXIV positivity as compared with those of ATRA-treated mice. Dotted line-encircled areas in A and B are shown magnified in C and D, respectively, to illustrate COXIV positivity in the cytoplasm of unilocular adipocytes. COXIV positive multilocular adipocytes as found in the inguinal depot of ATRA-treated mice are illustrated in F. Scale bars indicate 50 µm (A, B), 10 µm (C, D), and 20 µm (E, F).
DISCUSSION
White adipocytes in WAT depots are poor in mitochondria and have a low oxidative and thermogenic capacity that suits their energy storage function, while the opposite is true for brown adipocytes in BAT depots, which are specialized in regulated energy dissipation as heat (adaptive thermogenesis) through UCP1 activity. A positive effect of ATRA on WAT oxidative and thermogenic capacity, which eventually leads to the conversion of white adipocytes into more brown-like cells, has been described and linked to a fat-lowering effect of ATRA (11, 12, 15). However, until now, little has been known about the impact of ATRA on mitochondria in adipose tissue. Being the center of both oxidative metabolism and thermogenesis, mitochondria are the master organelles of cell fuel utilization and as such represent an obvious potential target of ATRA action. In this work, we show that ATRA strongly impacts mitochondria biogenesis and function in white adipocytes leading to increased OXPHOS capacity and mitochondrial content. This is revealed by transcriptome analysis of mature white adipocytes exposed to ATRA in vitro and confirmed by additional lines of evidence and in ATRA-treated mice.
Changes in gene expression in mature 3T3-L1 adipocytes induced by ATRA included the upregulation of genes coding for classical transcription factors governing mtDNA replication and transcription (Tfam, Mterf, and Tfb2m), nuclear coactivators (Ppargc1α and Ppargc1β), interacting transcription factors (Nrf2, Errα, Yy1, and Pparα) that control mitochondrial biogenesis and respiratory gene expression, and genes coding for protein involved in mitochondria fusion and fission. In fact, ∼60% of the genes in the OXPHOS pathway were upregulated at the mRNA level in the ATRA-treated adipocytes. Genes encoding mitochondrial proteins capable of uncoupling OXPHOS to generate heat, namely, Ucp1 (the molecular marker of brown adipocytes) and Ucp2, were also upregulated. Importantly, gene expression analysis on the adipose tissues of ATRA-treated mice revealed a similar picture, with an increased expression of genes linked to mitochondria and the OXPHOS pathway. In addition, we provide evidence that these ATRA-mediated regulations could be mainly driven by the RAR nuclear receptor, whereas PPARδ, another known ATRA-activated nuclear receptor (12) could be less active in our conditions. However, we cannot exclude that part of the regulations could also occur via activation of RXR consecutively to ATRA isomerization to 9-cis-RA.
Both in ATRA-exposed 3T3-L1 adipocytes and in WAT depots of ATRA-treated mice, the Ppargc1β gene was upregulated to a greater extent and more consistently than the Ppargc1α gene. The two PGC-1 isoforms coactivate transcription factors controlling genes required for mitochondria biogenesis and energy metabolism, located in both the mitochondrial and nuclear genomes (29). Mitochondrial biogenesis, in particular, is stimulated by the PGC-1-NRF1/2-TFAM pathway, where PGC-1 coactivates the NRF1/2 transcription factor to increase gene expression of TFAM, a final effector that enters the mitochondria to stimulate transcription and replication of mtDNA molecules (30). The best-known member of the PGC-1 family is PGC-1α, which participates in the control of multiple processes, including the activation of brown fat thermogenesis (31, 32). Less is known about the role of PGC-1β in adipose tissues. PGC-1β does not seem to be required for adrenergically stimulated brown fat thermogenesis, but it is needed together with PGC-1α for differentiation-dependent mitochondrial biogenesis and respiration during brown adipocyte differentiation (32). There is also evidence of a role for PGC-1β favoring mitochondrial biogenesis in white adipocytes, as silencing of PGC-1β inhibits the expression of mitochondrial genes, ATP synthesis, and adipogenesis in primary rat differentiating preadipocytes (33). Reciprocally, PGC-1β overexpression upregulates mitochondrial biosynthesis and adipogenesis marker genes during differentiation of 3T3-L1 preadipocytes (33). PGC-1β overexpression also enhances mitochondrial biogenesis and function in fully differentiated 3T3-L1 adipocytes, together with improved insulin sensitivity (34).
Another gene that was strongly and consistently upregulated in white adipocytes after ATRA treatment was the one encoding PPARα. This nuclear receptor transactivates many nuclear genes involved in cellular fatty acid uptake and oxidation (35) and, similar to PGC-1, plays a major role in tissues that normally use fatty acids for ATP or heat production, including BAT. In fact, high Pparα expression is considered a distinctive marker of the BAT with respect to the WAT phenotype (36). Importantly, overexpression or pharmacological activation of Pparα promotes mitochondrial biogenesis, fatty acid oxidation, and a BAT-like pattern of gene expression in white adipocytes (37–40).
In accordance with the higher expression levels of transcription factors and coactivators linked to mitochondria biogenesis, mitochondrial content was increased in the ATRA-treated 3T3-L1 adipocytes, as judged by the results of mtDNA content and MitoTracker® staining analysis performed. Similarly, stronger COXIV positivity upon immunohistochemical staining is suggestive of an increased density of mitochondria in the adipose tissue depots of ATRA-treated mice. The histochemical analysis also revealed a smaller size of the adipocytes in the WAT depots of ATRA-treated mice and an increased number of adipocytes with a multilocular distribution of intracellular lipids, which is typical of brown adipocytes as opposed to unilocular white adipocytes, in the subcutaneous (inguinal) WAT samples of these animals. Additionally, several beige/BRITE markers were found to be induced in WAT of ATRA-treated mice. Altogether, our results are in concordance with previous reports showing ATRA-induced remodeling toward increased oxidative metabolism in WAT depots (11, 12) and cultured 3T3-L1 adipocytes (15). Similar effects were seen in skeletal muscles of ATRA-treated obese mice, where oxidative metabolism capacities and mitochondrial content were found to be elevated (12). In keeping with increased overall oxidative metabolism and associated thermogenesis, ATRA treatment in mice results in an increase in rectal temperature (11, 12).
Enhancement of energy expenditure in WAT is a potential antiobesity strategy that can be achieved by inducing in white adipocytes UCP1-mediated thermogenesis and/or a futile substrate cycle consisting in hydrolysis of intracellular triacylglycerols and fatty acid reesterification (TAG/FA cycle) (1). We have shown previously that ATRA induces UCP1 in mouse embryo fibroblasts-derived adipocytes (41) and report here a clear induction of Ucp1 gene transcription by ATRA in 3T3-L1 adipocytes. Additionally, evidence has been provided that ATRA may stimulate a TAG/FA cycle in association with enhanced β-oxidation in 3T3-L1 adipocytes (15). Importantly, activation of either UCP1 or futile cycles should go hand in hand with the induction of mitochondrial β-oxidation of fatty acids and OXPHOS capacity for a meaningful effect on adipocyte lipid content and whole body energy expenditure to be achieved. It has been previously shown that ATRA enhances OXPHOS and oxygen consumption in SH-SY5Y human neuroblastoma cells, even without any increase in the number of mitochondria or measurable changes in the composition of the electron transport chain (42). Here, we present evidence of increased OXPHOS capacity and mitochondrial content in 3T3-L1 adipocytes following ATRA exposure, necessary for both UCP1-dependent and UCP1-independent energy dissipation mechanisms. Interestingly, whereas multilocular, UCP1-expressing brown adipocyte-like cells (i.e., beige/BRITE adipocytes) are readily induced in vivo upon ATRA treatment in the subcutaneous (inguinal) WAT of mice, in the visceral depots ATRA treatment increases OXPHOS capacity without inducing the appearance of such cells [this work and (11)]. Subcutaneous WAT is more prone to browning than visceral WAT (43), whereas rodent and human visceral adipocytes contain more mitochondria and have higher oxidative capacity as compared with subcutaneous adipocytes (44, 45). It would appear, therefore, that ATRA treatment is able to potentiate differential intrinsic capacities of each type of fat depot, thus enhancing energy expenditure by an UCP1-dependent mechanism in subcutaneous WAT and by an UCP1-independent mechanism in the visceral WAT depots.
One limitation of the present work is the relatively high dose of ATRA used in the mouse study (50 mg ATRA/kg animal per day). However, no signs of toxicity were found; such doses did not modify food intake or indicate hepatic damage (data not shown). In addition, we have to keep in mind that the route of administration (subcutaneous injection) implies a progressive release of the agent from the point of injection and a considerable dilution of the agent, which lowers the concentration of ATRA that actually reaches the adipose cells.
In summary, this work shows that ATRA impacts mitochondria in white adipocytes, leading to increased OXPHOS capacity and mitochondrial content. This knowledge might be of interest in the context of obesity management, as modulation of WAT metabolism could contribute to reduction of adiposity. Beyond obesity, knowledge of factors capable of modulating mitochondria might find application in the treatment of disorders caused by mitochondria malfunctioning.
Supplementary Material
Acknowledgments
The authors thank Enzo Ceresi for excellent technical assistance in immunohistochemical analysis.
Footnotes
Abbreviations:
- ATRA
- all-trans retinoic acid
- BAT
- brown adipose tissue
- GSEA
- gene set enrichment analysis
- mtDNA
- mitochondrial DNA
- OXPHOS
- oxidative phosphorylation
- PGC
- peroxisome proliferator-activated receptor gamma coactivator
- RA
- retinoic acid
- RAR
- retinoid acid receptor
- RXR
- retinoid X receptor
- UCP
- uncoupling protein
- WAT
- white adipose tissue
This work was supported by the Aix-Marseille Université, INRA, and EU FP7 project DIABAT. Authors belong to the Nutrigenomics-group, awarded as “Group of Excellence” of the Autonomous Community of the Balearic Islands (CAIB) and supported by “Direcció General d’Universitats, Recerca i Transferència del Coneixement” of Regional Government (CAIB) and FEDER funds (EU). CIBER de Fisiopatología de la Obesidad y Nutrición is an initiative of the ISCIII (Spanish Government). H.M. was the recipient of a fellowship from the government of the CAIB.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Flachs P., Rossmeisl M., Kuda O., Kopecky J. 2013. Stimulation of mitochondrial oxidative capacity in white fat independent of UCP1: a key to lean phenotype. Biochim. Biophys. Acta. 1831: 986–1003. [DOI] [PubMed] [Google Scholar]
- 2.Blomhoff R., Blomhoff H. K. 2006. Overview of retinoid metabolism and function. J. Neurobiol. 66: 606–630. [DOI] [PubMed] [Google Scholar]
- 3.Theodosiou M., Laudet V., Schubert M. 2010. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell. Mol. Life Sci. 67: 1423–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tourniaire F., Gouranton E., von Lintig J., Keijer J., Luisa Bonet M., Amengual J., Lietz G., Landrier J. F. 2009. beta-Carotene conversion products and their effects on adipose tissue. Genes Nutr. 4: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landrier J. F., Marcotorchino J., Tourniaire F. 2012. Lipophilic micronutrients and adipose tissue biology. Nutrients. 4: 1622–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puigserver P., Vazquez F., Bonet M. L., Pico C., Palou A. 1996. In vitro and in vivo induction of brown adipocyte uncoupling protein (thermogenin) by retinoic acid. Biochem. J. 317: 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonet M. L., Oliver J., Pico C., Felipe F., Ribot J., Cinti S., Palou A. 2000. Opposite effects of feeding a vitamin A-deficient diet and retinoic acid treatment on brown adipose tissue uncoupling protein 1 (UCP1), UCP2 and leptin expression. J. Endocrinol. 166: 511–517. [DOI] [PubMed] [Google Scholar]
- 8.Ribot J., Felipe F., Bonet M. L., Palou A. 2001. Changes of adiposity in response to vitamin A status correlate with changes of PPAR gamma 2 expression. Obes. Res. 9: 500–509. [DOI] [PubMed] [Google Scholar]
- 9.Felipe F., Bonet M. L., Ribot J., Palou A. 2004. Modulation of resistin expression by retinoic acid and vitamin A status. Diabetes. 53: 882–889. [DOI] [PubMed] [Google Scholar]
- 10.Felipe F., Mercader J., Ribot J., Palou A., Bonet M. L. 2005. Effects of retinoic acid administration and dietary vitamin A supplementation on leptin expression in mice: lack of correlation with changes of adipose tissue mass and food intake. Biochim. Biophys. Acta. 1740: 258–265. [DOI] [PubMed] [Google Scholar]
- 11.Mercader J., Ribot J., Murano I., Felipe F., Cinti S., Bonet M. L., Palou A. 2006. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology. 147: 5325–5332. [DOI] [PubMed] [Google Scholar]
- 12.Berry D. C., Noy N. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol. Cell. Biol. 29: 3286–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amengual J., Ribot J., Bonet M. L., Palou A. 2008. Retinoic acid treatment increases lipid oxidation capacity in skeletal muscle of mice. Obesity (Silver Spring). 16: 585–591. [DOI] [PubMed] [Google Scholar]
- 14.Amengual J., Ribot J., Bonet M. L., Palou A. 2010. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell. Physiol. Biochem. 25: 657–666. [DOI] [PubMed] [Google Scholar]
- 15.Mercader J., Madsen L., Felipe F., Palou A., Kristiansen K., Bonet M. L. 2007. All-trans retinoic acid increases oxidative metabolism in mature adipocytes. Cell. Physiol. Biochem. 20: 1061–1072. [DOI] [PubMed] [Google Scholar]
- 16.Landrier J. F., Gouranton E., El Yazidi C., Malezet C., Balaguer P., Borel P., Amiot M. J. 2009. Adiponectin expression is induced by vitamin E via a peroxisome proliferator-activated receptor gamma-dependent mechanism. Endocrinology. 150: 5318–5325. [DOI] [PubMed] [Google Scholar]
- 17.Marcotorchino J., Gouranton E., Romier B., Tourniaire F., Astier J., Malezet C., Amiot M. J., Landrier J. F. 2012. Vitamin D reduces the inflammatory response and restores glucose uptake in adipocytes. Mol. Nutr. Food Res. 56: 1771–1782. [DOI] [PubMed] [Google Scholar]
- 18.Gouranton E., Aydemir G., Reynaud E., Marcotorchino J., Malezet C., Caris-Veyrat C., Blomhoff R., Landrier J. F., Ruhl R. 2011. Apo-10’-lycopenoic acid impacts adipose tissue biology via the retinoic acid receptors. Biochim. Biophys. Acta. 1811: 1105–1114. [DOI] [PubMed] [Google Scholar]
- 19.Landrier J. F., Malezet-Desmoulins C., Reboul E., Marie Lorec A., Josephe Amiot M., Borel P. 2008. Comparison of different vehicles to study the effect of tocopherols on gene expression in intestinal cells. Free Radic. Res. 42: 523–530. [DOI] [PubMed] [Google Scholar]
- 20.Landrier J. F., Gouranton E., Reboul E., Cardinault N., El Yazidi C., Malezet-Desmoulins C., Andre M., Nowicki M., Souidi M., Borel P. 2010. Vitamin E decreases endogenous cholesterol synthesis and apo-AI-mediated cholesterol secretion in Caco-2 cells. J. Nutr. Biochem. 21: 1207–1213. [DOI] [PubMed] [Google Scholar]
- 21.Romier B., Tourniaire F., Marcotorchino J., Gouranton E., Astier J., Malezet C., Blouin E., Landrier J. F. 2013. Bioeffects of a combination of trace elements on adipocyte biology. Metallomics. 5: 524–531. [DOI] [PubMed] [Google Scholar]
- 22.Tourniaire F., Romier-Crouzet B., Lee J. H., Marcotorchino J., Gouranton E., Salles J., Malezet C., Astier J., Darmon P., Blouin E., et al. 2013. Chemokine expression in inflamed adipose tissue is mainly mediated by NF-kappaB. PLoS ONE. 8: e66515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamei N., Tobe K., Suzuki R., Ohsugi M., Watanabe T., Kubota N., Ohtsuka-Kowatari N., Kumagai K., Sakamoto K., Kobayashi M., et al. 2006. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 281: 26602–26614. [DOI] [PubMed] [Google Scholar]
- 24.Hollung K., Rise C. P., Drevon C. A., Reseland J. E. 2004. Tissue-specific regulation of leptin expression and secretion by all-trans retinoic acid. J. Cell. Biochem. 92: 307–315. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Matheny M., Zolotukhin S., Tumer N., Scarpace P. J. 2002. Regulation of adiponectin and leptin gene expression in white and brown adipose tissues: influence of beta3-adrenergic agonists, retinoic acid, leptin and fasting. Biochim. Biophys. Acta. 1584: 115–122. [DOI] [PubMed] [Google Scholar]
- 26.Felipe F., Bonet M. L., Ribot J., Palou A. 2003. Up-regulation of muscle uncoupling protein 3 gene expression in mice following high fat diet, dietary vitamin A supplementation and acute retinoic acid-treatment. Int. J. Obes. Relat. Metab. Disord. 27: 60–69. [DOI] [PubMed] [Google Scholar]
- 27.Han S., Sidell N. 2002. Peroxisome-proliferator-activated-receptor gamma (PPARgamma) independent induction of CD36 in THP-1 monocytes by retinoic acid. Immunology. 106: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J., Bostrom P., Sparks L. M., Ye L., Choi J. H., Giang A. H., Khandekar M., Virtanen K. A., Nuutila P., Schaart G., et al. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 150: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finck B. N., Kelly D. P. 2006. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez R., Checa M., Brun S., Vinas O., Mampel T., Iglesias R., Giralt M., Villarroya F. 2000. Both retinoic-acid-receptor- and retinoid-X-receptor-dependent signalling pathways mediate the induction of the brown-adipose-tissue-uncoupling-protein-1 gene by retinoids. Biochem. J. 345: 91–97. [PMC free article] [PubMed] [Google Scholar]
- 31.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 92: 829–839. [DOI] [PubMed] [Google Scholar]
- 32.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3: 333–341. [DOI] [PubMed] [Google Scholar]
- 33.Ji H., Lu R. H., Chang Z. G., Su S. S., Yang G. S. 2012. PGC-1beta modulates the expression of genes involved in mitochondrial function and adipogenesis during preadipocyte differentiation. Reprod. Domest. Anim. 47: 419–427. [DOI] [PubMed] [Google Scholar]
- 34.Gao C. L., Liu G. L., Liu S., Chen X. H., Ji C. B., Zhang C. M., Xia Z. K., Guo X. R. 2011. Overexpression of PGC-1beta improves insulin sensitivity and mitochondrial function in 3T3–L1 adipocytes. Mol. Cell. Biochem. 353: 215–223. [DOI] [PubMed] [Google Scholar]
- 35.Mandard S., Muller M., Kersten S. 2004. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61: 393–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villarroya F., Iglesias R., Giralt M. 2007. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2007: 74364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elliott R. 2005. Mechanisms of genomic and non-genomic actions of carotenoids. Biochim. Biophys. Acta. 1740: 147–154. [DOI] [PubMed] [Google Scholar]
- 38.Hondares E., Rosell M., Diaz-Delfin J., Olmos Y., Monsalve M., Iglesias R., Villarroya F., Giralt M. 2011. Peroxisome proliferator-activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC-1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J. Biol. Chem. 286: 43112–43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribet C., Montastier E., Valle C., Bezaire V., Mazzucotelli A., Mairal A., Viguerie N., Langin D. 2010. Peroxisome proliferator-activated receptor-alpha control of lipid and glucose metabolism in human white adipocytes. Endocrinology. 151: 123–133. [DOI] [PubMed] [Google Scholar]
- 40.Cabrero A., Alegret M., Sanchez R. M., Adzet T., Laguna J. C., Vazquez M. 2001. Bezafibrate reduces mRNA levels of adipocyte markers and increases fatty acid oxidation in primary culture of adipocytes. Diabetes. 50: 1883–1890. [DOI] [PubMed] [Google Scholar]
- 41.Mercader J., Palou A., Bonet M. 2010. Induction of uncoupling protein-1 in mouse embryonic fibroblast-derived adipocytes by retinoic acid. Obesity (Silver Spring). 18: 655–662. [DOI] [PubMed] [Google Scholar]
- 42.Xun Z., Lee D. Y., Lim J., Canaria C. A., Barnebey A., Yanonne S. M., McMurray C. T. 2012. Retinoic acid-induced differentiation increases the rate of oxygen consumption and enhances the spare respiratory capacity of mitochondria in SH-SY5Y cells. Mech. Ageing Dev. 133: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seale P., Lazar M. A. 2009. Brown fat in humans: turning up the heat on obesity. Diabetes. 58: 1482–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deveaud C., Beauvoit B., Salin B., Schaeffer J., Rigoulet M. 2004. Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol. Cell. Biochem. 267: 157–166. [DOI] [PubMed] [Google Scholar]
- 45.Kraunsøe R., Boushel R., Hansen C. N., Schjerling P., Qvortrup K., Stockel M., Mikines K. J., Dela F. 2010. Mitochondrial respiration in subcutaneous and visceral adipose tissue from patients with morbid obesity. J. Physiol. 588: 2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.