Abstract
We hypothesized that brains from vitamin E-deficient (E−) zebrafish (Danio rerio) would undergo increased lipid peroxidation because they contain highly polyunsaturated fatty acids, thus susceptible lipids could be identified. Brains from zebrafish fed for 9 months defined diets without (E−) or with (E+) added vitamin E (500 mg RRR-α-tocopheryl acetate per kilogram diet) were studied. Using an untargeted approach, 1-hexadecanoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine [DHA-PC 38:6, PC 16:0/22:6]was the lipid that showed the most significant and greatest fold-differences between groups. DHA-PC concentrations were approximately 1/3 lower in E− (4.3 ± 0.6 mg/g) compared with E+ brains (6.5 ± 0.9 mg/g, mean ± SEM, n = 10 per group, P = 0.04). Using lipidomics, 155 lipids in brain extracts were identified. Only four phospholipids (PLs) were different (P < 0.05) between groups; they were lower in E− brains and contained DHA with DHA-PC 38:6 at the highest abundances. Moreover, hydroxy-DHA-PC 38:6 was increased in E− brains (P = 0.0341) supporting the hypothesis of DHA peroxidation. More striking was the depletion in E− brains of nearly 60% of 19 different lysophospholipids (lysoPLs) (combined P = 0.0003), which are critical for membrane PL remodeling. Thus, E− brains contained fewer DHA-PLs, more hydroxy-DHA-PCs, and fewer lysoPLs, suggesting that lipid peroxidation depletes membrane DHA-PC and homeostatic mechanisms to repair the damage resulting in lysoPL depletion.
Keywords: α-tocopherol, 1-hexadecanoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine, Land’s cycle
Vitamin E was discovered because it is critical during pregnancy to prevent fetal resorption in rodents (1). We have used zebrafish to characterize the molecular consequences of vitamin E deficiency because zebrafish require dietary α-tocopherol, especially during embryonic development (2). Zebrafish also express the α-tocopherol transfer protein (α-TTP), which facilitates hepatic α-tocopherol secretion into the circulation in humans (3), and likely facilitates α-tocopherol delivery from the yolk to the developing zebrafish embryo (4). Importantly, when the α-TTP was knocked down in zebrafish embryos by 12–15 h postfertilization, the embryonic brain and eyes failed to form appropriately (4), indicating that α-tocopherol is necessary for nervous system development. Further, during embryonic growth over 72 h both DHA (22:6) and arachidonic acid (AA) (20:4) decreased at faster rates in vitamin E-deficient (E−) compared with vitamin E-sufficient (E+) zebrafish embryos (5). Moreover, adequate dietary ascorbic acid was necessary to prevent accelerated α-tocopherol deficiency and tissue damage (5).
We hypothesized that the devastating effects of severe α-tocopherol deficiency in zebrafish embryos (4) are a result of depletion and alteration of critical brain lipids. Hypothetically, the brain, which is highly enriched in DHA yet cannot synthesize DHA to meet its needs (6), is highly susceptible to lipid peroxidation in the α-tocopherol-deficient state. It has been recognized for decades in humans that vitamin E deficiency causes a progressive spinocerebellar ataxia (7). Moreover, Ulatowski et al. (8) have shown that vitamin E is necessary for preservation of Purkinje cells in brains from α-TTP knockout mice. The specific lipids depleted during vitamin E deficiency, however, are not known.
The objective of this study is to elucidate the molecular consequences of vitamin E deficiency on lipids in zebrafish brains. However, embryonic brains are too small for rigorous analysis; therefore, we used adult brains from E− compared with E+ zebrafish to evaluate changes in lipid composition and abundance, and applied a combination of targeted and untargeted methods to enable a comprehensive profiling of brain lipids to test this hypothesis. We chose zebrafish nearly 1 year of age because long-term vitamin E deficiency causes severe depletion of tissue α-tocopherol, resulting in behavioral and physiological damage (5, 9). It should be noted that the diets fed were adequate in ascorbic acid, but restricted in long chain PUFAs. These 1-year-old zebrafish are equivalent to middle aged (10, 11), but not elderly, adult humans eating a low vitamin E diet (12).
MATERIALS AND METHODS
Materials
1-Hexadecanoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine [DHA-PC, phosphatidylcholine (PC) 16:0/22:6] and 1,2-ditridecanoyl-sn-glycero-3-phosphocholine (DTD-PC, PC 13:0/13:0) were obtained from Avanti Polar Lipids Inc. (Alabaster, AL) and used without further purification.
Zebrafish housing and feeding
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Institutional Animal Care and Use Committee of Oregon State University approved the protocol (ACUP number 4344). Every effort was made to minimize suffering; prior to sampling, all fish were euthanized by an overdose of tricaine (MS 222; Argent Chemical Laboratories, Inc., Redmond, WA).
Tropical 5D strain zebrafish were housed in the Sinnhuber Aquatic Research Laboratory. Adult zebrafish were kept under standard laboratory conditions at 28.5°C with a 14 h light/10 h dark cycle. For 9 months, beginning at 50 days postfertilization, zebrafish were fed defined diets, which contained only fatty acids shorter than 18 carbons with two or three double bonds (2, 5, 9), were prepared in 300 g batches with the vitamin C source as StayC (500 mg/kg; Argent Chemical Laboratories Inc.) without (E−) or with (E+) added vitamin E (500 mg RRR-α-tocopheryl acetate/kg diet; ADM, Decatur, IL), as described previously (5). Diets were stored at −20°C until fed to the zebrafish.
Using high-pressure LC with electrochemical detection, α-tocopherol was measured in diets, brains, and the matching bodies (without brain), as described (13); vitamin C was measured in diet as described (14). Measured α-tocopherol concentrations in the E− and E+ diets were 1.6 ± 0.1 mg/kg and 334 ± 12 mg/kg, respectively; vitamin C was 143 ± 16 mg ascorbic acid/kg. This level of dietary vitamin C has been found to be adequate for the zebrafish (5, 15).
Zebrafish tissue extraction and lipidomic analysis by MS/MSALL TM
For survey experiments using MS/MSALL TM, the head was removed just behind the eye from whole frozen zebrafish (n = 5 per group). The heads were weighed, then homogenized in methanol:water (80:20, v/v). Samples were extracted using a modified Bligh and Dyer solvent system (16); the final ratio was chloroform:methanol:water (2:2:1.8). Samples were mixed vigorously for 20 s, and then centrifuged at 1,000 g for 10 min, the top layer was removed by aspiration and discarded. The bottom layer was collected and evaporated under nitrogen, then stored at −80°C less than 24 h before MS/MSALL TM analysis.
MS/MSALL TM analysis was performed using the TripleTOF 5600 mass spectrometer with direct infusion into ESI source (AB SCIEX, Framingham, MA). Each sample was reconstituted with 400 μl methanol:chloroform (1:2, v/v) containing 5 mM ammonium formate. Samples were delivered to the source at 15 μl/min using an isocratic flow of methanol:isopropanol (3:1, v/v) with 5 mM ammonium formate. A second isocratic pump delivered a solution of 98% isopropanol and 2% methanol containing 5 mM ammonium formate, as a make-up flow to the source through a T-junction at a rate of 60 μl/min. Total flow was 75 μl/min at point of entry into the ESI source. Source parameters included nebulizing gases: GS1 at 20 psi, GS2 at 15 psi, curtain gas at 20 psi, positive mode ion spray voltage at 5,500 V, negative mode ion spray voltage at −4,000 V, declustering potential at 40 V, and at an ESI source operating temperature of 400°C. Collision energy for each MS/MS step was 50 ± 30 eV and −40 ± 30 eV, respectively, for positive and negative ion mode experiments.
A fully automatic workflow was developed using the infusion MS/MSALL TM technique on the TripleTOF 5600 system with a flow injection sample introduction strategy. The MS/MSALL TM infusion method (17) allowed automated collection of TOF MS and TOF MS/MS of all lipids, using a step-wise sampling of precursor ions selected at unit resolution (1 amu) in quadrupole (Q1), followed by fragmentation in Q2, and then product ions were detected simultaneously using TOF. High-resolution TOF MS/MS spectra were collected with an accumulation time of 250 ms from lipid precursors from mass range m/z 200.050 to 1,200.049, followed by 1,000 product ion experiments with 1,000 precursors evenly spaced from m/z 200.050 to m/z 1,200.049. The data were accumulated for 100 ms for each fragment and collected in order from low to high m/z. The total time to carry out the MS/MSALL TM acquisition for one sample was 5.48 min. Each sample was run in positive and negative modes. To obviate carryover, there was a washing time (5 min) between samples. Sample carryover was assessed during the wash step by injecting sample blanks containing only solvent.
Brain tissue extraction for DHA-PC analysis by triple quadrupole LC-MS/MS
The brains from E− and E+ zebrafish (n = 10 per group) were dissected and weighed. Brains were transferred individually to Eppendorf tubes, homogenized under 4°C, and extracted individually by adding, in the following order: 250 μl methanol:water (1:1, v/v), DTD-PC as an internal standard (2.5 μg in 5 μl methanol), and 750 μl methylene chloride:isopropanol:methanol (25:10:65, v/v/v). Solvents were all at 4°C. Samples were mixed vigorously for 20 s, and then centrifuged at 13,000 g for 10 min. The extracts were transferred to a new tube and were stored less than 24 h at −80°C until analysis.
LC was carried out using a Shimadzu high-pressure LC system (Columbia, MD) with LC elution conditions as described previously (18). The system was coupled to an Applied Biosystems API 3000 triple quadrupole mass spectrometer with a TurboIon spray source operated with both mass analyzers set at unit resolution in positive mode (LC-MS/MS, Applied Biosystems/MDS Sciex API 3000, Foster City, CA). Nebulizer, curtain, and collision (CAD) gas parameters were set at 8, 10, and 2 psi, respectively. Heater gas was supplied at 6 l/min at 425°C. All gases were high purity nitrogen supplied by a custom liquid nitrogen system (Polar Cryogenics, Portland, OR). The ionizing voltage was 5,000 V, and the declustering, focusing, entrance, and exit potentials were 50, 200, 10, 35, and 12 V, respectively.
Multiple reaction monitoring (MRM) was used to quantify PC 38:6, which was identified as DHA-PC in brain extracts. Analytes were detected using MRM, which allowed for simultaneous quantification of specific isomers based upon unique transitions between the m/z of the precursor ion [M+H]+ in the positive mode and characteristic product ions for each species, as well as their characteristic LC retention times. Authentic standards of DHA-PC and DTD-PC eluted at 14.6 min and 11.2 min, respectively. Measurements of DHA-PC limit of detection and limit of quantitation were assessed using the area ratio of DHA-PC to DTD-PC. The limit of detection and the limit of quantitation were defined as the concentrations with a signal-to-noise ratio of at least 3 and 10, respectively. The DHA-PC detection limit was 0.59 pmol/injection and the quantitation limit was 1.76 pmol/injection. The LC-MRM standard curves relating peak area ratios of DHA-PC/DTD-PC versus known authentic DHA-PC amounts (0.2–10 μg injected) were used to quantitate DHA-PC; values are expressed per brain weight.
Ultra-performance LC-TOF-MS/MS lipidomic analyses
The brain tissue extraction procedure for untargeted lipidomics was the same as the extraction for DHA-PC, as described above. Brain weights were not significantly different between groups (E− 2.3 ± 0.3 g compared with E+ 2.2 ± 0.4 g). To identify lipid metabolites, LC was performed using a 1.8 μm particle 100 × 2.1 mm id HSS T3 column (Waters, Milford, MA) coupled to a quadrupole TOF mass spectrometer (AB SCIEX, TripleTOF 5600) operated in information-dependent MS/MS acquisition mode. The column was heated to 50°C in the column oven. A gradient system was used consisting of mobile phase A (60:40 acetonitrile:water containing 10 mM ammonium formate) and mobile phase B [90:10:5 (v/v/v) isopropanol:acetonitrile:water with 10 mM ammonium formate] with sample analysis performed over 17 min total run time. The initial starting conditions were 50% A and 50% B, then for 10 min the gradient was ramped in a linear fashion to 100% B and held at 100% B for 2 min. Subsequently, the system was switched to the initial ratio for 1 min, and equilibrated at the initial ratio for additional 4 min. The flow rate was 0.4 ml/min and the injection volume was 10 μl. TOF MS acquisition time was 0.25 s, and MS/MS acquisition time was 0.1 s. The scan range was m/z 70–1,200 for TOF MS and m/z 50–1,200 for MS/MS. Source parameters included nebulizing gases: GS1 at 45, GS2 at 50, curtain gas at 35, positive mode ion spray voltage at 5,500 V, negative mode ion spray voltage at −4,000 V, declustering potential at 80 V, and at an ESI source operating temperature of 500°C. Collision energy for each MS/MS step was 35 ± 10 eV and −35 ± 10 eV, respectively, for positive and negative ion mode experiments. Data were generated from the ultra-performance LC (UPLC) separation of the lipid extract of individual brains, TOF accurate mass detection, and MS/MS fragment characterization.
Data processing and statistical analyses
The data acquired during the Infusion MS/MSALL TM and LC-MS/MS experiments were processed in batch mode using LipidView software version 1.2 (AB SCIEX) for automated identification and quantitation of lipid species. The processed LipidView data files were imported into MarkerView software (AB SCIEX) for initial data processing, including feature detection, peak integration, principal component analysis, and discriminant analysis.
Student’s t-test comparisons were carried out between the two diet groups using MarkerView. Statistical significance was set at P < 0.05. To discover significant features with the greatest changes between the E− and E+ diet groups (n = 5 per group), the resulting P values (Student’s t-tests) were plotted against fold change (log 10), calculated by MarkerView. Features with both the largest fold changes and smallest P values were selected for identification.
For quantitative data from the LC-MS/MS experiments, statistical analyses of DHA-PC concentrations were performed using GraphPad Prism software (GraphPad, La Jolla, CA). Significance (P < 0.05) was determined using logarithmically transformed data by a one-way ANOVA followed by a Tukey post hoc test. To estimate overall significance of lysophospholipids (lysoPLs), the responses for each individual were summed and the sums compared using a t-test.
The data from UPLC-TOF-MS/MS lipidomic analyses were imported into PeakView software for relative quantification and lipid identification. Lipid species were confirmed by high-resolution MS, MS/MS fragmentation, and isotopic distribution, and then compared using the PeakView database. Peak intensities were used for relative quantification between E+ and E− zebrafish brains and were corrected for added internal standard (DTD-PC) and brain weight of each brain. Statistical differences between lipids were assessed as described for LC-MS/MS experiments.
RESULTS
Discovery of differentiating lipidomic metabolites and identification by MS/MSALL ™ analysis
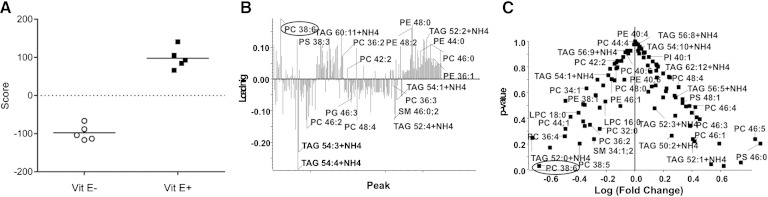
We used an untargeted shotgun lipidomics method consisting of the ordered acquisition of high-resolution accurate mass TOF detection to identify several characteristic lipid species, including PC, lysophosphatidylcholine (LPC), phosphatidylethanolamine (PE), SM, phosphatidylglycerol (PG), and TGs. Principal component analysis-discriminant analysis revealed significant differences between the lipid profiles of the extracts of E− and E+ zebrafish heads. The score plot provides a visual distribution of the features and degree of discrimination between the E− and E+ extracts (Fig. 1A), while the loading plot shows a number of lipids that lead to sample clustering (Fig. 1B). PC 38:6 was selected for further analysis because it both decreased in E− extracts (P < 0.05) and showed large fold differences between the two groups (Fig. 1C).
Fig. 1.
Identification of PC 38:6 as significantly depleted in heads from E− zebrafish. For 9 months beginning at 50 days postfertilization, zebrafish were fed defined diets (E+ or E−), which contained only fatty acids shorter than 18 carbons with two or three double bonds. For survey experiments using MS/MSALL TM, the head was removed just behind the eye from whole frozen zebrafish (n = 5 per group). The heads were weighed, then homogenized in methanol:water (80:20, v/v). Samples were extracted using a modified Bligh and Dyer solvent system (16); the final ratio was chloroform:methanol:water (2:2:1.8). Samples were mixed vigorously for 20 sec, and then centrifuged at 1,000 g for 10 min, the top layer was removed by aspiration and discarded. The bottom layer was collected and evaporated under nitrogen, then stored at −80°C less than 24 h before MS/MSALL TM analysis. A: Score plot provides a visual distribution of the features and degree of discrimination between individuals in E− compared with E+ groups. B: Loading plot shows the lipids that lead to sample clustering. Positive percent loading shows higher and negative percent loading shows lower intensities of lipid metabolites in the E+ group. C: Volcano plot of the Student’s t-test versus the log fold-change between groups. PC 38:6 (circled) was significantly decreased in E− extracts. TAG, triacylglyceride.
Brain DHA-PC (PC 16:0/22:6) and vitamin E concentrations
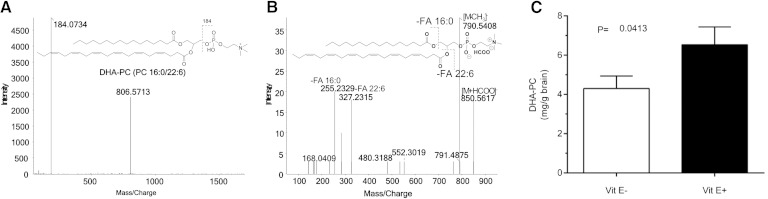
Brains were excised from individual fish; extracts were analyzed for PC 38:6, which was identified as DHA-PC (Fig. 2A, B). Brain PC 38:6 identification was based upon the retention time of an authentic standard and the detection of protonated molecular ion of m/z 806.7 [M+H]+ (data not shown). Moreover, the collision-induced dissociation of molecular ion m/z 806.7 produced a characteristic fragment ion of m/z 184 corresponding to the PC moiety (19). DTD-PC, the internal standard, also showed the same fragment ion of m/z 184 (Fig. 2A). The fragment ion, m/z 790.5408 shows loss of a –CH3 group from PC; the fragment ion, m/z 480.31882, is characteristic of 1-hexadecanoyl-sn-glycerophosphocholine (Fig. 2B). Additionally, the fragment ion peaks at m/z 225.2329 and 327.2315 are the fatty acyls 16:0 and 22:6, respectively. Thus, we confirmed the identification of DHA-PC.
Fig. 2.
Quantitation of PC 38:6 from E− and E+ brains. Fish were fed as described in Fig. 1. The brains from E− and E+ zebrafish (n = 10 per group) were dissected, weighed, and extracted individually at 4°C by adding, in the following order: 250 μl methanol:water (1:1, v/v), DTD-PC as an internal standard (2.5 μg in 5 μl methanol), and 750 μl methylene chloride:isopropanol:methanol (25:10:65, v/v/v). Samples were mixed vigorously for 20 s, and then centrifuged at 13,000 g for 10 min. The extracts were transferred to a new tube and were stored less than 24 h at −80°C until analysis. A: Chemical structure and positive fragmentation ion of DHA-PC based on MS/MSALL. B: Negative fragmentation ions of DHA-PC based on the TOF-MS/MS spectrum. C: Quantitation of DHA-PC (milligrams per gram of brain) in zebrafish brain extracts by LC-MS/MS in E− (open bar) and E+ (solid bar) (n = 10 per group). Error bars = SEM.
Quantitatively E− brains contained 30% less DHA-PC than did E+ brains (P < 0.05, Fig. 2C). Additionally, α-tocopherol concentrations in E− brains (3.4 ± 0.1 nmol/g, mean ± SEM) were 70 times lower than in the E+ brains (243 ± 32 nmol/g); α-tocopherol concentrations in E− bodies without brains (2.3 ± 0.3 nmol/g) were 60 times lower than in E+ bodies without brains (137 ± 13 nmol/g; two-way ANOVA: main effect of diet P < 0.0001, main effect of body part P = 0.0014). α-Tocopherol concentrations in E− brains (3.4 ± 0.1 nmol/g) were higher than in the rest of the body (2.3 ± 0.3 nmol/g, P < 0.05); as were α-tocopherol concentrations in E+ brains (234 ± 32 nmol/g) compared with E+ bodies without brains (137 ± 13 nmol/g, <0.05, paired t-test).
Untargeted lipidomics identifies alterations in brain lipids
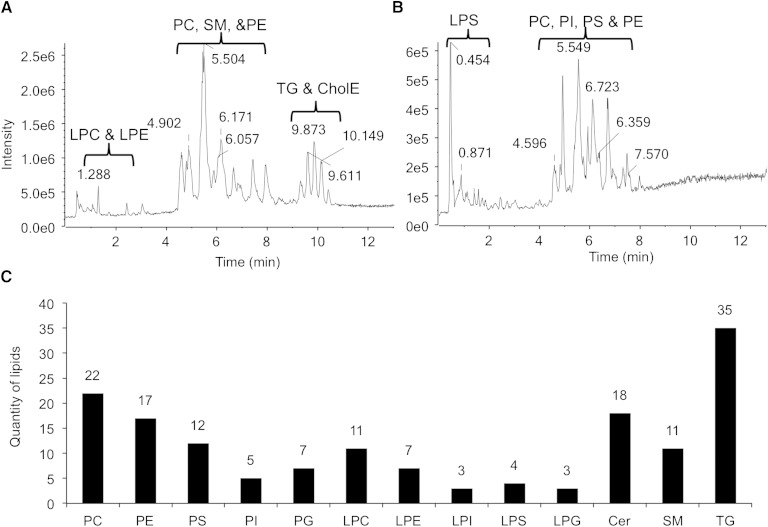
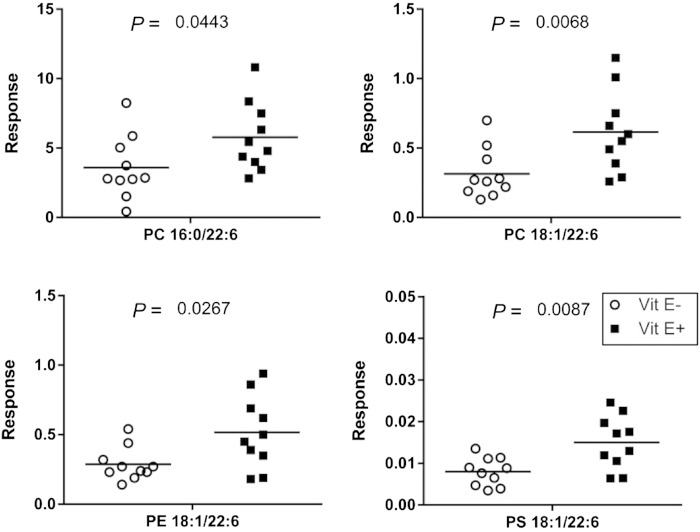
To further examine alterations in lipid distribution and relative abundances between E+ and E− zebrafish brains, we used an UPLC lipidomics technique (Fig. 3A, B). Four lipid classes including 13 lipid species with 155 specific lipids were identified (Fig. 3C). Lipid classes were detected in both positive [PC, ceramide, SM, PE, LPC, lysophosphatidylethanolamine (LPE), and TG] and in negative ion modes [phosphatidylserine (PS) and lysophosphatidylserine (LPS), PG, phosphatidylinositol (PI), and lysophosphatidylinositol (LPI)]; PC and PE were detected in both modes. E+ zebrafish brains contained several lipids (PC, LPC, LPE, LPS, LPI, and PS) at greater concentrations (P < 0.05) than in E− brains. PC 38:6 (16:0/22:6) was present at the highest intensities of any of the lipids that were significantly different between groups (Fig. 4). Additionally, three PLs containing both DHA and oleic acid (18:1) (PC 40:7, PE 40:7, and PS 40:7) were significantly different between the two groups (Fig. 4).
Fig. 3.
Lipid identification from UPLC-TOF-MS chromatograms. UPLC-TOF-MS chromatograms of zebrafish brain extracts (n = 10 per group) with precise separations during a run time of 15 min using positive (A) and negative (B) ion mode; different lipid classes elute in different elution time windows. CholE, Cholesterol ester. C: 13 lipid species and 155 individual lipids were identified in extracts of zebrafish brains, example of chromatogram. Cer, ceramide.
Fig. 4.
Four specific PLs containing DHA are significantly lower in E− brains. Data were generated from the UPLC separation of the lipid extracts of individual brains from E− and E+ fish, extracted as described in Fig. 2. Lipid species were confirmed by high-resolution MS, MS/MS fragmentation, and isotopic distribution, and then compared using the PeakView database; PeakView software was also used for relative quantification and lipid identification. Peak intensities were used for relative quantification between E+ and E− zebrafish brains. Shown are the individual values (E−, circles; E+, squares; n = 10 per group) with the median indicated with a line.
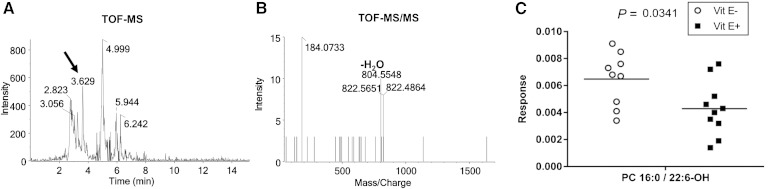
To determine whether loss of PC 38:6 (16:0/22:6) in E− brains could be a result of lipid peroxidation, the relative abundances of oxidized-DHA-PC in the extracts were assessed (Fig. 5). 1-Hexadecanoyl-2-(hydroxydocosahexaenoyl)-sn-glycero-3-phosphocholine [PC 16:0/22:6(OH)] was identified by the fragment ion, m/z 804.5548, which shows loss of a [H2O] group from the DHA fatty acyl chain and the fragment ion, m/z 184.0733, which is characteristic of PC. The increased hydroxy-DHA-PC concentrations in E− brains (P = 0.0341) support the hypothesis of greater DHA peroxidation.
Fig. 5.
Identification and quantitation of oxidized PC [16:0/22:6(OH)]. Data were generated from the UPLC separation of the lipid extracts of individual brains from E− and E+ fish, shown in Fig. 4. UPLC-TOF MS chromatogram (A) and MS/MS spectrum (B) of zebrafish brain extract using positive ion mode. C: Hydroxy-DHA-PC, shown are individual values of PC [16:0/22:6(OH)] in zebrafish brain extracts analyzed and corrected for internal standard and brain weight (P = 0.0341). [An outlier value in E− brains was excluded (Grubbs’, P < 0.05) from E− data set; otherwise P = 0.0175.]
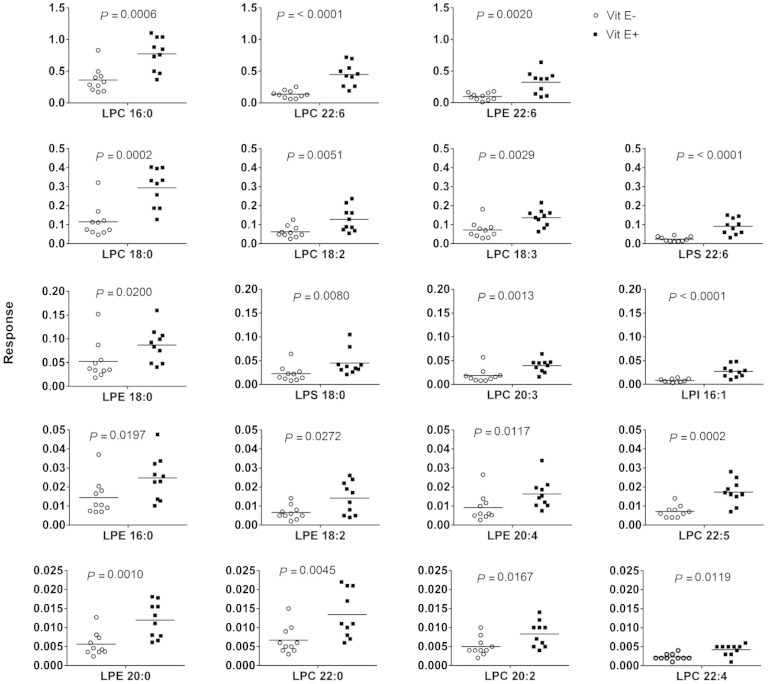
Remarkably, 19 lysoPLs were present at significantly higher concentrations in the E+ compared with E− brains (Fig. 6). These lysoPLs have been identified and confirmed using exact mass matching and MS/MS fragment patterns. Depending upon collision energy, a head group and/or one or more fatty acid chains were lost from the lipid molecules, which allowed us to identify the lipid class and fatty acid chains. PE generated the characteristic losses of 43 and 140 Da known to be associated with PE-head group losses; PS showed −87 and −184 Da loss. The lysoPL abundances varied with regard to fatty acid, but were uniformly higher in E+ brains. lysoPLs that were significantly higher in E+ and present at the highest abundances included LPC 16:0, LPC 22:6, and LPE 22:6 (top row, Fig. 6). There were several lysoPLs with saturated fatty acids significantly higher in E+ compared with E− brains; these not only included LPC 16:0, but also LPC 18:0, LPE 18:0, and LPS 18:0 and at lower abundances LPE 16:0 and LPE 20:0. Overall, the lysoPLs decreased in E− brains by nearly 60% (P = 0.0003).
Fig. 6.
lysoPLs are significantly depleted in E− zebrafish brains. Data were generated from the UPLC separation of the lipid extracts of individual brains from E− and E+ fish, shown in Fig. 4. Shown are the individual lysoPL species (vitamin E−, circles; vitamin E+, squares), which were significantly different between groups with the medians indicated (n = 10 per group). The data are displayed with those lysoPLs present at the highest abundances near the top of the figure, and the lowest near the bottom. Fatty acids present in the lysoPLs are indicated on the x axis, position could not be determined by this analysis.
DISCUSSION
Our discoveries in zebrafish show that low brain α-tocopherol concentrations are associated with a nearly 60% depletion of a total of 19 brain lysoPLs (combined P = 0.0003). The wide variety of lysoPLs that is depleted suggests that the entire lysoPL population is affected. lysoPLs are needed for PL remodeling during membrane synthesis, repair, and replacement (20, 21). Because rates of DHA synthesis in the brain are insufficient to meet brain DHA requirements (6, 22, 23), it has been proposed that the brain acquires DHA as docosahexaenoyl-sn-glycero-3-phosphocholine (lyso-DHA-PC) (24). Recently, a transporter from the major facilitator superfamily, major facilitator superfamily d2a (MFSD2a), which has high specificity toward lyso-DHA-PC (25), was shown to function as a DHA delivery mechanism facilitating brain docosahexaenoyl-sn-glycero-3-phospholipid (lyso-DHA-PL) uptake (25). Additionally, the MFSD2a transporter is critical to maintain the blood-brain barrier (26).
These major effects of vitamin E status on brain lysoPLs are surprising because lysoPLs with both saturated and unsaturated fatty acids are depleted in the E− brains. Generally, PLs have saturated fatty or monounsaturated acids located at the sn-1-acyl moiety and PUFAs are located at the sn-2-acyl moiety; the latter are released by phospholipase A (PLA)2 (27). Hypothetically, an oxidized DHA would be released by this mechanism, generating a lysoPL with a saturated or monounsaturated fatty acid. This mechanism would explain a higher level of lysoPLs with saturated fatty acids in E− brains; however, we observed the opposite, lower levels in E− brains of all the lysoPLs that were different between the two groups. An alternative explanation is the involvement of PLA1 enzymes releasing the sn-1-acyl moiety of PLs (28). Kuge et al. (29) investigated the role of PLA1 in regulating the localized distribution of PL species in the plasma membrane of cultured neurons and suggest that PLA1 may also contribute to the dynamic remodeling of the neural membrane.
Although our lipidomics analyses do not allow determination of the position of the fatty acids, we found three DHA-containing lysoPLs (LPC, LPE, and LPS) that were present at significantly lower concentrations in the E− brains. In dogs, supplements of fish oil (0.45, 0.9, or 1.35% fat) or vitamin E (dl-α-T-acetate, 640 IU/kg diet) increased serum 1-docosahexaenoyl-sn-glycero-3-phosphocholine concentrations (30). This finding of DHA specifically in the sn-1 position further supports our findings of decreases in LPC-DHA in E− adult zebrafish brains and the role of lyso-DHA-PC as the form DHA is transported from the liver to the brain (25).
DHA-PC was the lipid that showed the largest fold-differences between brains from E+ and E− zebrafish. Independent quantitation of DHA-PC showed that it was 30% lower in E− brains. Moreover, hydroxy-DHA-PC was elevated (Fig. 5). We have not identified whether the hydroxy-DHA was in the 1 or the 2 position because we lack authentic compounds; however, the high accuracy mass spectrometry data supports the identification of hydroxy-DHA-PC. Based on the relative responses of DHA-PC (16:0/22:6, Fig. 4) and hydroxy-DHA-PC (Fig. 5C), hydroxy-DHA-PC is less than 0.2% of the DHA-PC.
Our findings suggest that increased lipid peroxidation due to inadequate α-tocopherol leads to the depletion of a highly abundant brain PL, DHA-PC, as well as three other PLs containing both DHA and oleic acid (18:1) (PC, PE and PS, Fig. 4). Given α-tocopherol’s role as a peroxyl radical scavenger (31), it is expected that PLs containing the highly unsaturated DHA would be depleted from the E− brains. For example, Xu, Davis, and Porter (32) reported the rate constants for autoxidation of the propagation of several unsaturated lipids in benzene solution, showing that the rate of DHA oxidation (334 M−1 s−1) is far greater than that of AA (197 M−1 s−1). Previously, we identified a more rapid depletion of both DHA and AA from E− zebrafish embryos (5), but in these adult brains, we found little evidence for AA depletion. This difference may be a result of the greater likelihood for oxidation of DHA (32). Alternatively, the brain may be able to synthesize sufficient AA, but not DHA, for its needs (33). Additionally, 7-dehydrocholesterol, an intermediate in cholesterol synthesis, is more highly oxidizable than either of these fatty acids, but using our lipidomics data, we did not detect either 7-dehydrocholesterol or its oxidation product. It is likely that adult brain does not synthesize cholesterol extensively and therefore 7-dehydrocholesterol was not detectable.
Neural membrane PL remodeling liberates both AA and DHA, which serve as precursors for lipid mediators and signaling molecules. For example, AA can be metabolized to inflammatory eicosanoids via cyclooxygenases to yield prostaglandins and thromboxanes or via lipoxygenases to yield leukotrienes (34). By contrast, DHA can be metabolized to the anti-inflammatory docosanoids via lipoxygenase enzymes to yield D-series resolvins and other neuroprotective compounds (e.g., neuroprotectin D1) (35). Given the high brain DHA concentrations, as well as DHA’s role in supporting neurological health across the lifespan (36, 37), it is remarkable that PL-containing DHA, but not AA, was different between adult E− and E+ zebrafish brains. These results suggest that α-tocopherol is required within the nervous system to protect DHA and thereby maintain the brain’s homeostatic balance between anti-inflammatory and inflammatory compounds. These results also show that there was not artifactual nonspecific lipid peroxidation of the E− samples.
We are pursuing vitamin E deficiency effects in development of zebrafish embryos. Notably, Huang et al. (38) have published that PC 38:6 is decreased in 24 and 48 h embryos compared with earlier time points. These findings suggest that PC 38:6 is an important target to evaluate in the E− embryo.
We report herein that α-tocopherol deficiency depletes four DHA-containing PLs and 19 lysoPLs. A unique biological role has been proposed for vitamin E in membrane repair (39). Potentially, the alterations in lysoPLs we observed in the E− brains are a result of a depletion of DHA-containing PLs and increased requirements for DHA for PL synthesis and lysoPLs for remodeling for membrane repair (Fig. 7). Presumably, inadequate α-tocopherol concentrations allow lipid peroxidation to deplete not only brain DHA-PC, but DHA throughout the body, thereby limiting DHA delivery to the brain. Our data delineate that critical lipids are protected by α-tocopherol and suggest why α-tocopherol is needed as a vitamin.
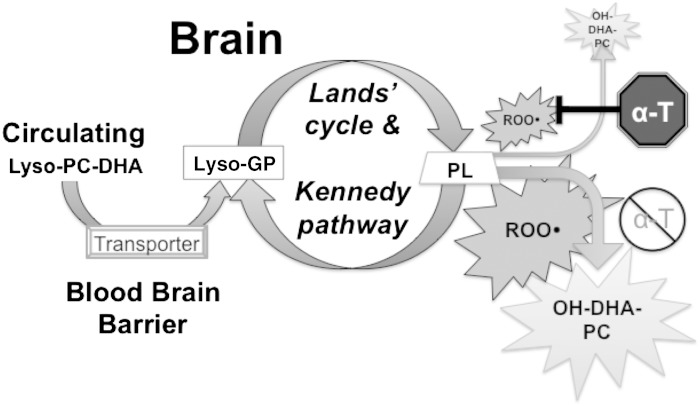
Fig. 7.
Hypothesis linking lysoPLs, DHA-PC, and vitamin E deficiency in zebrafish brains. To maintain the membranes in the brain, lysoPLs, especially lyso-DHA-PC (with DHA in the 1 position), are transported into the brain from the circulation via a transporter, likely MFSD2a (25), where these lysoPLs are converted via the Land’s cycle and Kennedy pathway to various PLs, including DHA-PC (with DHA in the 2 position). A peroxyl radical (ROO•) attacks a membrane PL (e.g., DHA-PC, likely forming an intermediate [DHA-(OOH)-PC], which is reduced to DHA-(OH)-PC (hydroxy-DHA-PC was detected in this study). α-Tocopherol (α-T) limits this process. Inadequate vitamin E allows lipid peroxidation to damage the membrane, which must be repaired. In the face of inadequate α-T, more lysoPLs are needed for membrane repair, but sufficient amounts, especially of lyso-DHA-PC, may not be available (possibly due to increased lipid peroxidation in the rest of the body). The increased membrane remodeling depletes various lysoPLs in the brains of E− zebrafish.
Acknowledgments
The authors thank Carrie L. Barton and Jane La Du for providing excellent technical assistance.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- α-TTP
- α-tocopherol transfer protein
- DHA-PC (38:6)
- 1-hexadecanoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine
- DTD-PC
- 1,2-ditridecanoyl-sn-glycero-3-phosphocholine
- E−
- vitamin E-deficient
- E+
- vitamin E-sufficient
- LPC
- lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- LPI
- lysophosphatidylinositol
- LPS
- lysophosphatidylserine
- MFSD2a
- major facilitator superfamily d2a
- MRM
- multiple reaction monitoring
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PLA
- phospholipase A
- PS
- phosphatidylserine
- UPLC
- ultra-performance LC
This work was supported by National Institutes of Health Grants S10RR027878, NICHD HD062109 (M.G.T. and R.L.T.), and NIEHS ES000210.
REFERENCES
- 1.Evans H. M., Bishop K. S. 1922. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science. 56: 650–651. [DOI] [PubMed] [Google Scholar]
- 2.Miller G. W., Labut E. M., Lebold K. M., Floeter A., Tanguay R. L., Traber M. G. 2012. Zebrafish (Danio rerio) fed vitamin E-deficient diets produce embryos with increased morphologic abnormalities and mortality. J. Nutr. Biochem. 23: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traber M. G. 2013. Mechanisms for the prevention of vitamin E excess. J. Lipid Res. 54: 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller G. W., Ulatowski L., Labut E. M., Lebold K. M., Manor D., Atkinson J., Barton C. L., Tanguay R. L., Traber M. G. 2012. The alpha-tocopherol transfer protein is essential for vertebrate embryogenesis. PLoS ONE. 7: e47402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebold K. M., Lohr C. V., Barton C. L., Miller G. W., Labut E. M., Tanguay R. L., Traber M. G. 2013. Chronic vitamin E deficiency promotes vitamin C deficiency in zebrafish leading to degenerative myopathy and impaired swimming behavior. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 157: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapoport S. I., Igarashi M. 2009. Can the rat liver maintain normal brain DHA metabolism in the absence of dietary DHA? Prostaglandins Leukot. Essent. Fatty Acids. 81: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulatowski L., Manor D. 2013. Vitamin E trafficking in neurologic health and disease. Annu. Rev. Nutr. 33: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulatowski L., Parker R., Warrier G., Sultana R., Butterfield D. A., Manor D. 2014. Vitamin E is essential for Purkinje neuron integrity. Neuroscience. 260: 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebold K. M., Jump D. B., Miller G. W., Wright C. L., Labut E. M., Barton C. L., Tanguay R. L., Traber M. G. 2011. Vitamin E deficiency decreases long-chain PUFA in zebrafish (Danio rerio). J. Nutr. 141: 2113–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishi S., Uchiyama J., Baughman A. M., Goto T., Lin M. C., Tsai S. B. 2003. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp. Gerontol. 38: 777–786. [DOI] [PubMed] [Google Scholar]
- 11.Keller E. T., Murtha J. M. 2004. The use of mature zebrafish (Danio rerio) as a model for human aging and disease. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 138: 335–341. [DOI] [PubMed] [Google Scholar]
- 12.Traber M. G. 2014. Vitamin E inadequacy in humans: causes and consequences. Adv. Nutr. 5: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podda M., Weber C., Traber M. G., Packer L. 1996. Simultaneous determination of tissue tocopherols, tocotrienols, ubiquinols, and ubiquinones. J. Lipid Res. 37: 893–901. [PubMed] [Google Scholar]
- 14.Frei B., England L., Ames B. N. 1989. Ascorbate is an outstanding antioxidant in human-blood plasma. Proc. Natl. Acad. Sci. USA. 86: 6377–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkwood J. S., Lebold K. M., Miranda C. L., Wright C. L., Miller G. W., Tanguay R. L., Barton C. L., Traber M. G., Stevens J. F. 2012. Vitamin C deficiency activates the purine nucleotide cycle in zebrafish. J. Biol. Chem. 287: 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 17.Simons B., Duchoslav E., Burton L., Bonner R. 2011. Molecular Characterization and Quantitation of Lipids with High Resolution Accurate Mass Tandem MS Techniques. AB SCIEX, Framingham, MA. Available from: http://www.absciex.com/Documents/Downloads/Literature/Lipid20Profiling_Infusion%20MSMSALL_TripleTOF%205600_4430211-01.pdf ABScience Publication Number: 4430211-01.
- 18.Monette J. S., Gomez L. A., Moreau R. F., Bemer B. A., Taylor A. W., Hagen T. M. 2010. Characteristics of the rat cardiac sphingolipid pool in two mitochondrial subpopulations. Biochem. Biophys. Res. Commun. 398: 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu F. F., Turk J. 2003. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: the fragmentation processes. J. Am. Soc. Mass Spectrom. 14: 352–363. [DOI] [PubMed] [Google Scholar]
- 20.Shindou H., Hishikawa D., Harayama T., Eto M., Shimizu T. 2013. Generation of membrane diversity by lysophospholipid acyltransferases. J. Biochem. 154: 21–28. [DOI] [PubMed] [Google Scholar]
- 21.Hishikawa D., Hashidate T., Shimizu T., Shindou H. 2014. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 55: 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igarashi M., Chang L., Ma K., Rapoport S. I. 2013. Kinetics of eicosapentaenoic acid in brain, heart and liver of conscious rats fed a high n-3 PUFA containing diet. Prostaglandins Leukot. Essent. Fatty Acids. 89: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapoport S. I., Igarashi M., Gao F. 2010. Quantitative contributions of diet and liver synthesis to docosahexaenoic acid homeostasis. Prostaglandins Leukot. Essent. Fatty Acids. 82: 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagarde M., Bernoud N., Brossard N., Lemaitre-Delaunay D., Thies F., Croset M., Lecerf J. 2001. Lysophosphatidylcholine as a preferred carrier form of docosahexaenoic acid to the brain. J. Mol. Neurosci. 16: 201–204, discussion 215–221. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen L. N., Ma D., Shui G., Wong P., Cazenave-Gassiot A., Zhang X., Wenk M. R., Goh E. L., Silver D. L. 2014. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 509: 503–506. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Zvi A., Lacoste B., Kur E., Andreone B. J., Mayshar Y., Yan H., Gu C. 2014. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 509: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennis E. A., Cao J., Hsu Y. H., Magrioti V., Kokotos G. 2011. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111: 6130–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond G. S., Smith T. K. 2011. Phospholipases A(1). Int. J. Mol. Sci. 12: 588–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuge H., Akahori K., Yagyu K., Honke K. 2014. Functional compartmentalization of the plasma membrane of neurons by a unique acyl chain composition of phospholipids. J. Biol. Chem. 289: 26783–26793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall J. A., Brockman J. A., Jewell D. E. 2011. Dietary fish oil alters the lysophospholipid metabolomic profile and decreases urinary 11-dehydro thromboxane B(2) concentration in healthy Beagles. Vet. Immunol. Immunopathol. 144: 355–365. [DOI] [PubMed] [Google Scholar]
- 31.Traber M. G., Atkinson J. 2007. Vitamin E, antioxidant and nothing more. Free Radic. Biol. Med. 43: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L., Davis T. A., Porter N. A. 2009. Rate constants for peroxidation of polyunsaturated fatty acids and sterols in solution and in liposomes. J. Am. Chem. Soc. 131: 13037–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L. E., Chen C. T., Hildebrand K. D., Liu Z., Hopperton K. E., Bazinet R. P. 2015. Chronic dietary n-6 PUFA deprivation leads to conservation of arachidonic acid and more rapid loss of DHA in rat brain phospholipids. J. Lipid Res. 56: 390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frisardi V., Panza F., Seripa D., Farooqui T., Farooqui A. A. 2011. Glycerophospholipids and glycerophospholipid-derived lipid mediators: a complex meshwork in Alzheimer’s disease pathology. Prog. Lipid Res. 50: 313–330. [DOI] [PubMed] [Google Scholar]
- 35.Bazan N. G. 2009. Cellular and molecular events mediated by docosahexaenoic acid-derived neuroprotectin D1 signaling in photoreceptor cell survival and brain protection. Prostaglandins Leukot. Essent. Fatty Acids. 81: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H. Y. 2007. Novel metabolism of docosahexaenoic acid in neural cells. J. Biol. Chem. 282: 18661–18665. [DOI] [PubMed] [Google Scholar]
- 37.André A., Juanéda P., Sebédio J. L., Chardigny J. M. 2006. Plasmalogen metabolism-related enzymes in rat brain during aging: influence of n-3 fatty acid intake. Biochimie. 88: 103–111. [DOI] [PubMed] [Google Scholar]
- 38.Huang S. M., Xu F., Lam S. H., Gong Z., Ong C. N. 2013. Metabolomics of developing zebrafish embryos using gas chromatography- and liquid chromatography-mass spectrometry. Mol. Biosyst. 9: 1372–1380. [DOI] [PubMed] [Google Scholar]
- 39.Howard A. C., McNeil A. K., McNeil P. L. 2011. Promotion of plasma membrane repair by vitamin E. Nat. Commun. 2: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]