Abstract
Interleukin 17A (IL-17A) is a proinflammatory cytokine involved in the pathogenesis of chronic inflammatory diseases. In the field of immunometabolism, we have studied the impact of IL-17A on the lipid metabolism of human in vitro-generated monocyte-derived dendritic cells (DCs). Microarrays and lipidomic analysis revealed an intense remodeling of lipid metabolism induced by IL-17A in DCs. IL-17A increased 2–12 times the amounts of phospholipids, cholesterol, triglycerides, and cholesteryl esters in DCs. Palmitic (16:0), stearic (18:0), and oleic (18:ln-9c) acid were the main fatty acid chains present in DCs. They were strongly increased in response to IL-17A while their relative proportion remained unchanged. Capture of extracellular lipids was the major mechanism of lipid droplet accumulation, visualized by electron microscopy and Oil Red O staining. Besides this foamy phenotype, IL-17A induced a mixed macrophage-DC phenotype and expression of the nuclear receptor NR1H3/liver X receptor-α, previously identified in the context of atherosclerosis as the master regulator of cholesterol homeostasis in macrophages. These IL-17A-treated DCs were as competent as untreated DCs to stimulate allogeneic naive T-cell proliferation. Following this first characterization of lipid-rich DCs, we propose to call these IL-17A-dependent cells “foamy DCs” and discuss the possible existence of foamy DCs in atherosclerosis, a metabolic and inflammatory disorder involving IL-17A.
Keywords: immunology, monocytes, inflammation, microarrays, lipidomics, interleukin 17A, lipid, immunometabolism
The emerging field of research called immunometabolism results from mutual interactions between metabolism and immune system. Chronic inflammation and changes in immune cells participate in metabolic disorders such as atherosclerosis, type 2 diabetes mellitus, and obesity (1), which are now consequently considered both metabolic and chronic inflammatory diseases. Conversely, the physiopathological remodeling of cell-intrinsic metabolic pathways modulates the functions of immune cells (1, 2).
Macrophages and dendritic cells (DCs) are antigen-presenting cells, distributed in the tissues as sentinels of the immune system. They play major roles in many pathological conditions, in line with their ability to produce cytokines and chemokines, run aggressive enzymatic attacks, and activate lymphocytes of innate and adaptive responses. Remodeling of lipid metabolism has been documented in macrophages in the context of atherosclerosis. When newly recruited monocytes engulf oxidized LDLs, they differentiate into lipid-laden foamy macrophages involved in both inflammatory responses and tissue remodeling within the arterial intima (3, 4). A subpopulation of lipid-laden foam cells was suggested to be derived from DCs in the ldlr−/− mouse model of atherosclerosis (5); however, very little is known about lipid accumulation in DCs. In recent years, immunogenic DCs with high endogenous lipid content have been characterized at homeostasis in the liver (6), while tolerogenic lipid-laden DCs have been identified in the malignant microenvironment (7).
Interleukin (IL) 17A is a proinflammatory cytokine produced during innate response by lymphoid or nonlymphoid cells and during adaptive response by TH17 cells (8, 9). IL-17A is also involved in several chronic inflammatory disorders including rheumatoid arthritis, multiple sclerosis, Crohn’s disease, psoriasis and Langerhans cell histiocytosis [see review (10)], but also in immunometabolic diseases. Obesity promotes expansion of TH17 cells (11) while IL-17A inhibits adipocyte development (12). Atherogenesis correlates to the high number of IL-17A-producing cells, although the exact role of those cells in this disease remains unclear (13). Target cells of IL-17A usually express IL-17RA and IL-17RC chains that form the IL-17A receptor. They include epithelial cells, endothelial cells, fibroblasts, and immune cells (14). IL-17A induces the production of proinflammatory mediators, metalloproteases, and antimicrobial peptides (15). We also demonstrated that IL-17A promotes long-term survival of monocyte-derived DCs and their fusion into multinucleated giant cells (16, 17). In vitro, the short DC life span was extended above 12 days by exposure of DCs to IL-17A, suggesting that IL-17A-treated DCs may contribute to the development of chronic inflammation resulting from multiple DC-T-cell cross talks, in vivo. For more than 20 years, the biology of DCs has been studied on in vitro-generated monocyte-derived DCs obtained with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4. This model, although being restrictive compared with the numerous DC subpopulations existing in vivo, shows functional properties consistent with their counterpart in vivo. The existence of DCs fully differentiated from monocytes with a prominent role in initiating adaptive immunity was demonstrated in mice (18), while in humans, Segura et al. (19) identified a population of human DCs present in inflammatory environments that were most likely derived from monocytes.
For the first time, we report that IL-17A strongly impacts the lipid metabolism of human in vitro-generated monocyte-derived DCs. IL-17A led to the generation of liver X receptor (LXR)-α+ foamy DCs, highly competent in fatty acid capture and still able to stimulate allogeneic T-cell proliferation, as demonstrated by phenotypic and functional analysis. Previously published literature incites discussion of the physiological relevance of IL-17A-dependent foamy DCs in vivo, in the field of atherosclerosis.
MATERIALS AND METHODS
Reagents
Antibodies directed against i) CD3 (HIT3a), CD19 (HIB19), both from Ozyme (Saint-Quentin-en-Yvelines, France), and CD56 (ERIC-1) from AbD Serotec (Colmar, France) were used for negative magnetic depletion; ii) CD1a (BL6), CD14 (RMO52), HLA-DR (Immu-357), CD83 (HB15a), CD86 (HAS.2B7), and isotype controls from Beckman coulter (Villepinte, France), CD68, CD163, and CD206 from Becton Dickinson (Le Pont de Claix, France), and C-type lectin domain family 9A (CLEC9A; clone 8F9) and isotype control from Miltenyi Biotec (Paris, France) were used for flow cytometry; and iii) LXR-α, (PPZ0412; R and D Systems, Cambridge, UK), APOE (D6E12; Abcam, Paris, France), perilipin 2 (PLIN2) (AP125; Progen, Heidelberg, Germany), and β-actin (Cell Signaling, Danvers, MA) were used for Western blotting. Recombinant human cytokines included IL-4, IL-17A, and IFN-γ purchased from PeproTech (Neuilly sur Seine, France) and GM-CSF and macrophage colony-stimulating factor (M-CSF) from AbCys (Courtaboeuf, France). Fluorescent reagents included Bodipy 493/503 (Bodipy) and fatty acid Bodipy-FL-C16 purchased from Molecular Probes/Life Technologies (Cergy Pontoise, France) and Carboxyfluorescein_succinimidyl_ester (CFSE) from Interchim (Montluçon, France).
Monocyte purification, macrophage and DC differentiation, and cultures
Blood donation was obtained from healthy adult volunteers (Etablissement français du sang, Lyon Gerland, France). The local ethics committee (Research Committee for the Hospices Civils de Lyon) approved this study, and we obtained written informed consent from each subject (national procedure used for blood donations). Briefly, CD14+ monocytes were purified (>95% CD14+) from the peripheral blood by Ficoll and Percoll gradients, followed by negative magnetic depletion of cells expressing CD3, CD56, or CD19. HLA-DRlow CD1a+ CD83− CD86− immature DCs (>98%) were generated from monocytes seeded at 4,800 cells/mm2 and cultured for 6 days with 100 ng/ml GM-CSF and 10 ng/ml IL-4 in complete RPMI: RPMI 1640 supplemented with 10 mM HEPES, 2 mM l-glutamine, 40 μg/ml gentamicin (Gibco/Life Technologies, Cergy Pontoise, France), and 10% heat-inactivated FCS (Eurobio, Courtaboeuf, France) (20). Macrophages were generated from monocytes cultured for 7 days with 50 ng/ml M-CSF in complete RPMI. Immature DCs were harvested, washed, and seeded at 4,800 cells/mm2 (day 0) in complete RPMI without (DC) or with (DC-17) 2 ng/ml IL-17A. IL-17A was replenished once a week.
RNA extraction and microarrays
Cell lysis and RNA extraction were performed in Trizol (Invitrogen Life Technologies, Cergy Pontoise, France). RNA quality was checked with an Agilent bioanalyzer [RNA integrity number (RIN) >9]. RNA profiling was performed using a high-density oligonucleotide array covering the whole human genome (Genechip human genome U133 Plus 2.0; Affymetrix, Santa Clara, CA). Sample processing and array hybridization was performed according to the manufacturer’s protocols. Expression values and absent/present/marginal calls were calculated using the GCOS v1.4 software (Affymetrix). Absolute expression transcript levels were normalized for each chip by globally scaling all probe sets to a target signal intensity of 500.
Analysis of microarray data
Data were filtered on the detection call so that probe sets with an absent call among all samples were excluded from the analysis. Statistical analyses were performed on 33,253 probe sets with linear models for microarray data (LIMMA) package (21) in R/Bioconductor. LIMMA uses moderated t-statistics, which provides for greater power at small sample sizes. Probe sets with a Benjamini-Hochberg-corrected P value ≤0.01 and a fold change ≥2 or ≤0.5 in DC-17 versus DC were considered as differentially expressed. The regulated gene ontology (GO) biological pathways were identified using DAVID (Database for Annotations, Visualization and Integrated Discovery) (22). The data set is available from the Gene Expression Omnibus database (GSE53163).
Reverse transcription quantitative PCR
First-strand cDNA were synthesized from 250 ng of total RNA in the presence of 100 U Superscript II (Life Technologies) and a mixture of random hexamers and oligo(dT) primers (Promega, Charbonnières-les-Bains, France). Reverse transcription quantitative PCR (RT-qPCR) assays were performed using a Rotor-Gene 6000 (QIAGEN, Courtaboeuf, France). For quantification, a standard curve was generated for each target gene and for the housekeeping gene TATA-binding protein (TBP), with six different amounts (150 to 30,000 molecules/tube) of purified target cDNA cloned in the pGEM plasmid (Promega). For each gene of interest, the amount of mRNA determined from the appropriate standard curve was divided by the amount of TBP mRNA to obtain a normalized value. Primer sequences are available upon request.
Western blotting
Cells were lysed for 15 min at 4°C with RIPA buffer containing a protease inhibitor cocktail (Roche, Indianapolis, IN). Cellular debris were pelleted by centrifugation (10,000 g, 15 min at 4°C), and protein extracts (50 µg per lane) were loaded onto a 12% SDS-polyacrylamide gel and blotted on polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked with 5% milk in PBS/0.5% Tween 20 (PBS-T) for 1 h and then incubated overnight at 4°C with 2 µg/ml anti-LXR-α, 2 µg/ml anti-APOE, and 0.5 µg/ml anti-PLIN2, all in PBS-T plus 5% milk. After three washes with PBS-T, membranes were incubated for 2 h with 0.4 µg/ml HRP-conjugated goat anti-mouse antibody (Promega) in PBS-T plus 5% milk. Detection was performed using Luminata Classico Western HRP Substrate (Millipore, Molsheim, France). Membranes were stripped with Restore Western Blot Stripping buffer (Thermoscientific, Courtaboeuf, France) for 45 min at 60°C under agitation and reprobed with anti-β-Actin (1/5,000).
Lipid content analysis
Total lipids were extracted twice from cells with ethanol-chloroform (1:2, v/v). Before extraction, 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine,1,2-diheptadecanoyl-sn-glycero3phosphoethanolamine, stigmasterol, cholesteryl ester 17:0, and tri-17:0 triglyceride (all from Sigma-Aldrich) were added as internal standards. The organic phases were dried under nitrogen, and the different lipids classes were then separated by thin-layer chromatography using the solvent mixture hexane-diethylether-acetic acid (80:20:1, v/v/v) as eluent. Lipids were detected by UV light after spraying with 0.2% dichlorofluorescein in ethanol and identified by comparison with standards. Silica gel was scraped off. Triacylglycerols, cholesteryl esters, and phospholipids were transmethylated, and the fatty acid methylesters were analyzed by gas chromatography. Briefly, each fraction was treated separately with toluene-methanol (2:3, v/v) and 14% boron trifluoride in methanol. Transmethylation was carried out at 100°C for 90 min in screw-capped tubes. The reaction was terminated by cooling the tubes to 0°C and by the addition of 1.5 ml K2CO3 in 10% water. The resulting fatty acid methyl esters were extracted by isooctane and analyzed by GC with a DELSI instrument model DI 200 equipped with a fused silica capillary SP-2380 column (60 × 0.22 mm), with helium as a carrier gas. Cholesterol was extracted by a mixture of ethanol-chloroform (1:2, v/v). The dry residue was derivatized with 100 μl N,O-bis-trimethylsilyl-trifluoroacetamide for 20 min at 60°C. Derivatized cholesterol was then analyzed by GC/MS using positive chemical ionization mode.
Transmission electron microscopy on DCs
Fixation was initiated by adding an equal volume of fixative solution, previously warmed to 37°C to the cells, either untreated or treated for 10 days with IL-17A. The fixative solution contained 5% glutaraldehyde (Electron Microscopy Sciences, Euromedex, Strasbourg, France) in a 0.1 M sodium cacodylate buffer (both Merck, Darmstadt, Germany) (305 milliosmoles pH 7.3). After 10 min the mixture was centrifuged, the supernatant was discarded, and the pellet resuspended in the fixative solution containing 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for 45 min at room temperature. The cells were then washed in 0.1 M sodium cacodylate buffer, postfixed for 1 h at 4°C with 1% osmium tetroxide (Merck) in the same buffer, and stained for 1 h at 4°C in 4% uranyl acetate. After further washing in distilled water, the cells were dehydrated in graded (50, 70, 80, 95, and 100%) ethanol solutions, incubated for 1 h in Epon (Electron Microscopy Sciences):absolute alcohol (1:1. v/v), then overnight in Epon and embedded in Epon. Ultrathin sections, stained with lead citrate (Leica, Bron, France) and uranyl acetate (Merck), were examined under a Philips CM 120 BioTwin electron microscope (120 kV).
Oil Red O and Hoechst DNA staining
Cells were fixed with 4% formaldehyde for 15 min at room temperature and subsequently stained with a solution of 0.4% Oil Red O dissolved in isopropanol (Sigma-Aldrich) for 20 min and gently shaken at room temperature. After three washes in water, DNA was stained with 10 μg/ml of Hoechst 33342 (Sigma-Aldrich) for 30 min at room temperature. Pictures were analyzed using a Leica DMiRB microscope equipped with ×40/0.30 NA or ×40/0.55 NA objective lenses (Leica) a Leica DC300F camera and the Leica FW400 software.
Flow cytometry analysis
Cell suspensions were labeled according to standard procedures using antibodies directly coupled to fluorochrome for a 30 min incubation in 1% BSA (BSA) and 3% human serum-phosphate-buffered saline (PBS). After three washes in this buffer, cells were analyzed on a FACSCalibur (Becton Dickinson). For bodipy staining, cells were incubated with 1µg/ml of Bodipy diluted in PBS with 1% BSA for 20 min at room temperature, washed twice, and resuspended in PBS with 1% BSA. Fluorescence was quantified on a LSRII (Becton Dickinson) and analyzed using FlowJo software.
Bodipy-FL-C16 capture
Cells (105) were resuspended in 100 µl PBS with or without 0.5 µg/ml of Bodipy-FL-C16 (Invitrogen) and incubated at 37°C or at 4°C for 10 min. Cells were washed five times by centrifugation at 450 g for 5 min in ice-cold PBS containing 0.2% BSA. Fluorescence was measured by flow cytometry on a LSRII and analyzed using the FlowJo software. The intracellular Bodipy-FL-C16 fluorescence was estimated from the shift in the mean fluorescence intensity between 37°C and 4°C.
Alloreactivity measurement
T CD4+ cells were suspended at 107 cells/ml in α-MEM medium containing 2% FCS. After 13 min of incubation in the presence of 10 μM of CFSE, the CFSE incorporation was blocked by the addition of a large excess of α-MEM medium containing 2% FCS. T cells were then washed twice by centrifugation at 1,500 rpm for 10 min at 4°C in α-MEM medium containing 2% FCS. Flow cytometry was used to survey that 100% of T cells have been labeled by CFSE. Monocyte-derived DCs were cultured in various numbers (10–105 DCs per well) for 5 days, in the presence of a constant number of CFSE+ T cells (105 cells/well) purified from a different donor (allogeneic), in α-MEM medium containing 10% FCS. Cells were then harvested after 5 days of culture, and expression of CFSE was quantified on an LSRII and analyzed using FlowJo software. The total number of CFSE-diminished daughter T cells per well was quantified by a time-monitored flow cytometry analysis during 2 min at high speed (1 µl/s).
Statistical analysis
Statistical analysis of the differences between DCs and DC-17s were performed using LIMMA (21), with Benjamini-Hochberg-corrected P ≤ 0.01 considered statistically significant.
RESULTS
Gene expression profile of IL-17A-treated DCs reveals intense remodeling of lipid metabolism
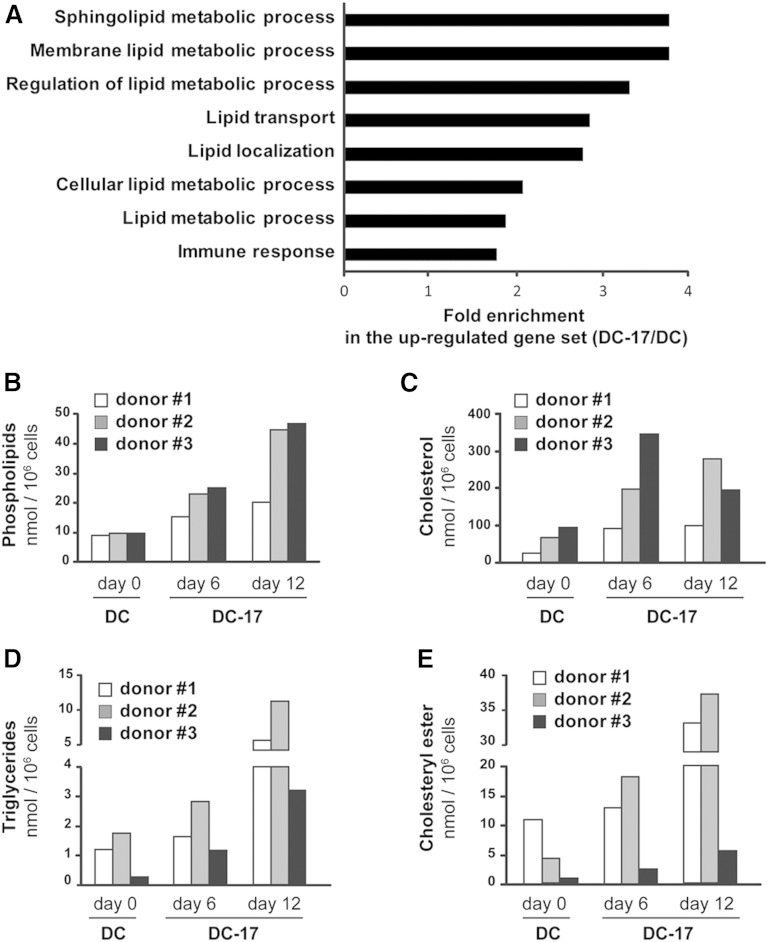
In order to get a comprehensive picture of genes regulated by IL-17A in DCs, we compared the gene expression profile of in vitro-generated monocyte-derived DCs treated (DC-17) or not (DC) with IL-17A using whole human genome microarrays. Analysis performed with LIMMA identified 1,184 significantly upregulated probe sets (fold-change ≥2 and P ≤ 0.01) and 937 significantly downregulated probe sets (fold-change ≤0.5 and P ≤ 0.01) in DC-17s compared with DCs (supplementary Table 1). To identify if any GO classes were enriched in these two sets of genes, analysis were performed using DAVID. We found a total of eight significantly enriched GO “biological process” categories with a P value cutoff of 0.05 in the upregulated gene set (Fig. 1A). Strikingly, only one out of the eight highlighted biological processes was linked to the “immune response.” The remaining seven significantly enriched pathways were all directly related to lipid metabolism, arguing for a strong modification of lipid metabolism in DC-17s. Regarding the downregulated gene set, none of the GO classes were enriched with a P value cutoff of 0.05 (data not shown). The seven lipid-related GO biological pathways included 117 probe sets corresponding to 70 genes (supplementary Table 2). The top 50% of genes showing the greatest fold change in DC-17s compared with DCs are shown in Table 1. Those genes encoded proteins involved in various aspects related to lipid metabolic processes and their regulation, lipid transport and localization, as well as sphingolipid metabolism. These results reveal a new and unexpected strong impact of IL-17A on lipid metabolism.
Fig. 1.
Role of IL-17A treatment on lipid metabolism of DCs. A: GO “biological processes” with significant overrepresentation among the genes significantly upregulated in DC-17s (n = 5) versus untreated DCs (n = 4) (P ≤ 0.01 and expression fold change ≥2). Assignment to GO groups was possible for 853 out of 1,184 probe sets overexpressed in DC-17s versus DCs. The fold enrichment for the eight significantly enriched pathways (P < 0.05) is presented. B–E: Total lipids were extracted from DCs of three different donors, either untreated at day 0 (DC) or treated with IL-17A at indicated time points (DC-17). The different lipid classes were separated by thin-layer chromatography, and the amount of phospholipids, cholesterol, triglycerides, and cholesteryl ester was analyzed by GC or by GC/MS in DCs.
TABLE 1.
Upregulation of lipid-related genes in DC-17
| Probeset ID | Gene Symbol | Gene Name | GO Pathways | Mean Expression ± SD in DCs | Mean Expression ± SD in DC-17s | Fold Change |
| 211748_x_at | PTGDS | Prostaglandin D2 synthase | (1, 2) | 219 ± 41 | 31,871 ± 9,162 | 146 |
| 212884_x_at | APOE | Apolipoprotein E | (1–3, 5, 6) | 725 ± 618 | 35,527 ± 7,100 | 49 |
| 207092_at | LEP | Leptin | (1–3) | 5 ± 2 | 196 ± 184 | 40 |
| 213553_x_at | APOC1 | Apolipoprotein C-I | (1–3, 5, 6) | 1,107 ± 1,334 | 43,123 ± 11,867 | 39 |
| 203979_at | CYP27A1 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | (1) | 273 ± 232 | 10,520 ± 1,104 | 39 |
| 208792_s_at | CLU | Clusterin | (1, 5, 6) | 151 ± 35 | 4,454 ± 2,000 | 30 |
| 218922_s_at | LASS4 | LAG1 homolog, ceramide synthase 4 | (1, 2, 4, 7) | 6 ± 1 | 166 ± 71 | 28 |
| 202575_at | CRABP2 | Cellular retinoic acid binding protein 2 | (1, 2) | 647 ± 934 | 13,916 ± 10,024 | 22 |
| 203920_at | NR1H3 | Nuclear receptor subfamily 1, group H, member 3 | (1–3) | 414 ± 316 | 8,599 ± 3,457 | 21 |
| 228716_at | THRB | Thyroid hormone receptor, β | (3) | 49 ± 30 | 607 ± 359 | 13 |
| 211026_s_at | MGLL | Monoglyceride lipase | (1) | 666 ± 436 | 7,799 ± 1,620 | 12 |
| 223432_at | OSBP2 | Oxysterol binding protein 2 | (1, 5, 6) | 17 ± 7 | 175 ± 104 | 11 |
| 204561_x_at | APOC2 | Apolipoprotein C-II | (1–3, 5, 6) | 783 ± 650 | 8,095 ± 4,286 | 10 |
| 203423_at | RBP1 | Retinol binding protein 1, cellular | (1, 2) | 154 ± 90 | 1,433 ± 460 | 9 |
| 202481_at | DHRS3 | Dehydrogenase/reductase (SDR family) member 3 | (1, 2) | 203 ± 146 | 1,865 ± 621 | 9 |
| 223952_x_at | DHRS9 | Dehydrogenase/reductase (SDR family) member 9 | (1, 2) | 509 ± 521 | 4,517 ± 2,390 | 9 |
| 209785_s_at | PLA2G4C | Phospholipase A2, group IVC | (1, 2) | 324 ± 338 | 2,730 ± 879 | 8 |
| 209122_at | PLIN2 | Perilipin 2 | (5, 6) | 1,808 ± 681 | 14,885 ± 5,347 | 8 |
| 225847_at | KIAA1363 | Arylacetamide deacetylase-like 1 | (1) | 950 ± 922 | 7,426 ± 1,463 | 8 |
| 210942_s_at | SIAT10 | ST3 β-galactoside α-2,3-sialyltransferase 6 | (1, 2, 4) | 275 ± 178 | 2,061 ± 412 | 8 |
| 1555416_a_at | ALOX15B | Arachidonate 15-lipoxygenase, type B | (1, 2) | 63 ± 15 | 439 ± 262 | 7 |
| 218099_at | HT008 | Testis expressed 2 | (1, 2, 4, 7) | 742 ± 153 | 5,003 ± 1,727 | 7 |
| 205934_at | PLCL1 | Phospholipase C-like 1 | (1) | 270 ± 135 | 1,788 ± 841 | 7 |
| 228713_s_at | DHRS10 | Hydroxysteroid (17-β) dehydrogenase 14 | (1) | 201 ± 71 | 1,238 ± 448 | 6 |
| 238524_at | CEPT1 | Choline/ethanolamine phosphotransferase 1 | (1, 2) | 137 ± 103 | 840 ± 172 | 6 |
| 235678_at | GM2A | GM2 ganglioside activator | (1, 2, 4–7) | 3,285 ± 2,814 | 18,962 ± 5,607 | 6 |
| 227379_at | OACT1 | Membrane-bound O-acyltransferase domain containing 1 | (1, 2) | 285 ± 144 | 1,601 ± 899 | 6 |
| 201050_at | PLD3 | Phospholipase D family, member 3 | (1) | 3,941 ± 3,361 | 21,463 ± 2,991 | 5 |
| 200785_s_at | LRP1 | Low-density lipoprotein-related protein 1 | (1) | 463 ± 368 | 2,436 ± 806 | 5 |
| 220675_s_at | C22orf20 | Patatin-like phospholipase domain containing 3 | (1, 2) | 45 ± 13 | 182 ± 37 | 4 |
| 207275_s_at | ACSL1 | Acyl-CoA synthetase long-chain family member 1 | (1–3) | 1,368 ± 590 | 5,473 ± 486 | 4 |
| 1552637_at | PTPN11 | Protein tyrosine phosphatase, nonreceptor type 11 | (1, 2) | 12 ± 4 | 47 ± 9 | 4 |
| 208771_s_at | LTA4H | Leukotriene A4 hydrolase | (1, 2) | 4,728 ± 716 | 17,889 ± 3,425 | 4 |
| 203505_at | ABCA1 | ATP-binding cassette, subfamily A (ABC1), member 1 | (1, 5, 6) | 1,965 ± 476 | 7,339 ± 783 | 4 |
| 221675_s_at | CHPT1 | Choline phosphotransferase 1 | (1, 2) | 1,600 ± 343 | 5,964 ± 604 | 4 |
List of the top 50% of human genes from microarray analyses, showing the greatest mRNA expression fold change in DC-17s compared with untreated DCs among all genes found in the seven lipid-related GO pathways. The GO pathways are as follows: (1) lipid metabolic process, (2) cellular lipid metabolic process, (3) regulation of lipid metabolic process, (4) membrane lipid metabolic process, (5) lipid localization, (6) lipid transport, and (7) sphingolipid metabolic process. When several probe sets were available for a given gene, the probe set with the most significant overexpression in DC-17s versus DCs was selected. The fold change DC-17/DC was calculated from the mean expression in DC-17s (n = 5) and DCs (n = 4).
All lipid species are increased in DC-17s
To further explore the biological relevance of transcriptomic analysis, we analyzed the lipid content of DC-17s for 6 and 12 days using thin-layer chromatography and GC or GC/MS. DC-17s from three different donors had marked increased amounts of all the lipids species analyzed (i.e., phospholipids, cholesterol, triglycerides, and cholesteryl esters) compared with DCs (Fig. 1B–E). Kinetic study revealed that after 6 days of culture with IL-17A, all lipid species were increased compared with DCs from the three donors. In addition, between day 6 and 12, phospholipids, triglycerides, and cholesteryl esters were further augmented in DC-17s from all donors (Fig. 1B, D, E) while the amount of cholesterol was differentially regulated from one donor to another, showing stability, increase, or decrease in donor 1, 2, and 3, respectively (Fig. 1C). Overall, at the optimum time point and depending on the donor, phospholipid levels were increased 2–5 times, cholesterol levels 2–4 times, triglyceride levels 5–12 times, and cholesteryl ester levels 3–9 times.
Quantities of palmitic, stearic, and oleic acid are strongly increased in DC-17s while their relative proportion is stable
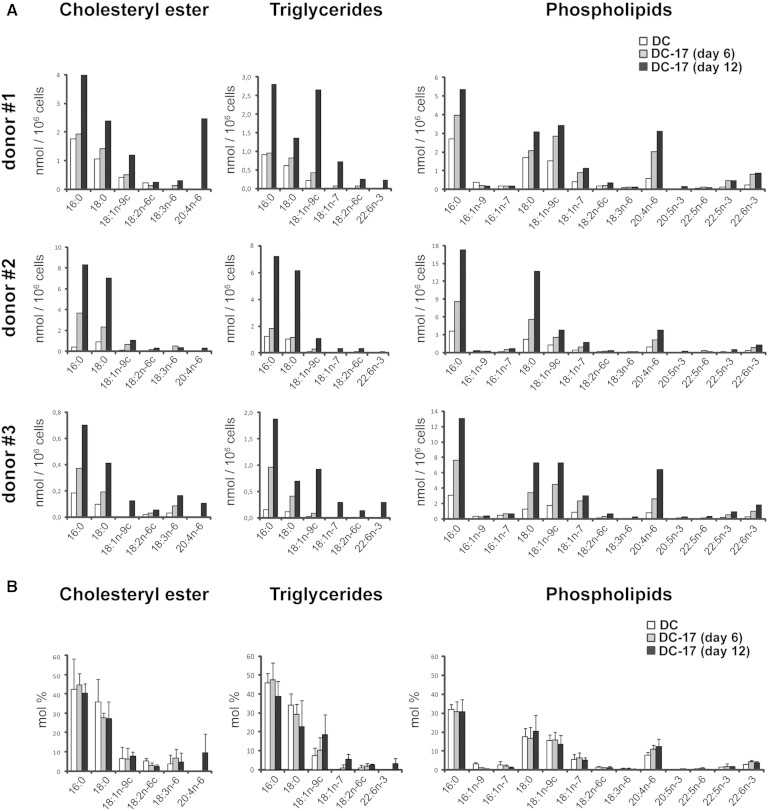
We then investigated whether IL-17A could influence the fatty acid composition of phospholipids, triglycerides, and cholesteryl esters. Similarly to DCs, palmitic (16:0), stearic (18:0), and oleic (18:ln-9c) acid were the main fatty acid chains present in DC-17s from three donors (Fig. 2A), each accounting for ∼40%, 25%, and 10% of total fatty acids, respectively (Fig. 2B). There was at least a 2-fold increase (ranging from 2-fold in phospholipids of donor 1 to 19-fold in cholesteryl ester from donor 2) in the amount of palmitic acid (16:0) in DC-17s compared with DCs from the three donors, regardless of the class of lipid (Fig. 2A). Similarly, an increase in the amount of stearic acid (18:0), ranging from 1.8-fold in phospholipids of donor 1 to 8-fold in cholesteryl ester from donor 2, was observed in DC-17s from the three donors (Fig. 2A). Oleic acid (18:ln-9c) was augmented in all three DC-17s, in particular in triglycerides where it was induced 10 to 30 times depending on the donor (Fig. 2A). Although the amounts of palmitic, stearic, and oleic acids were increased in response to IL-17A, overall, their relative proportion remained unchanged (Fig. 2B). Therefore, this lipidomic analysis demonstrates that IL-17A highly increases the amount of all fatty acids present in DCs with a conserved composition and a variable intensity depending on the donor.
Fig. 2.
Role of IL-17A treatment on fatty acid composition of DCs. Total lipids were extracted from DCs of three different donors, either untreated at day 0 (DC) or treated with IL-17A for 6 and 12 days (DC-17). The different lipid classes were separated by thin-layer chromatography, and the amount of phospholipids, cholesterol, triglycerides, and cholesteryl ester was analyzed by GC or by GC/MS. A: Results are expressed as nmol of the fatty acid normalized per million cells. B: Results are expressed as mole percentages (mol %) of the selected fatty acids in cholesteryl ester, triglycerides, and phopholipids and are means ± SD of results from the three donors presented in A. Only the main fatty acids are presented.
DC-17s become lipid-laden foamy DCs
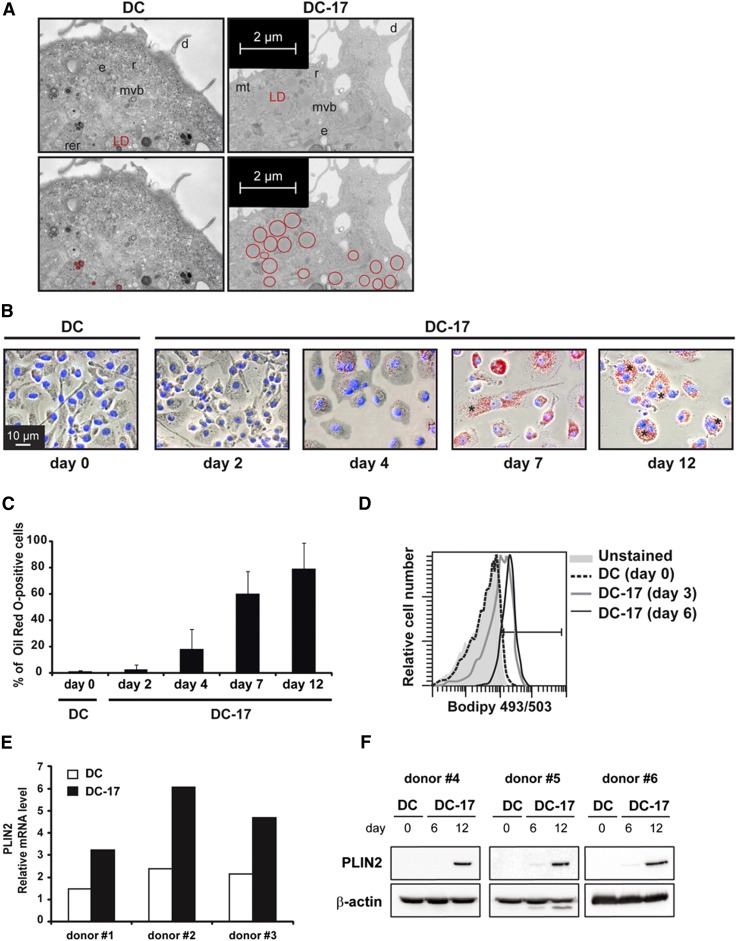
Based on the increased amount of neutral lipids and phospholipids in DC-17s, we suspected that exposure to IL-17A would lead to the generation of foamy cells. Electron microscopy analysis of monocyte-derived DCs cultured for 10 days with IL-17A revealed the presence of numerous lipid droplets (LDs) in the cytoplasm while very few LDs were visible in untreated DCs (Fig. 3A). The LDs found in DC-17s were bigger (mean diameter = 0.4 µm) than those of untreated DCs (mean diameter = 0.1 µm). To investigate the kinetics of LD formation, cells were observed at days 0, 2, 4, 7, and 12 of culture with IL-17A following staining with Oil Red O, a dye specific for neutral lipids. In agreement with electron microscopy data, the vast majority of DCs contained few LDs at day 0 (Fig. 3B, C). After 2 days of culture with IL-17A, 2.5% of DCs accumulated Oil Red O-positive LDs. The amount of LDs inside cells, as well as the percentage of Oil Red O-positive cells, gradually increased over time: ∼80% of the cells were full of LDs after 12 days of culture (Fig. 3C). High amounts of LDs were found in binucleated cells (Fig. 3B) and multinucleated giant cells with more than two nuclei (not shown, <10% of total cells at day 12) resulting from IL-17A-induced cell fusion. Binucleated cells accounted for 10% and 20% of total cells at days 4 and 12, respectively. The increased amount of lipids in DC-17s and percentages of lipid-rich DCs were further confirmed by flow cytometry, using the lipophilic fluorescent dye Bodipy 493/503 (Fig. 3D). In agreement with Oil Red O staining data, IL-17A treatment markedly increased the lipid content after 3 and 6 days of culture. Perilipins are a family of proteins that associate with the surface of LDs. The PLIN 2 gene was upregulated in DC-17s versus DCs, as observed in microarray data (Table 1) and validated by real-time RT-PCR (Fig. 3E). PLIN2 was also induced at the protein level after 12 days of culture with IL-17A (Fig. 3F). In addition, we analyzed the expression of cytokines known to regulate the formation of foamy macrophages to look for cytokine-mediated indirect effect of IL-17A on DCs. IL-1β was 4-fold upregulated in DC-17s versus DCs, but with a weak expression level, and no significant change was observed for TNF-α, IL-6, transforming growth factor (TGF)-β1, IL-10, IFN-γ, and IL-33 expression (supplementary Table 3). Therefore, IL-17A induces the generation of foamy DCs full of LDs without inducing expression of other cytokines involved in the regulation foam cell formation, except a weak level of IL-1β.
Fig. 3.
Characterization of LD content in DC-17s. A: Electron microscopy pictures of DCs versus DC-17s following 10 days of treatment with IL-17A. The upper and lower panels represent the same images, with LDs circled in red in the lower ones. d, dendrite; e, endosome; mt, mitochondria; mvb, multivesicular body; r, ribosome area; rer, rough endoplasmic reticulum. Representative of three experiments. B: Representative images of Oil Red O staining from a kinetic study of untreated DCs at day 0 (DC) or DC-17s at indicated time points (DC-17). Red, Oil Red O-positive cells; blue, Hoechst nucleus staining. Representative of five experiments. * indicates binucleated cells. C: Percentage of Oil Red O-positive DCs at day 0 (DC) or at indicated time points during IL-17A treatment of DCs. The bars represent means ± SD of five independent experiments. D: Flow cytometry analysis of untreated DCs at day 0 (DC) or DC-17s, before (Unstained) or after Bodipy staining realized at indicated time points. Representative of three experiments. E: Relative mRNA expression of PLIN2 in DC-17s treated with IL-17A for 12 days compared with untreated DCs from three different donors. mRNAs were quantified by RT-qPCR. F: Western blot analysis of PLIN2 (48 kDa) in untreated DCs at day 0 (DC) or DC-17s treated with IL-17A for 6 and 12 days on three independent donors. β-actin (45 kDa) was used as a loading control.
DC-17s uptake lipids from the microenvironment
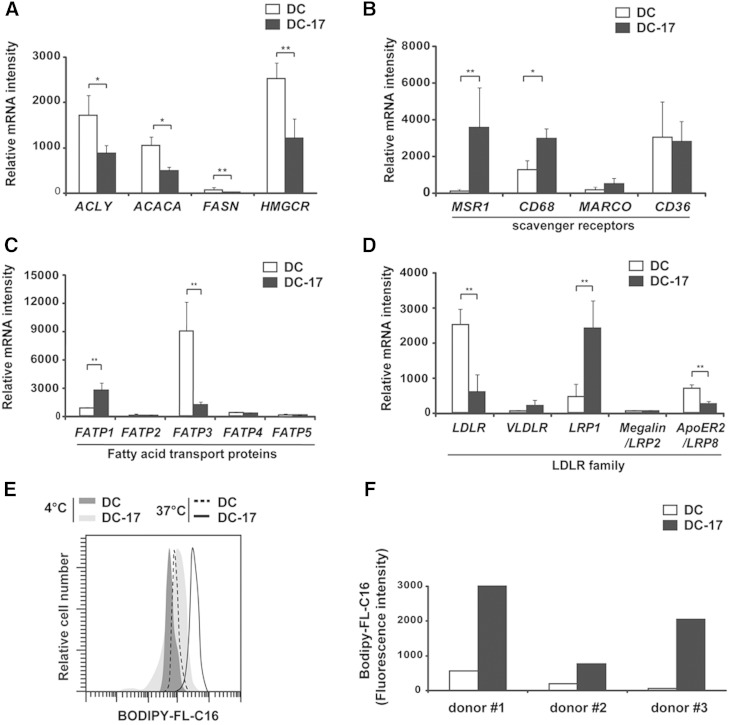
We then investigated the origin of the lipids accumulated in DC-17s. As shown in Fig. 4A, genes encoding the key metabolic enzymes of lipid synthesis [i.e., ATP citrate lyase (ACLY), acetyl-CoA carboxylase α (ACACA), fatty acid synthase (FASN), and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR)] were significantly downregulated in DC-17s versus DCs. This suggested that lipid synthesis was not the main mechanism responsible for IL-17A-induced lipid accumulation in human DCs. Thus, we postulated that DC-17s have acquired increased ability to uptake lipids from the microenvironment. Using Affymetrix microarrays, we found that the scavenger receptors MSR1 and CD68 were significantly overexpressed in DC-17s compared with DCs (Fig. 4B). MARCO and CD36 were not significantly upregulated in DC-17s. Among the six fatty acid transport proteins (FATPs) and seven members of the low density lipoprotein receptor (LDLR) family, FATP1 and LDLR-related protein 1 (LRP1) were significantly overexpressed in DC-17s versus DCs (Fig. 4C, D).
Fig. 4.
Functional phenotype of molecules involved in lipid anabolism and lipid uptake in DC-17s. A–D: mRNA expression measured by Affymetrix microarray in untreated DCs at day 0 (n = 4) and DC-17s treated with IL-17A for 12 days (n = 5). Results are means ± SD. Statistical significance was determined with LIMMA.* P < 0.05; ** P < 0.01 for enzymes of fatty acid synthesis (A), scavenger receptors (B), fatty acid transport proteins (C), and LDLR-family members (D). E–F: Flow cytometry analysis of fatty acid uptake in DCs from three donors. Untreated DCs at day 0 (DC) or DC-17s cultured for 5 days with IL-17A were incubated with fluorescently labeled C16 fatty acid (Bodipy-FL-C16) for 30 min at 4°C or 37°C. E: Representative staining for donor 1. F: Shift in the Bodipy-FL-C16 mean fluorescence intensity between 37°C and 4°C for three donors in separate experiments.
To investigate if DC-17s had an increased ability to capture lipids from the microenvironment, we measured cellular uptake of fatty acids using the fluorescently labeled palmitic acid Bodipy-FL-C16. Untreated DCs or DCs treated for 5 days with IL-17A were incubated with Bodipy-FL-C16 for 10 min at 37°C or 4°C, and intracellular fluorescence was measured by flow cytometry. DC-17s displayed higher amounts of intracellular Bodipy-FL-C16 than DCs (Fig. 4E). The ability of DCs to uptake the fatty acid Bodipy-FL-C16 was estimated on three different donors from the shift in the mean fluorescence intensity between 4°C and 37°C, which was markedly enhanced by IL-17A treatment (Fig. 4F). Altogether, those data suggest that increased lipid uptake is the main mechanism responsible for lipid accumulation in IL-17A-induced foamy DCs.
IL-17A-induced foamy DCs acquire macrophage markers and keep their immunogenic properties
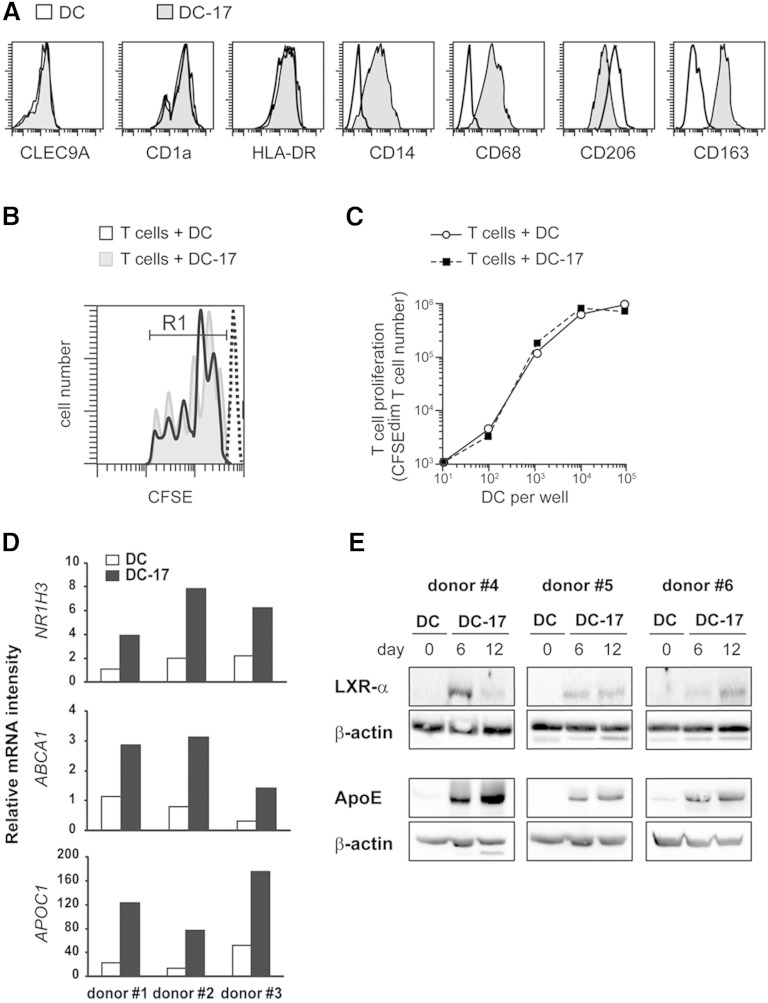
Then, we performed a phenotypic, genetic, and functional characterization of human monocyte-derived foamy DCs obtained with IL-17A. At day 6 of IL-17A treatment, DC-17s did not express CLEC9A, but they were positive for CD1a, HLA-DR, CD14, CD68, CD206, and CD163 expression (Fig. 5A). Compared with expression of these markers on monocytes, monocyte-derived macrophages, and monocyte-derived DCs, we concluded that DC-17s concomitantly expressed DCs and macrophage markers (Table 2), thus raising the question of the appropriate name for these foamy myeloid cells. DCs have been originally differentiated from macrophages by their ability to activate naïve T-cell proliferation in coculture with allogeneic T cells (23). In cocultures of DCs and CFSE+ T cells from different donors, DC-17s activated the proliferation of T cells, which underwent up to five cell cycles (Fig. 5B). Numeration of CFSE-diminished daughter T cells demonstrated that allogeneic T-cell proliferation obtained with IL-17A-induced foamy DCs was similar as that obtained with untreated DCs (Fig. 5C). Therefore, we propose the name of foamy DCs for the IL-17A-treated DCs characterized by a mixed DC/macrophage phenotype and the ability to stimulate allogeneic T cell proliferation.
Fig. 5.
Analyses of phenotype, specific genetic program, and immunogenicity of DC-17s. A: Flow cytometry analysis of the expression of CLEC9A, CD1a, HLA-DR, CD14, CD68, CD206, and CD163 in DCs and after 6 days of culture with IL-17A. Representative of n > 5 experiments. B, C: Untreated DCs versus DC-17s treated with IL-17A for 5 days were cultured for 5 additional days in the presence of CFSE-labeled T cells purified from allogeneic donors. At day 5, the decrease of CFSE fluorescence in T cells was measured by flow cytometry and compared with parental CFSE+ T cells at day 0 (dashed line). B: Individualized pics for each cell division are shown. C: The number of CFSE-diminished T cells represents the progeny of CFSE+ T cells, in the presence of increased number of allogeneic DC. Results are those of one experiment representative of two. D: Relative mRNA expression of NR1H3, ABCA1, and APOC1 in DC-17s treated with IL-17A for 12 days compared with untreated DCs at day 0 from three different donors. mRNAs were quantified by RT-qPCR. E: Western blot analysis of LXR-α (from NR1H3 gene, 50 kDa) and APOE (38 kDa) in untreated DCs at day 0 or DC-17s treated with IL-17A for 6 and 12 days on three independent donors. β-actin (45 kDa) was used as a loading control.
TABLE 2.
Phenotype of in vitro-generated myeloid cells
| Mo | MP | DC | DC-17 | |
| CLEC9A | - | - | - | - |
| CD1a | - | - | ++ | ++ |
| HLA-DR | + | + | ++ | ++ |
| CD14 | ++ | + | - | + |
| CD68 | + | ++ | - | + |
| CD206 | - | ++ | ++ | + |
| CD163 | + | + | - | + |
Monocytes (Mo), monocyte-derived macrophages (MP), and monocyte-derived DCs before (DC) and after (DC-17) 6 days of treatment with IL-17A. Expression of the indicated markers was analyzed on Mo, MP, DC, and DC-17 by flow cytometry. Representative of n > 3 experiments. -, absence of marker expression compared with isotype control; + and ++, low versus high positive expression, according to the mean fluorescence intensity.
IL-17A activates the LXR-α genetic program in DCs
The nuclear receptor LXR-α/NR1H3 is a key regulator of foamy macrophage function. LXR-α controls transcriptional programs involved in the regulation of lipid homeostasis in response to rapid changes in cellular lipids and inflammation (24). Interestingly, one of the top 50% of genes upregulated in DC-17s versus DCs was NR1H3 (Table 1). NR1H3 was 21-fold higher in DC-17s (mean value = 8,599) than that in untreated DCs (mean value = 414; Table 1). Affymetrix data were confirmed by RT-qPCR (Fig. 5D) and by Western blot (Fig. 5E) on three independent donors for each experiment. LXR-α protein expression was induced after 6 days of culture with IL-17A and still maintained at day 12 (Fig. 5E). Furthermore, the expression of several NR1H3 target genes such as ABCA1, a cholesterol transporter, or APO, the structural components of lipoprotein particles, was also increased in DC-17s versus DCs (Table 1). Those data were also validated at the mRNA level (ABCA1 and APOC1; Fig. 5D) and at the protein level (APOE; Fig. 5E). Thus, the LXR-α genetic program is active in IL-17A-induced foamy DCs, as previously established in foamy macrophages.
DISCUSSION
Immunometabolism is an emerging field of investigation at the interface between immunological and metabolic processes. Deregulation of intracellular lipid metabolism has been extensively studied in foamy macrophages in the context of atherosclerosis (4). However, much less is known regarding DCs. Here we show for the first time that in vitro-generated monocyted-derived DCs respond to the proinflammatory cytokine IL-17A by modulating their lipid metabolism thus generating foamy DCs, in vitro.
We report an intense remodeling of lipid metabolism induced by IL-17A in DCs: i) several genes involved in lipid metabolism were upregulated; ii) all the analyzed lipid species were quantitatively increased with a qualitatively stable composition of fatty acid chains; and iii) LDs accumulated in the cytoplasm. Regarding those intracellular metabolic aspects, foamy DCs resemble foamy macrophages characterized in atheroma. In atherosclerosis, lipid overload under the form of LDL is a risk factor because chronic inflammation oxidizes LDLs that are specifically captured by macrophages through the scavenger receptors MSR1 and CD36 (4), converting those cells into foamy macrophages. In DCs, IL-17A upregulated the scavenger receptors MSR1 and CD68 but also the fatty acid transporter FATP1, suggesting that the main mechanism supporting lipid accumulation was an increased capture of lipids from the microenvironment. This hypothesis was validated by the enhanced uptake of the fatty acid FL-C16 by DCs in response to IL-17A. Thus, foamy DC formation in response to IL-17A comes from capture of extracellular lipids. IL-17A-induced foamy DCs were generated in long-term cultures and may be indirectly mediated by a cytokine cascade. Several cytokines were previously shown to modulate the formation of foamy macrophages generated in the presence of aggregated, acetylated or oxidized LDL. IFN-γ, IL-1β, and TNF-α promote macrophage foam cell formation (25–28), while IL-6, TGF-β1, and IL-33 (29–31) have been described as anti-foam cell cytokines in humans and IL-10 facilitates both cholesterol uptake and efflux in macrophages (32). Only IL-1β expression was weakly affected by IL-17A treatment, suggesting a role for this cytokine, which may be further enhanced by inflammasome activation, in vivo. However, it was recently demonstrated that fatty acid-induced mitochondrial uncoupling abrogated IL-1β secretion, deviating the cholesterol crystal-elicited response toward selective production of IL-1α (33). Therefore, IL-17A induction of foamy DCs rather results from direct genetic reprogramming of lipid metabolism than on indirect cascade of cytokines, though we cannot totally exclude indirect effects via other unidentified products secreted by IL-17A-stimulated DCs.
The nuclear receptor LXR-α/NR1H3 is a sterol sensor that controls the regulation of lipid homeostasis. LXR-α activation is a hallmark of foamy macrophages (24). DC expression of LXR-α was dramatically increased by IL-17A, both at the mRNA and protein levels. LXR-α target genes such as ABCA1 and APOC1, APOC2, and APOE (34) were strongly upregulated, indicating that the LXR-α transcriptional function is active when DCs are treated by IL-17A. LXR-α activation takes place upon ligand binding that leads to a molecular switch replacing a corepressor by a coactivator complex (35). Previous studies showed that adding exogenous synthetic LXR ligands activated the LXR program in monocyte-derived DCs (36, 37). However, our data provide first evidence that LXR-α can be transcriptionally active in the presence of IL-17A without exogenously added synthetic ligands. This implies that natural endogenous ligands, which remain to be determined, are probably generated in response to IL-17A.
Interestingly, the role of IL-17A has been investigated in two mouse models of atherosclerosis, the Apoe−/− and the Ldlr−/− model, with opposite conclusions. In the Apoe−/− model, in vivo administration of an antibody blocking IL-17A decreased atherosclerotic phenotype suggesting that IL-17A is proatherogenic, independently of APOE (38). In Ldlr−/− mice, neutralizing anti-IL-17A antibodies had no effect. Based on prior studies demonstrating that Socs3 negatively regulates IL-17A expression in T cells (39), Socs3−/−Ldlr−/− chimeric mice were generated and had reduced atherogenesis (40). Anti-IL-17A antibody treatment or IL-17A deficiency increased plaque formation in those mice, suggesting that IL-17A may be antiatherogenic when the Apoe gene is functional (41, 42). Assuming that IL-17A-dependent foamy DCs are physiologically relevant, our in vitro data provide knowledge to solve the apparent discrepancies on the role of IL-17A in atherosclerosis mouse models. We demonstrate that IL-17A strongly induces APOE, an apolipoprotein involved in HDL formation allowing the reverse transport of cholesterol to the liver and thereby limiting atherosclerosis. Accordingly, IL-17A may sustain two antagonistic functions in atherogenesis: the proinflammatory role of IL-17A would promote plaque formation while the IL-17A-induced APOE expression would counteract plaque formation. In the Apoe−/− mice, only the first function can be active and may explain the major proatherogenic role of IL-17A. In the Ldlr−/− mouse model, IL-17A would exert both functions and the second function may counteract proinflammatory one.
The origin of foamy cells in atherosclerosis should be questioned: do they belong to macrophage or DC lineage? Historically, DCs have been functionally defined by their original ability to efficiently stimulate allogeneic T-cell proliferation (23). As this property is maintained in IL-17A-induced foamy cell generated in vitro from monocyte-derived DCs, we propose to call these cells “foamy DCs.” However, we show that IL-17A induces the expression of the macrophage markers CD14, CD68, and CD163 on foamy DCs. In addition, the M2 macrophage marker CD206 is expressed on both DCs and DC-17s. Finally, CLEC9A (also known as DNGR-1), a marker of the BDCA3+ human conventional DC subset, is not expressed by monocyte-derived DCs, as previously described (43). In vitro microarray studies showed that in response to oxidized LDL, monocyte-derived foamy macrophages may acquire a DC-like gene expression pattern (44). So, the exact nature of foamy myeloid cells in atherosclerosis remains an intriguing question, which cannot be solved by in vitro experiments. In vivo, foam cell formation and atherosclerotic plaque growth in the artery was first attributed to foamy macrophages defined as fat-laden myeloid cells expressing macrophage markers (F4/80 in mice and CD68 in humans) (45). However, a recent study using the Ldlr−/− mouse model have demonstrated that the majority of intimal lipids in nascent lesions were located within foam cells that express CD11c (5), a marker widely used as a specific marker for murine DCs. CD11c is in fact also expressed by several tissue macrophages (46) as well as monocytes in models of atherosclerosis (47). CD11c+ circulating monocytes can be activated by intracellular lipid accumulation prior to their recruitment to athero-prone regions of the vasculature, confusing the issue of what are CD11c+ foam cells (47). It is not possible to determine whether foamy cells originate from macrophages or DC lineage based on phenotypical analysis. To understand whether foamy DCs exist in vivo, it would be necessary to perform foam cell purification in mouse model of atherosclerosis, treated or not with anti-IL-17A, for ex vivo evaluation of their alloreactive properties. Our in vitro data on the generation of LXR-α+ APOE+ alloreactive foamy DCs in the presence of IL-17A, together with the role of IL-17A in different mouse models of atherosclerosis in vivo, suggest that different subpopulations of foamy macrophages and foamy DCs exist in atherosclerosis. Knowing that TH17 and other IL-17A-positive cells participate in the atherosclerotic-associated inflammation in human arteries (13), IL-17A may participate in the generation of foamy DCs in atherosclerosis. Whether these foamy DCs really exist in vivo as a separate and relevant myeloid entity will have to be addressed in the future.
IL-17A is involved in the pathogenesis of several chronic inflammatory diseases, not only those associated with metabolic disorders such as atherosclerosis, type 2 diabetes mellitus, and obesity, but also cancer and tuberculosis where foam cells were characterized (7, 48). In addition to its proinflammatory functions, IL-17A may participate in the generation of foamy DCs in various chronic inflammatory contexts, in vivo.
Supplementary Material
Acknowledgments
For stimulating discussions on interfacing disciplines, in memoriam of C. Rabourdin-Combe. The authors thank UMS3444/US8 for the platforms PLATIM imaging and flow cytometry, ProfileXpert for array analysis (http://www.profilexpert.fr), and Hélène Valentin for helpful discussions and critical reading of the manuscript.
Footnotes
Abbreviations:
- CFSE
- carboxyfluorescein_succinimidyl_ester
- CLEC9A
- C-type lectin domain family 9A
- DC
- dendritic cell
- DC-17
- IL-17A-treated DC
- FATP
- fatty acid transport protein
- GO
- gene ontology
- IL
- interleukin
- LD
- lipid droplet
- LDLR
- LDL receptor
- LIMMA
- linear models for microarray data
- LXR
- liver X receptor
- PLIN2
- perilipin 2
This work was supported by grants from (France) CNRS, INSERM, Université de Lyon 1, Institut Universitaire de France, Fondation pour l’Innovation et la Valorisation en Infectiologie, ANR Microbiologie-Immunologie-Environnement, LyonBiopole, Etablissement Français du Sang EFS-Alsace, and ARMESA (Association de Recherche et de Développement en Médecine et Santé Publique); and from (Italy) Associazione Italiana Ricerca Istiocitosi (AIRI).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Nikolajczyk B. S., Jagannathan-Bogdan M., Denis G. V. 2012. The outliers become a stampede as immunometabolism reaches a tipping point. Immunol. Rev. 249: 253–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathis D., Shoelson S. E. 2011. Immunometabolism: an emerging frontier. Nat. Rev. Immunol. 11: 81–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller Y. I., Chang M. K., Binder C. J., Shaw P. X., Witztum J. L. 2003. Oxidized low density lipoprotein and innate immune receptors. Curr. Opin. Lipidol. 14: 437–445. [DOI] [PubMed] [Google Scholar]
- 4.Hansson G. K., Robertson A. K., Soderberg-Naucler C. 2006. Inflammation and atherosclerosis. Annu. Rev. Pathol. 1: 297–329. [DOI] [PubMed] [Google Scholar]
- 5.Paulson K. E., Zhu S. N., Chen M., Nurmohamed S., Jongstra-Bilen J., Cybulsky M. I. 2010. Resident intimal dendritic cells accumulate lipid and contribute to the initiation of atherosclerosis. Circ. Res. 106: 383–390. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim J., Nguyen A. H., Rehman A., Ochi A., Jamal M., Graffeo C. S., Henning J. R., Zambirinis C. P., Fallon N. C., Barilla R., et al. 2012. Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in mouse and human liver. Gastroenterology. 143: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herber D. L., Cao W., Nefedova Y., Novitskiy S. V., Nagaraj S., Tyurin V. A., Corzo A., Cho H. I., Celis E., Lennox B., et al. 2010. Lipid accumulation and dendritic cell dysfunction in cancer. Nat. Med. 16: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cua D. J., Tato C. M. 2010. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10: 479–489. [DOI] [PubMed] [Google Scholar]
- 9.Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., et al. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miossec P., Kolls J. K. 2012. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 11: 763–776. [DOI] [PubMed] [Google Scholar]
- 11.Winer S., Paltser G., Chan Y., Tsui H., Engleman E., Winer D., Dosch H. M. 2009. Obesity predisposes to Th17 bias. Eur. J. Immunol. 39: 2629–2635. [DOI] [PubMed] [Google Scholar]
- 12.Shin J. H., Shin D. W., Noh M. 2009. Interleukin-17A inhibits adipocyte differentiation in human mesenchymal stem cells and regulates pro-inflammatory responses in adipocytes. Biochem. Pharmacol. 77: 1835–1844. [DOI] [PubMed] [Google Scholar]
- 13.Butcher M., Galkina E. 2011. Current views on the functions of interleukin-17A-producing cells in atherosclerosis. Thromb. Haemost. 106: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright J. F., Bennett F., Li B., Brooks J., Luxenberg D. P., Whitters M. J., Tomkinson K. N., Fitz L. J., Wolfman N. M., Collins M., et al. 2008. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J. Immunol. 181: 2799–2805. [DOI] [PubMed] [Google Scholar]
- 15.Iwakura Y., Ishigame H., Saijo S., Nakae S. 2011. Functional specialization of interleukin-17 family members. Immunity. 34: 149–162. [DOI] [PubMed] [Google Scholar]
- 16.Coury F., Annels N., Rivollier A., Olsson S., Santoro A., Speziani C., Azocar O., Flacher M., Djebali S., Tebib J., et al. 2008. Langerhans cell histiocytosis reveals a new IL-17A-dependent pathway of dendritic cell fusion. Nat. Med. 14: 81–87. [DOI] [PubMed] [Google Scholar]
- 17.Olsson Åkefeldt S., Maisse C., Belot A., Mazzorana M., Salvatore G., Bissay N., Jurdic P., Arico M., Rabourdin-Combe C., Henter J. I., et al. 2013. Chemoresistance of human monocyte-derived dendritic cells is regulated by IL-17A. PLoS ONE. 8: e56865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheong C., Matos I., Choi J. H., Dandamudi D. B., Shrestha E., Longhi M. P., Jeffrey K. L., Anthony R. M., Kluger C., Nchinda G., et al. 2010. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 143: 416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segura E., Touzot M., Bohineust A., Cappuccio A., Chiocchia G., Hosmalin A., Dalod M., Soumelis V., Amigorena S. 2013. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 38: 336–348. [DOI] [PubMed] [Google Scholar]
- 20.Fugier-Vivier I., Servet-Delprat C., Rivailler P., Rissoan M. C., Liu Y. J., Rabourdin-Combe C. 1997. Measles virus suppresses cell-mediated immunity by interfering with the survival and functions of dendritic and T cells. J. Exp. Med. 186: 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 23.Steinman R. M., Inaba K. 1985. Stimulation of the primary mixed leukocyte reaction. Crit. Rev. Immunol. 5: 331–348. [PubMed] [Google Scholar]
- 24.Bensinger S. J., Tontonoz P. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 454: 470–477. [DOI] [PubMed] [Google Scholar]
- 25.Brand K., Mackman N., Curtiss L. K. 1993. Interferon-gamma inhibits macrophage apolipoprotein E production by posttranslational mechanisms. J. Clin. Invest. 91: 2031–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N., McLaren J. E., Michael D. R., Clement M., Fielding C. A., Ramji D. P. 2010. ERK is integral to the IFN-gamma-mediated activation of STAT1, the expression of key genes implicated in atherosclerosis, and the uptake of modified lipoproteins by human macrophages. J. Immunol. 185: 3041–3048. [DOI] [PubMed] [Google Scholar]
- 27.Persson J., Nilsson J., Lindholm M. W. 2008. Interleukin-1beta and tumour necrosis factor-alpha impede neutral lipid turnover in macrophage-derived foam cells. BMC Immunol. 9: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei L., Xiong Y., Chen J., Yang J. B., Wang Y., Yang X. Y., Chang C. C., Song B. L., Chang T. Y., Li B. L. 2009. TNF-alpha stimulates the ACAT1 expression in differentiating monocytes to promote the CE-laden cell formation. J. Lipid Res. 50: 1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao H. S., Matsumoto A., Itakura H., Doi T., Honda M., Kodama T., Geng Y. J. 1999. Transcriptional inhibition by interleukin-6 of the class A macrophage scavenger receptor in macrophages derived from human peripheral monocytes and the THP-1 monocytic cell line. Arterioscler. Thromb. Vasc. Biol. 19: 1872–1880. [DOI] [PubMed] [Google Scholar]
- 30.Draude G., Lorenz R. L. 2000. TGF-beta1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am. J. Physiol. Heart Circ. Physiol. 278: H1042–H1048. [DOI] [PubMed] [Google Scholar]
- 31.McLaren J. E., Michael D. R., Salter R. C., Ashlin T. G., Calder C. J., Miller A. M., Liew F. Y., Ramji D. P. 2010. IL-33 reduces macrophage foam cell formation. J. Immunol. 185: 1222–1229. [DOI] [PubMed] [Google Scholar]
- 32.Han X., Kitamoto S., Lian Q., Boisvert W. A. 2009. Interleukin-10 facilitates both cholesterol uptake and efflux in macrophages. J. Biol. Chem. 284: 32950–32958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freigang S., Ampenberger F., Weiss A., Kanneganti T. D., Iwakura Y., Hersberger M., Kopf M. 2013. Fatty acid-induced mitochondrial uncoupling elicits inflammasome-independent IL-1alpha and sterile vascular inflammation in atherosclerosis. Nat. Immunol. 14: 1045–1053. [DOI] [PubMed] [Google Scholar]
- 34.Mak P. A., Laffitte B. A., Desrumaux C., Joseph S. B., Curtiss L. K., Mangelsdorf D. J., Tontonoz P., Edwards P. A. 2002. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J. Biol. Chem. 277: 31900–31908. [DOI] [PubMed] [Google Scholar]
- 35.Nagy L., Schwabe J. W. 2004. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 29: 317–324. [DOI] [PubMed] [Google Scholar]
- 36.Geyeregger R., Zeyda M., Bauer W., Kriehuber E., Saemann M. D., Zlabinger G. J., Maurer D., Stulnig T. M. 2007. Liver X receptors regulate dendritic cell phenotype and function through blocked induction of the actin-bundling protein fascin. Blood. 109: 4288–4295. [DOI] [PubMed] [Google Scholar]
- 37.Töröcsik D., Barath M., Benko S., Szeles L., Dezso B., Poliska S., Hegyi Z., Homolya L., Szatmari I., Lanyi A., et al. 2010. Activation of liver X receptor sensitizes human dendritic cells to inflammatory stimuli. J. Immunol. 184: 5456–5465. [DOI] [PubMed] [Google Scholar]
- 38.Erbel C., Chen L., Bea F., Wangler S., Celik S., Lasitschka F., Wang Y., Bockler D., Katus H. A., Dengler T. J. 2009. Inhibition of IL-17A attenuates atherosclerotic lesion development in apoE-deficient mice. J. Immunol. 183: 8167–8175. [DOI] [PubMed] [Google Scholar]
- 39.Kinjyo I., Inoue H., Hamano S., Fukuyama S., Yoshimura T., Koga K., Takaki H., Himeno K., Takaesu G., Kobayashi T., et al. 2006. Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-beta 1. J. Exp. Med. 203: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taleb S., Romain M., Ramkhelawon B., Uyttenhove C., Pasterkamp G., Herbin O., Esposito B., Perez N., Yasukawa H., Van Snick J., et al. 2009. Loss of SOCS3 expression in T cells reveals a regulatory role for interleukin-17 in atherosclerosis. J. Exp. Med. 206: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taleb S., Tedgui A., Mallat Z. 2010. Interleukin-17: friend or foe in atherosclerosis? Curr. Opin. Lipidol. 21: 404–408. [DOI] [PubMed] [Google Scholar]
- 42.Danzaki K., Matsui Y., Ikesue M., Ohta D., Ito K., Kanayama M., Kurotaki D., Morimoto J., Iwakura Y., Yagita H., et al. 2012. Interleukin-17A deficiency accelerates unstable atherosclerotic plaque formation in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 32: 273–280. [DOI] [PubMed] [Google Scholar]
- 43.Schreibelt G., Klinkenberg L. J., Cruz L. J., Tacken P. J., Tel J., Kreutz M., Adema G. J., Brown G. D., Figdor C. G., de Vries I. J. 2012. The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-)presentation by human blood BDCA3+ myeloid dendritic cells. Blood. 119: 2284–2292. [DOI] [PubMed] [Google Scholar]
- 44.Cho H. J., Shashkin P., Gleissner C. A., Dunson D., Jain N., Lee J. K., Miller Y., Ley K. 2007. Induction of dendritic cell-like phenotype in macrophages during foam cell formation. Physiol. Genomics. 29: 149–160. [DOI] [PubMed] [Google Scholar]
- 45.Kzhyshkowska J., Neyen C., Gordon S. 2012. Role of macrophage scavenger receptors in atherosclerosis. Immunobiology. 217: 492–502. [DOI] [PubMed] [Google Scholar]
- 46.Geissmann F., Gordon S., Hume D. A., Mowat A. M., Randolph G. J. 2010. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 10: 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H., Gower R. M., Wang H., Perrard X. Y., Ma R., Bullard D. C., Burns A. R., Paul A., Smith C. W., Simon S. I., et al. 2009. Functional role of CD11c+ monocytes in atherogenesis associated with hypercholesterolemia. Circulation. 119: 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell D. G., Cardona P. J., Kim M. J., Allain S., Altare F. 2009. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10: 943–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.