Abstract
HDL is typically isolated ultracentrifugally at 40,000 rpm or greater, however, such high centrifugal forces are responsible for altering the recovered HDL particle. We demonstrate that this damage to HDL begins at approximately 30,000 rpm and the magnitude of loss increases in a rotor speed-dependent manner. The HDL is affected by elevated ultracentrifugal fields resulting in a lower particle density due to the shedding of associated proteins. To circumvent the alteration of the recovered HDL, we utilize a KBr-containing density gradient and a lowered rotor speed of 15,000 rpm to separate the lipoproteins using a single 96 h centrifugation step. This recovers the HDL at two density ranges; the bulk of the material has a density of about 1.115 g/ml, while lessor amounts of material are recovered at >1.2 g/ml. Thus, demonstrating the isolation of intact HDL is possible utilizing lower centrifuge rotor speeds.
Keywords: high density lipoprotein/structure, lipoproteins/assembly, lipoproteins/kinetics, analytical ultracentrifugation, density gradient ultracentrifugation, high density lipoprotein isolation, shedding, ultracentrifugation
Lipoproteins are a child of the ultracentrifuge. This instrument, invented in the 1920s (1), could subject solutions to enormous centrifugal fields, with current instruments capable of reaching one million earth gravities with a rotor spinning at 150,000 rpm (2). The development of the ultracentrifuge allowed for the identification of lipoproteins.
John Gofman was a codiscoverer of lipoproteins, particles that float in water or density-enhanced solutions, using NaCl or KBr to increase the density (3). Because lipid is lighter than water, and by adding sufficient lipid to a denser particle, the particle will migrate in the opposite direction from the plasma proteins. This formed the basis for the sequential flotation of lipoproteins: flotation of chylomicrons and VLDL from plasma (<1.00 g/ml) for 28 h, adjustment to 1.063 g/ml and centrifugation to recover LDL (28 h), and finally adjustment to 1.21 g/ml with centrifugation for 38 h to isolate HDL using a 40.3 or Ti70 rotor at ≥40,000 rpm (4, 5). It was also John Gofman who proposed that elevated levels of lipoproteins cause atherosclerosis, the deposition of lipid in arteries and blood vessels that eventually leads to the premature deaths of so many people in the developed world (6). It was noticed that HDL could reverse this trend (7–9), which spurred an intense research focus on the mechanism of HDL as an atheroprotective particle (10–15), as well as identifying its other functions (16–20).
Few reports have attempted to characterize the stability of lipoproteins in a centrifugal field. Kunitake and Kane (21) studied human lipoproteins and found that apoAI was lost during each spin until one-third of the apoAI was lost with five spins. Additionally, Ståhlman et al. (22) demonstrated that using reduced ionic strength solutions allowed for greater recovery of exchangeable apolipoproteins. Increased ionic strength solutions were found to improve the retention of apoAI on the HDL (21), whereas this decreased the retention of the exchangeable apolipoproteins (22). Thus, for different apolipoproteins, their mechanism of dissociation from HDL differs. The assembly of HDL is hinted at by less disruptive characterization methods, such as nondenaturing gradient gel electrophoresis. Li et al. (23) discovered that native gel electrophoresis produced a series of 14 distinct HDL peaks, which they labeled in the order in which they appeared, although most individuals only have about six. LeBoeuf et al. (24), characterized mouse lipoproteins from a variety of inbred mouse strains and found that mouse lipoproteins were roughly similar in size, although there were differences in the electrophoretic position of the major peak. Thus, depending upon the method of isolation, the pattern of lipoproteins recovered differs.
As there are many contemporary publications that utilize high ultracentrifuge rotor speeds, from 65,000 to 120,000 rpm, in order to reduce the time frame needed to isolate lipoproteins (25–31), and many characterizations of HDL utilize material which was purified via ultracentrifugation (22, 32–35), this may exacerbate the alteration of their recovered HDL; as in this report, we demonstrate that centrifugation of HDL at rotor speeds in excess of 30,000 rpm results in alteration to the recovered particle. Minimally altered HDL can be recovered from plasma samples using a reduced rotor speed of 15,000 rpm and a KBr-containing gradient in a similar time frame as the traditional ultracentrifuge-based isolation.
MATERIALS AND METHODS
Mice
Male retired breeder C57BL/6J mouse plasma samples were purchased from Jackson Laboratories. Prior to blood collection, the mice were fasted overnight. EDTA was used as the anti-coagulant and blood was collected through cardiac puncture. The samples were stored at 4°C prior to overnight shipment on freezer packs (sample not frozen). Samples were subjected to HDL purification within 24 h of receipt.
Native gel electrophoresis and visualization
The 4–30% native gradient gels were purchased from CBS Scientific. Every other lane was used when using a 13-well comb. Briefly, samples were prepared by diluting plasma 1:1 with native loading buffer (Bio-Rad). After loading 20 μl of diluted plasma, the gels were run at 125 V for 30 min. Then a second and a third load was made with 30 min of electrophoresis between loading steps. Following the addition of the third aliquot, the gels were allowed to run for an additional 48 h at 125 V at 4°C. After disassembly of the apparatus, the gels were stained with Sudan black B overnight. Prior to photography, the gels were destained and soaked in water to allow them to return to their original shape. Following this, the gels were subsequently stained with Coomassie blue (Bio-Rad) in order to visualize the native molecular mass markers. Alternatively, gels containing purified lipoprotein samples (no plasma samples present) were directly stained with Coomassie blue.
Antibody affinity chromatography
Antibodies directed against mouse apoAI were generated in a goat (Cocalico Biological, Reamstown, PA), similar to McVicar et al. (36). The goat plasma was passed through a column onto which mouse apoAI was bound. After washing, a select amount of antibody was eluted using 0.5 M acetic acid. This antibody, which can dissociate from apoAI under low pH conditions, was used to construct the antibody affinity column used for rapidly isolating mouse HDL from mouse plasma samples. Briefly, mouse plasma was passed through the column to allow apoAI-containing species to bind to the column. After washing the column with 30 ml of TBS containing 0.05% EDTA, the HDL was eluted with a 0.5 M acetic acid solution directly into a 2 M Tris solution to neutralize the pH. The resulting HDL sample was desalted and exchanged into a TBS solution containing 0.05% EDTA using 10,000 MWCO centrifugal ultrafiltration devices (Amicon, Millipore).
Stability of mouse HDL in the preparatory ultracentrifuge
Affinity-purified mouse HDL was placed on top of a 1.2 g/ml KBr-containing TBS solution, supplemented with 0.05% EDTA. A Ti70.1 rotor was used (Beckman Instruments, Palo Alto, CA), with centrifuge speeds of 40,000, 50,000, or 60,000 rpm. Samples were centrifuged for 48 h prior to fractionation. Following fractionation, the samples were subjected to UV/Vis spectroscopy (HP8453; Agilent, Santa Clara, CA) to quantify the amount of material present in each fraction.
Analytical ultracentrifugation
HDL samples were examined by analytical ultracentrifugation in a Beckman XLA using the An60 rotor (Beckman Instruments). HDL samples were loaded into 1.2 cm path length charcoal-filled Epon double sector cells. Measurements were recorded at 280 nm to monitor the behavior of the HDL sample at various rotor speeds.
Ultracentrifugation separation of mouse lipoproteins using the preparatory ultracentrifuge
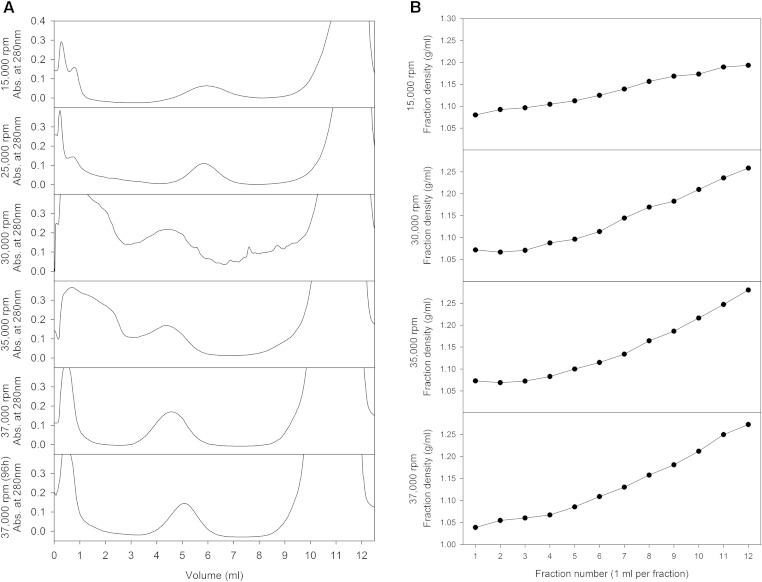
We loaded 200–500 μl of plasma, adjusted to a density of 1.35 g/ml with KBr, at the bottom of six centrifuge tubes of a swinging bucket rotor (SW-41 Ti; Beckman Instruments). Above this, we formed a density gradient of KBr ranging from 1.0171 to 1.26 g/ml using a gradient mixer. Throughout the gradient was 0.05% EDTA to protect against oxidation. The rotors were spun at 15,000 rpm for 96 h to isolate the HDL, which floated upward toward their equilibrium density of about 1.10 g/ml midway up the tube. For other runs demonstrating the ability of higher rotor speeds to affect the recovered HDL, speeds of 25,000–37,000 rpm were used while retaining the same gradient. We collected 1 ml fractions using an ISCO fraction collector (ISCO, Lincoln, NE), which also recorded optical density throughout the tube, as shown in Fig. 5A.
Fig. 5.
Density gradient ultracentrifugation of HDL. Density gradient ultracentrifugation was performed on C57BL/6J plasma at various rotor speeds. Absorbance profiles of the distribution of material following centrifugation (A) and its density (B) are plotted. Representative samples are shown.
RESULTS
Size range of native HDL
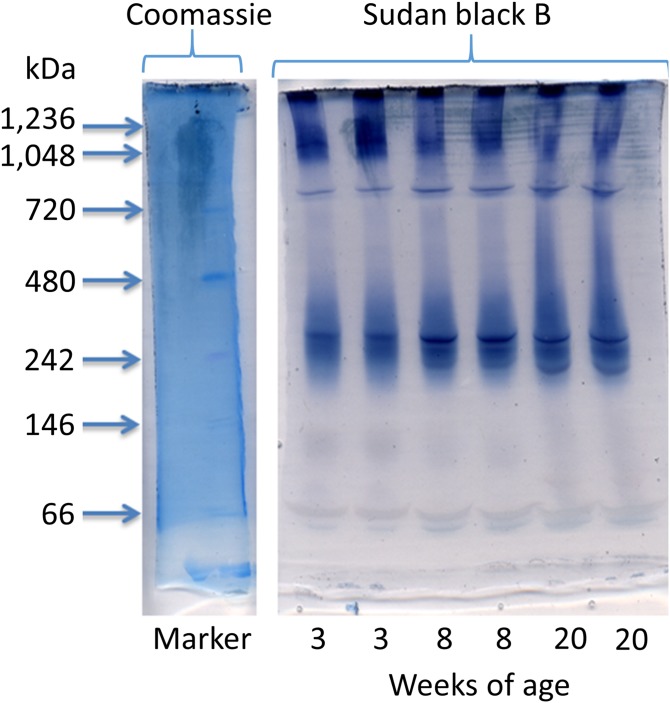
The various subclasses of HDL present in a plasma sample may be evaluated through a variety of methods. One such mildly perturbing separation method is native gradient gel electrophoresis. As seen in Fig. 1, the HDL pattern seen in C57BL/6J male mice appeared to encompass a molecular mass range of ∼180–480 kDa. Additional Sudan black B-stained species, indicative of lipoproteins, were found at 800 kDa and larger, as well as 150 kDa and smaller. While this method is suitable for identifying the molecular mass of material without resulting in the damage to the complex, it does not yield information such as the density of the HDL. Therefore, while this method does not disrupt the HDL, it will not be able to supplant ultracentrifugation for clinical assays regarding the density of HDL or for purification of HDL for subsequent characterization.
Fig. 1.
Native gel electrophoresis of unperturbed plasma samples. Samples were subjected to native gel electrophoresis gels prior to staining with either Coomassie blue or Sudan black B. Plasma samples were from male C57BL/6J mice of various ages.
Loss of protein from HDL
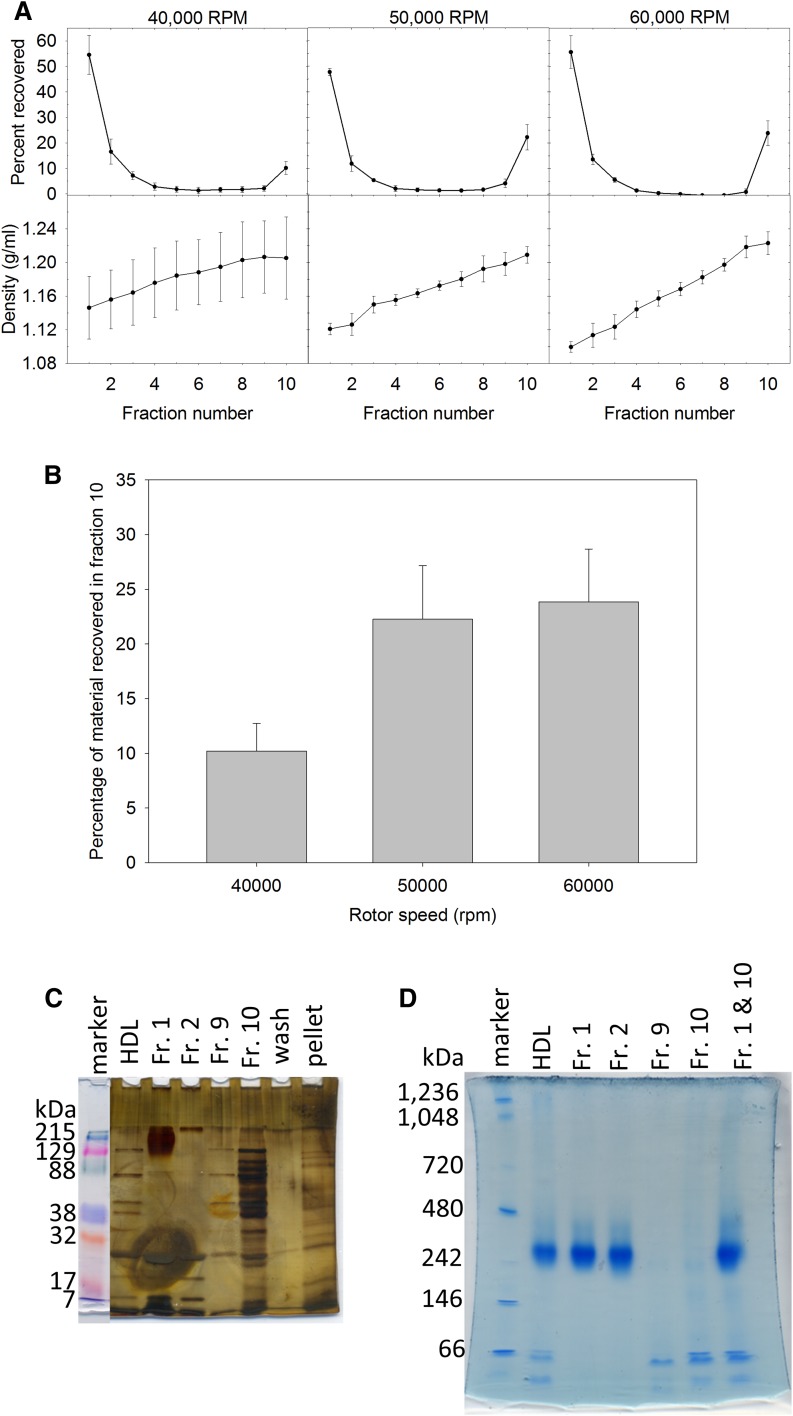
We then characterized the susceptibility to damage observed with the HDL when it was centrifuged in a preparatory ultracentrifuge. HDL was first purified from plasma samples using antibody affinity chromatography directed against mouse apoAI (36). The isolated HDL was placed on top of a 1.2 g/ml KBr solution and subjected to centrifugation at 40,000, 50,000, or 60,000 rpm for 48 h (Fig. 2A). Following the termination of the centrifuge run, fractions were collected and the absorbance at 280 nm was recorded. The amount recovered in each fraction was normalized to the total absorbance for all the recorded fractions and, therefore, are plotted as percentages. The fraction density was measured and suggested the formation of a gradient during the centrifuge run, because the top fraction was still approximately 1.1 g/ml; intact HDL should remain near the top of the gradient. The relative amount of material found at the bottom of the tube was plotted in Fig. 2B. Increased amounts of material were found at the tube bottom with increased rotor speed. As free protein would completely sediment under these conditions within 25.5 h, even at 40,000 rpm, these increases were not due to unassociated protein mixed with the purified HDL sample. This suggests that elevated rotor speed results in the loss of material from the lipoprotein. Additionally, SDS-PAGE was performed on the samples recovered following centrifugation at 60,000 rpm (Fig. 2C). This suggests that material dissociating from the HDL was a mixture of different HDL-associated proteins and not just apoAI alone, as there were a wide range of different molecular mass proteins rather than an enrichment of material consistent with the molecular mass of apoAI. As these proteins were found at densities greater than 1.2 g/ml, this suggests that the proteins have detached from the HDL complex and are associated with minimal amounts of lipid in order to reach the bottom of the centrifuge tube under these conditions. Experiments in the preparatory ultracentrifuge showed that a greater amount of material was found at the bottom of the centrifuge tube (rho >1.2 g/ml) with an increased rotor speed (Fig. 2A, B). This shows that HDL is indeed modified by the ultracentrifuge and this modification occurs in a rotor speed-dependent manner. As the amount of material increased from 10% of the material loaded for the 40,000 rpm runs to nearly 25% for both the 50,000 and 60,000 rpm runs, this indicates that rotor speed affects the degree of HDL damage. As there were no statistical differences in the amount of material lost in the 50,000 rpm versus the 60,000 rpm runs, this may indicate the maximum amount of material which can readily dissociate from the HDL. That is, the material still associated with the HDL particle following these high speed centrifugation runs is stably retained onto its surface. A native gel was run using material from before and after centrifugation at 50,000 rpm, the gel showed a minimal size change in the main HDL band with centrifugation (Fig. 2D). However, intensities for the 66 kDa band increased following centrifugation, relative to the original affinity-purified HDL sample. The native gels on fractions recovered following high speed ultracentrifugation indicate the formation of greater amounts of material at 66 kDa during the run. As the intensity of the 66 kDa band increased in fractions 9 and 10 relative to the original HDL sample, it is clear the centrifuge generated the material found at this position. Curiously, when the fragmented halves of HDL were mixed back together (fractions 1 and 10 from Fig. 2D), the 66 kDa particle did not reassemble onto the larger HDL fragment, suggesting a gross alteration to the structure of HDL preventing the reassociation with the smaller fragment.
Fig. 2.
Loss of material from HDL as a function of rotor speed. A: Percentages of material recovered (top) and the density (bottom) of the fractions recovered after centrifugation are plotted with the standard deviation. B: Average amounts of material lost to the bottom of the centrifuge tube are plotted with the standard deviation. C: The recovered fractions (Fr.) from the 60,000 rpm run were subjected to 10% SDS-PAGE then silver stained. D: Affinity-purified HDL before centrifugation (lane 2) and after centrifugation at 50,000 rpm were subjected to native gel electrophoresis (4–30% polyacrylamide-agarose gels) then stained with Coomassie blue.
Changes to HDL density in the ultracentrifuge
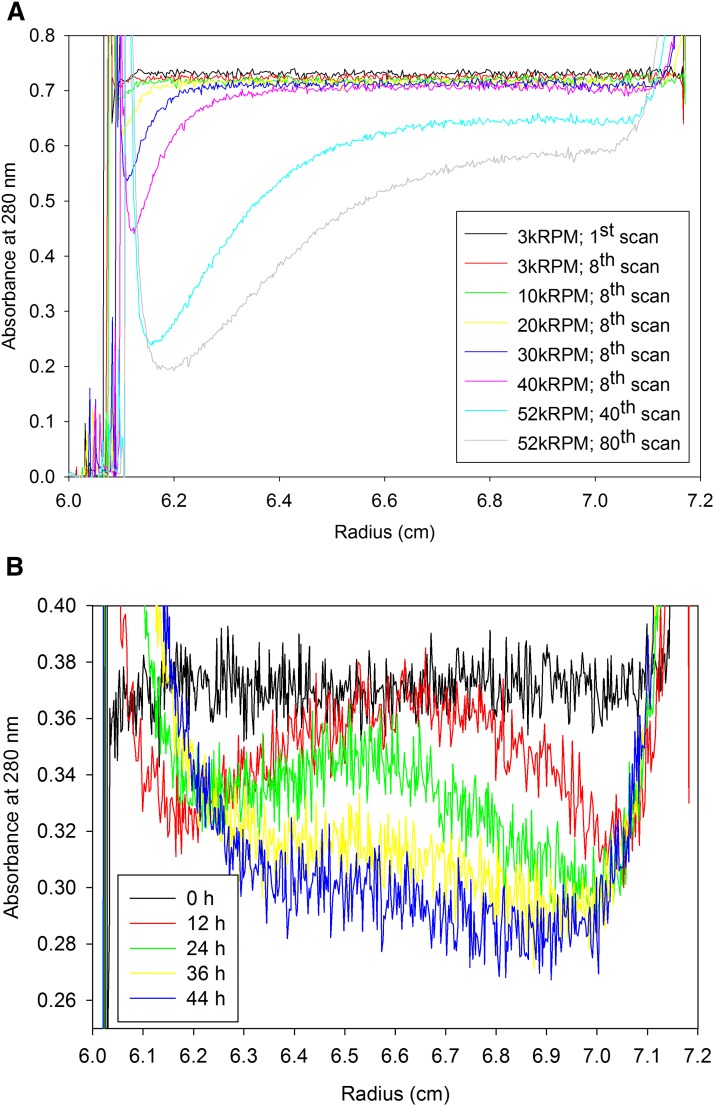
We next examined the loss of HDL carefully using the analytical ultracentrifuge. Isolated material was placed in a buffer density of approximately 1.10 g/ml KBr in order to band the HDL within the middle of the ultracentrifuge cell. If the HDL becomes altered during the ultracentrifuge run, presumably there will be a corresponding change in the density of the complex, resulting in a shift in the banding position of the material in the analytical cell. The rotor speed was increased in a step-wise fashion from 20,000 to 40,000 rpm in 10,000 rpm increments (Fig. 3A). At each step, 30 min of scans were taken. During these scans, no observable loss was detected at 20,000 rpm, but at 40,000 rpm observable amounts of material appeared to be lost. We further increased the rotor speed to 52,000 rpm and observed the loss of material in the plateau region of the scans over the course of 320 min. The decrease in the plateau absorbance exceeded the drop in absorbance expected due to radial dilution, indicating the density of the HDL was being altered during the run as material was being displaced from the center of the cell. We further tested the stability of the HDL at lower rotor speeds, but for longer time frames. A sample of HDL, which had been ultracentrifugally isolated at 30,000 rpm was examined in the analytical ultracentrifuge at the same rotor speed (Fig. 3B). Within 12 h, the HDL began to band within the analytical cell. However, by 24 h, the position of the peak had shifted to the left, suggesting the HDL sample was becoming less dense, indicating damage to the HDL. By 36 h, the height of the peak had dropped, indicating the loss of HDL. Approximately 20% of the 280 nm-absorbing material was displaced during this 44 h run. This indicated that even a reduced rotor speed of 30,000 rpm can result in the loss of HDL under a time frame normally employed to purify HDL. The mechanism behind the damage to ultracentrifugally isolated HDL appears to first occur through the shedding of associated proteins, followed by a disruption of the lighter HDL fragment.
Fig. 3.
HDL stability in the analytical ultracentrifuge. A: HDL was centrifuged at 3,000 rpm (black, red), 10,000 rpm (green), 20,000 rpm (yellow), 30,000 rpm (blue), and 40,000 rpm (pink) for 30 min at each speed. The rotor speed was then increased to 52,000 rpm and monitored for an additional 320 min (turquoise, gray). B: HDL was monitored in the analytical ultracentrifuge at 30,000 rpm for 44 h. Scans of the cell were made at 280 nm, and plotted at approximately 12 h intervals (0 h, black; 12 h, red; 24 h, green; 36 h, yellow; and 44 h, blue).
Reduced rotor speeds isolate intact HDL
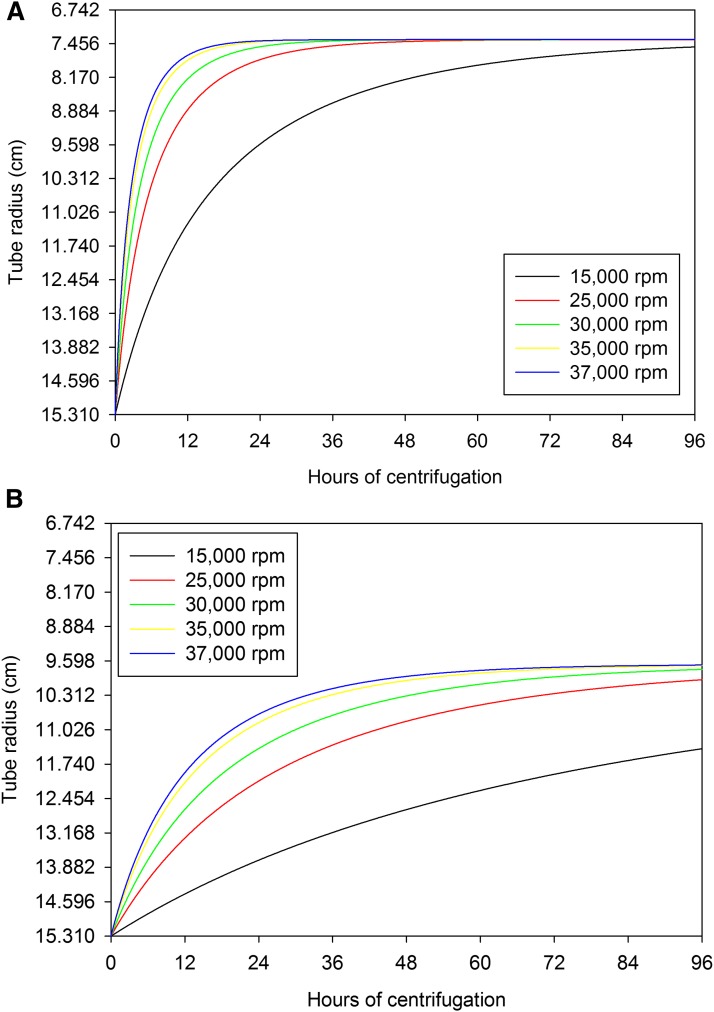
Next, to show that intact HDL can be isolated using the ultracentrifuge, we attempted to reduce the rotor speed used in the isolation process. The data from the analytical ultracentrifuge run suggested that rotor speeds below 30,000 rpm would be sufficient to minimize the degradation of the recovered HDL sample. We calculated the time necessary to separate the various lipoprotein components from the plasma proteins as a function of rotor speed (Fig. 4A, B). The estimations show that LDL will have been separated from the HDL and plasma protein contaminants within 24 h, however, to migrate HDL a far enough distance from the tube bottom and the plasma protein contaminants requires substantially more time with a rotor speed of 15,000 rpm. We kept a time frame of 96 h, similar enough to the conventional HDL isolation procedure, which allowed for a greatly reduced rotor speed of 15,000 rpm to be sufficient to separate VLDL, LDL, HDL, and plasma proteins in a KBr-containing TBS gradient (1.0171–1.26 g/ml) if the sample is placed at the bottom of the gradient (at a density of 1.35 g/ml). We then performed the experiments using the SW-41 rotor. Following centrifugation, the tube was fractionated using an ISCO fraction collector and the absorbance at 280 nm was recorded (Fig. 5A). Additional runs were made at different rotor speeds (25,000, 30,000, 35,000, and 37,000 rpm) using the same gradient. The location of the HDL peak varied depending upon the run speed due to the intensity of the centrifugal field (ω2t) and degradation of HDL induced by elevated centrifugal fields. If the ultracentrifuge run causes the HDL complex to lose a portion of the protein components attached to it (with or without a minimal amount of lipid), then the resulting HDL particle should be observed undergoing flotation faster than expected for a 1.115 g/ml particle, as the actual particle density is now smaller. The densities of the fractions were estimated using a Mettler analytical balance (AE50) and plotted in Fig. 5B. The steepness of the gradient was similar between the 30,000 and 37,000 rpm runs. However, the steepness of the gradient in the 15,000 rpm runs was reduced. This will not affect the purification procedure, as the isopycnic density of HDL is still around the center of the tube. Ultracentrifuge runs at 30,000 rpm or above exhibited peak locations further up the tube than expected for intact HDL. This shows that faster speeds affected the HDL particle while lower speeds can recover a denser HDL particle.
Fig. 4.
Estimated rate of movement for lipoproteins in a preparatory ultracentrifuge. To demonstrate the ability of a greatly reduced rotor speed being capable of separating the various components in a plasma sample under the conditions we utilized, we estimated the movement of LDL (A) and HDL (B) over 96 h in a 1.0171–1.26 g/ml gradient as a function of rotor speed for a SW-41 rotor.
Rotor speed influences rate of loss
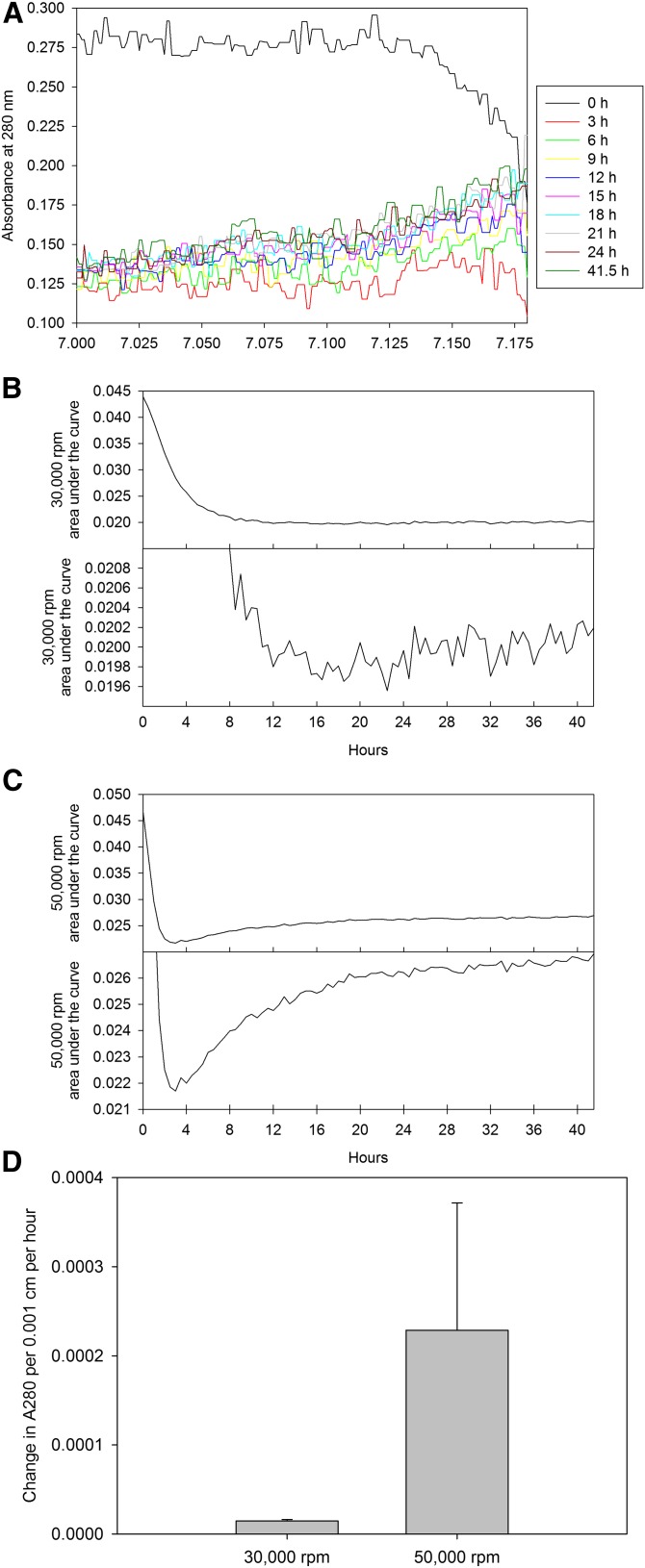
To characterize how the 15,000 rpm purified HDL behaves when subjected to increased centrifugal fields, we again turned to experiments in the analytical ultracentrifuge. The peak HDL fraction, as purified during a 96 h 15,000 rpm isolation in the preparatory ultracentrifuge was placed in the analytical ultracentrifuge without adjustment to the fraction density. As the HDL is close to, but not at, its isopycnic density, the intact lipoprotein will continue to float in the analytical ultracentrifuge. However, we can monitor the shedding of material as a function of time. In Fig. 6, we observed the behavior of the HDL when subjected to speeds of 30,000 or 50,000 rpm in the analytical ultracentrifuge. Initially, the lipoprotein floated toward the meniscus, clearing the bottom of the cell, which resulted in little material at the bottom of the cell by 6 h at 50,000 rpm (Fig. 6A). However, with increasing time, material observed in this region was consistent with a molecular mass range of the proteins associated with HDL. This is indicative of material being lost from the HDL during centrifugation. We plotted the area under the curve as a function of time, for the 30,000 (Fig. 6B) and 50,000 rpm (Fig. 6C) runs. These plots are representative of the material detected toward the bottom of the ultracentrifuge cell during the centrifuge run. Using the linear portion of this curve, we can observe a rate of increase in material at the bottom of the cell (Fig. 6D) which is markedly increased at 50,000 rpm (nearly 16-fold over that observed at 30,000 rpm). This suggests that not only can we observe material being lost from HDL at 30,000 rpm within 48 h, but increased rotor speeds of 50,000 rpm have greatly intensified the rate of alteration to the HDL.
Fig. 6.
Kinetics of HDL loss in the analytical ultracentrifuge. A: Increased levels of material appeared at the bottom of the centrifuge cell with increasing time, and the rate of appearance of material in this region varied between 30,000 rpm (B) and 50,000 rpm (C). D: Subjecting samples to 50,000 rpm resulted in a 15.7-fold increased rate of appearance of material over 30,000 rpm centrifugation runs.
DISCUSSION
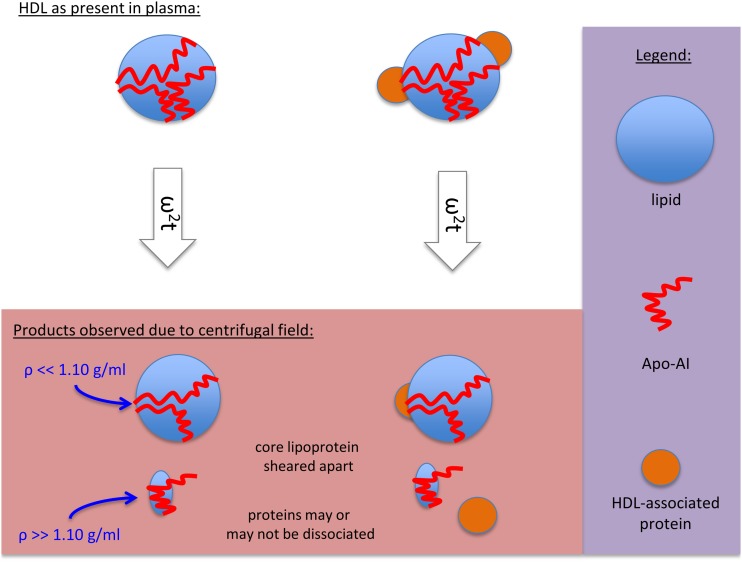
While isolating HDL by using sequential centrifugation steps is the classic method for recovering this lipoprotein complex, there are caveats to this method. Figure 7 depicts the mechanism of HDL loss when subjected to elevated centrifugal fields. Either apoAI or apoAI with an HDL-associated protein can be removed from the original HDL particle. As this material contains little lipid, it is dense enough to sediment in the gradients normally employed to isolate HDL. Figures 2D and 6A demonstrate how the traditional sequential flotation of lipoproteins causes this alteration to the HDL particle and how this process of HDL degradation was previously undetected. As rotor speeds as low as 30,000 rpm can disrupt the HDL, and these fragments do not reassociate, the small dense fragment is incapable of being separated from the plasma protein fraction and is discarded with the plasma proteins after recovery of the floating HDL component. Subsequent characterizations of the HDL recovered by this method may under-detect the original amount of, as well as types of, associated proteins bound to the HDL. This study has identified the formation of this HDL degradation product and has shown that excessive centrifugal fields generate it. It should be noted that for experiments utilizing the affinity-purified mouse HDL, there exists the possibility that this chromatographic separation method may have altered the recovered HDL, thereby increasing its susceptibility to ultracentrifuge-induced damage.
Fig. 7.
Purported model for HDL breakage at elevated rotor speeds. The intact HDL, with a density of 1.11 g/ml and a molecular mass of 280–450 kDa, can dissociate into multiple species when purified through high speed centrifugation. The resultant particles are a lipid-enriched fraction smaller in molecular mass and lower in density than the original HDL particle, as well as a smaller lipid-poor fraction of approximately 66 kDa with a density of >1.2 g/ml.
Possible methods to separate HDL from plasma samples include size exclusion chromatography, native gel electrophoresis, affinity chromatography, and ultracentrifugation. As size exclusion chromatography and native gel electrophoresis are molecular sieving methods, they may be unable to separate HDL away from similarly sized protein complexes present in plasma, leading to ambiguous results concerning which proteins are associated with HDL. Additionally, native gel electrophoresis is of limited utility as an isolation method due to the small amounts of material able to be recovered from the gel. Affinity antibody chromatography can selectively purify apoAI-containing material, however, as this method selectively removes material containing apoAI, regardless of the amount of associated lipid, this may recover additional material not traditionally classified as HDL. Ultracentrifugation is amenable to small single donor samples or can be scaled up to accommodate larger volumes of pooled plasma. This method is traditionally used with the sequential flotation method of lipoprotein isolations, however, to minimize the time necessary to perform these isolations, researchers have used ever increasing rotor speeds. This paper demonstrates that elevated rotor speeds result in the loss of material from the isolated HDL. We have described a density gradient isolation using reduced rotor speed to recover HDL containing more protein. As this does not require changes to laboratory instrumentation, only the protocols used to isolate HDL, adoption of this method will be relatively straightforward.
The low speed ultracentrifugation density gradient protocol described for isolating HDL may be adapted for isolation of VLDL and LDL. By adjusting the parameters of the run to utilize a density gradient of 1.0063–1.10 g/ml, VLDL would migrate rapidly to the tube top and LDL to the mid-portion of the tube. HDL and the plasma protein contaminants would likely remain at the tube bottom. The estimated migration of LDL is presented in supplementary Fig. 1.
All the characterizations described within this report have been performed using mouse HDL, which may not necessarily translate to human HDL. As the trend observed in the case of mouse HDL is quite clear, this work provides a cautionary example to other researchers utilizing a mouse model system. Even though reports from Ståhlman et al. (22), as well as Kunitake and Kane (21), demonstrate ultracentrifugally induced changes to human HDL, the exact mechanism of this damage should be further explored to determine whether it occurs in the same fashion as with mouse HDL.
This study has shown that HDL can be isolated in an intact fashion containing a greater variety of associated proteins using a method similar to the traditional ultracentrifuge isolation method. Instead of sequential flotation steps at 40,000 rpm or greater rotor speeds, we employ a KBr-based density gradient and a much reduced rotor speed of 15,000 rpm. This allows for the VLDL and LDL to rapidly migrate to the meniscus and for the HDL to migrate to the middle portion of the tube by the end of 96 h. As demonstrated in the calculated migration rates of the different components present in plasma samples (Fig. 4B), the HDL approaches its isopycnic density within the gradient, but does not fully reach it at the end of the centrifugation run. While this is sufficient to separate it from the plasma protein contaminants, such as the immunoglobulins and albumin, this prevents a direct measure of the density of the recovered HDL peaks. Thus, this method is useful for recovering intact HDL in a relatively straight-forward manner, with minimal changes to the instrumentation or methodology for labs currently isolating lipoprotein ultracentrifugally. The more gentle 15,000 rpm ultracentrifuge isolation may allow for a greater recovery of the HDL-associated proteins. This may be beneficial for identifying new proteins associated with the HDL particle and suggest additional functions for HDL.
Supplementary Material
Footnotes
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Svedberg T., Pedersen K. O. 1940. The Ultracentrifuge. Clarendon Press, Oxford. [Google Scholar]
- 2.Beckman Coulter. 2011. Instructions for Use Optima MAX-XP Ultracentrifuge. Beckman Coulter, Brea, CA. [Google Scholar]
- 3.Gofman J. W., Lindgren F. T., Elliott H. 1949. Ultracentrifugal studies of lipoproteins of human serum. J. Biol. Chem. 179: 973–979. [PubMed] [Google Scholar]
- 4.De Lalla O. F., Gofman J. W. 1954. Ultracentrifugal analysis of serum lipoproteins. Methods Biochem. Anal. 1: 459–478. [DOI] [PubMed] [Google Scholar]
- 5.Schumaker V. N., Puppione D. L. 1986. Sequential flotation ultracentrifugation. Methods Enzymol. 128: 155–170. [DOI] [PubMed] [Google Scholar]
- 6.Gofman J. W., Lindgren F., Elliot H., Mantz W., Hewitt J., Strisower B., Herring V., Lyon T. P. 1950. The role of lipids and lipoproteins in atherosclerosis. Science. 111: 166–171,186. [DOI] [PubMed] [Google Scholar]
- 7.Moore R. E., Kawashiri M. A., Kitajima K., Secreto A., Millar J. S., Practico D., Rader D. J. 2003. Apolipoprotein A-I deficiency results in markedly increased atherosclerosis in mice lacking the LDL receptor. Arterioscler. Thromb. Vasc. Biol. 23: 1914–1920. [DOI] [PubMed] [Google Scholar]
- 8.Benoit P., Emmanuel F., Caillaud J., Bassinet L., Castro G., Gallix P., Fruchart J., Branellec D., Denefle P., Duverger N. 1999. Somatic gene transfer of human apoA-I inhibits atherosclerosis progression in mouse models. Circulation. 99: 105–110. [DOI] [PubMed] [Google Scholar]
- 9.Miller G. J., Miller N. E. 1975. Plasma-high-density-lipoprotein concentration and development of ischæmic heart-disease. Lancet. 1: 16–19. [DOI] [PubMed] [Google Scholar]
- 10.Salonen J. T., Salonen R., Seppänen K., Rauramaa R., Tuomilehto J. 1991. HDL, HDL2, and HDL3 subfractions, and the risk of acute myocardial infarction. A prospective population study in eastern Finnish men. Circulation. 84: 129–139. [DOI] [PubMed] [Google Scholar]
- 11.van der Gaag M. S., van Tol A., Vermunt S. H., Scheek L. M., Schaafsma G., Hendriks H. F. 2001. Alcohol consumption stimulates early steps in reverse cholesterol transport. J. Lipid Res. 42: 2077–2083. [PubMed] [Google Scholar]
- 12.Nakamura Y., Kotite L., Gan Y., Spencer T., Fielding C., Fielding P. 2004. Molecular mechanism of reverse cholesterol transport: reaction of pre-beta-migrating high-density lipoprotein with plasma lecithin/cholesterol acyltransferase. Biochemistry. 43: 14811–14820. [DOI] [PubMed] [Google Scholar]
- 13.Rust S., Rosier M., Funke H., Real J., Amoura Z., Piette J., Deleuze J., Brewer H., Duverger N., Denèfle P., et al. 1999. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22: 352–355. [DOI] [PubMed] [Google Scholar]
- 14.Acton S., Rigotti A., Landschulz K., Xu S., Hobbs H., Krieger M. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 15.Fielding P. E., Fielding C. J. 1980. A cholesteryl ester transfer complex in human plasma. Proc. Natl. Acad. Sci. USA. 77: 3327–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaisar T., Pennathur S., Green P., Gharib S., Hoofnagle A., Cheung M., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hager K. M., Hajduk S. L. 1997. Mechanism of resistance of African trypanosomes to cytotoxic human HDL. Nature. 385: 823–826. [DOI] [PubMed] [Google Scholar]
- 18.Shiflett A. M., Bishop J. R., Pahwa A., Hajduk S. L. 2005. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J. Biol. Chem. 280: 32578–32585. [DOI] [PubMed] [Google Scholar]
- 19.Shih D. M., Gu L., Xia Y. R., Navab M., Li W. F., Hama S., Castellani L. W., Furlong C. E., Costa L. G., Fogelman A. M., et al. 1998. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 394: 284–287. [DOI] [PubMed] [Google Scholar]
- 20.Eichinger A., Nasreen A., Kim H., Skerra A. 2007. Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein D. J. Biol. Chem. 282: 31068–31075. [DOI] [PubMed] [Google Scholar]
- 21.Kunitake S. T., Kane J. P. 1982. Factors affecting the integrity of high density lipoprotein in the ultracentrifuge. J. Lipid Res. 23: 936–940. [PubMed] [Google Scholar]
- 22.Ståhlman M., Davidsson P., Kanmert I., Rosengren B., Borén J., Fagerberg B., Camejo G. 2008. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 49: 481–490. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., McNamara J., Ordovas J., Schaefer E. 1994. Analysis of high density lipoproteins by a modified gradient gel electrophoresis method. J. Lipid Res. 35: 1698–1711. [PubMed] [Google Scholar]
- 24.LeBeouf R. C., Puppione D. L., Schumaker V. N., Lusis A. J. 1983. Genetic control of lipid transport in mice. I. Structural properties and polymorphisms of plasma lipoproteins. J. Biol. Chem. 258: 5063–5070. [PubMed] [Google Scholar]
- 25.Yee M. S., Pavitt D. V., Tan T., Venkatesan S., Godsland I. F., Richmond W., Johnston D. G. 2008. Lipoprotein separation in a novel iodixanol density gradient, for composition, density, and phenotype analysis. J. Lipid Res. 49: 1364–1371. [DOI] [PubMed] [Google Scholar]
- 26.Gardner R. S., Ogden N. H., Cripps P. J., Billington D. 2003. Separation of bovine plasma lipoproteins by a rapid ultracentrifugation method. J. Comp. Pathol. 128: 15–23. [DOI] [PubMed] [Google Scholar]
- 27.Graham J. M., Higgins J. A., Gillott T., Taylor T., Wilkinson J., Ford T., Billington D. 1996. A novel method for the rapid separation of plasma lipoproteins using self-generating gradients of iodixanol. Atherosclerosis. 124: 125–135. [DOI] [PubMed] [Google Scholar]
- 28.Sawle A., Higgins M., Olivant M., Higgins J. 2002. A rapid single-step centrifugation method for determination of HDL, LDL, and VLDL cholesterol, and TG, and identification of predominant LDL subclass. J. Lipid Res. 43: 335–343. [PubMed] [Google Scholar]
- 29.Tong H., Knapp H., VanRollins M. 1998. A low temperature flotation method to rapidly isolate lipoproteins from plasma. J. Lipid Res. 39: 1696–1704. [PubMed] [Google Scholar]
- 30.Chung B. H., Wilkinson T., Geer J. C., Segrest J. P. 1980. Preparative and quantitative isolation of plasma lipoproteins: rapid, single discontinuous density gradient ultracentrifugation in a vertical rotor. J. Lipid Res. 21: 284–291. [PubMed] [Google Scholar]
- 31.Larner C. D., Henriquez R. R., Johnson J. D., Macfarlane R. D. 2011. Developing high performance lipoprotein density profiling for use in clinical studies relating to cardiovascular disease. Anal. Chem. 83: 8524–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Zychlinski A., Williams M., McCormick S., Kleffmann T. 2014. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteomics. 106: 181–190. [DOI] [PubMed] [Google Scholar]
- 33.Lepedda A. J., Nieddu G., Zinellu E., De Muro P., Piredda F., Guarino A., Spirito R., Carta F., Turrini F., Formato M. 2013. Proteomic analysis of plasma-purified VLDL, LDL, and HDL fractions from atherosclerotic patients undergoing carotid endarterectomy: identification of serum amyloid A as a potential marker. Oxid. Med. Cell. Longev. 2013: 385214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segrest J. P., Cheung M. C., Jones M. K. 2013. Volumetric determination of apolipoprotein stoichiometry of circulating HDL subspecies. J. Lipid Res. 54: 2733–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sreckovic I., Birner-Gruenberger R., Obrist B., Stojakovic T., Scharnagl H., Holzer M., Scholler M., Philipose S., Marsche G., Lang U., et al. 2013. Distinct composition of human fetal HDL attenuates its anti-oxidative capacity. Biochim. Biophys. Acta. 1831: 737–746. [DOI] [PubMed] [Google Scholar]
- 36.McVicar J. P., Kunitake S. T., Hamilton R. L., Kane J. P. 1984. Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I. Proc. Natl. Acad. Sci. USA. 81: 1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.