Abstract
Most new genes are thought to evolve from preexisting genes but duplications of entire genes or shuffling of preexisting exons provides only a limited repertoire of new sequences that can be presented to a cell. Only pieces that previously existed can be used in the construction and any further divergence depends on the slow accumulation of mutations. We show here the presence of a small, in-frame intron in a ciliate phosphoglycerate kinase gene and the insertion of an unusually random amino acid sequence at the same position in trypanosome phosphoglycerate kinase. The unusual sequences in trypanosomes were likely to have originally been introns that have been subsequently captured by the protein and have now been incorporated as part of the coding sequence. Via this path a truly unique sequence can be incorporated into an existing protein, leading in time to the evolution of a new, functionally distinct protein.
Full text
PDF
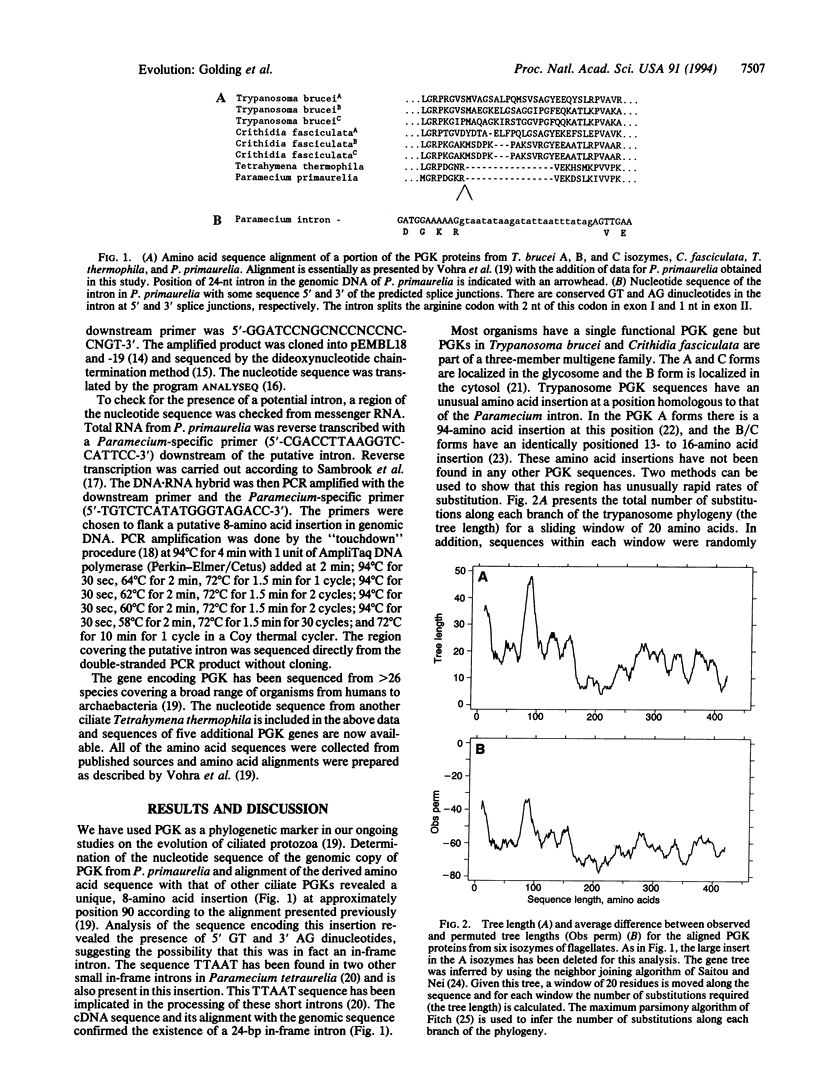
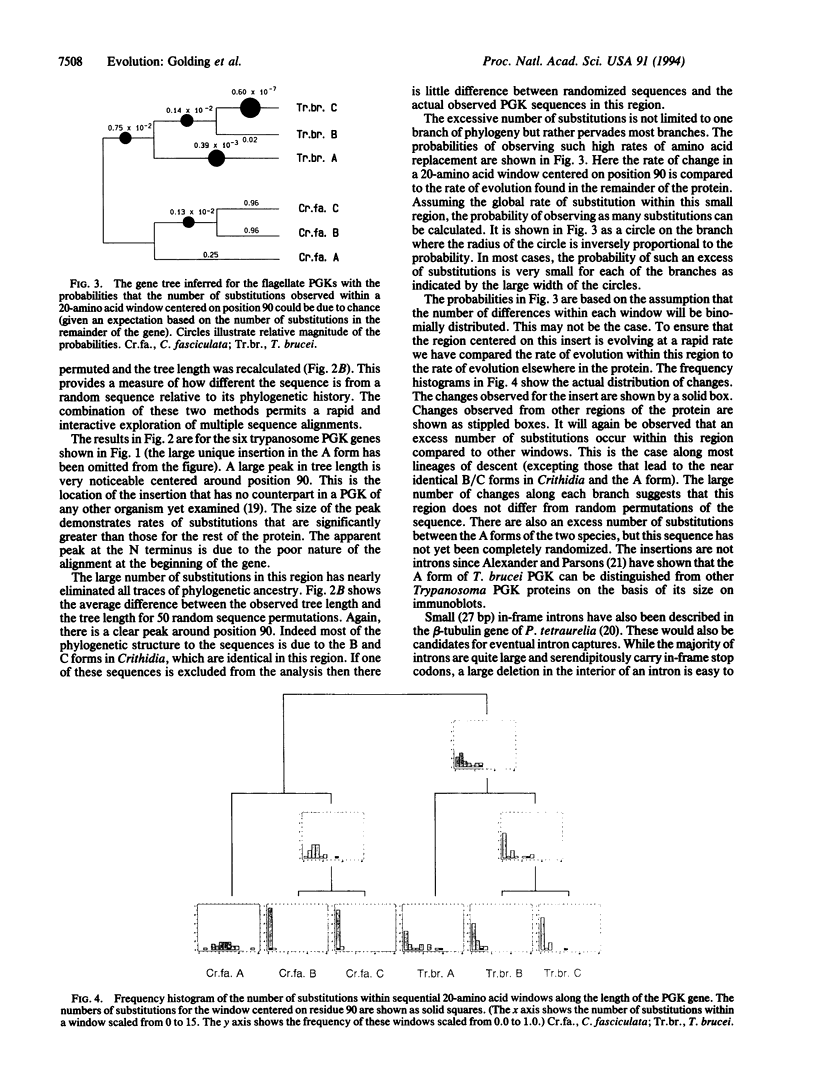

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander K., Parsons M. A phosphoglycerate kinase-like molecule localized to glycosomal microbodies: evidence that the topogenic signal is not at the C-terminus. Mol Biochem Parasitol. 1991 May;46(1):1–10. doi: 10.1016/0166-6851(91)90193-a. [DOI] [PubMed] [Google Scholar]
- Blake C. Exons--present from the beginning? Nature. 1983 Dec 8;306(5943):535–537. doi: 10.1038/306535a0. [DOI] [PubMed] [Google Scholar]
- Butticè G., Kaytes P., D'Armiento J., Vogeli G., Kurkinen M. Evolution of collagen IV genes from a 54-base pair exon: a role for introns in gene evolution. J Mol Evol. 1990 Jun;30(6):479–488. doi: 10.1007/BF02101102. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985 May 23;315(6017):283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Cox P. T., Wainwright B. J., Baker K., Mattick J. S. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991 Jul 25;19(14):4008–4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorit R. L., Schoenbach L., Gilbert W. How big is the universe of exons? Science. 1990 Dec 7;250(4986):1377–1382. doi: 10.1126/science.2255907. [DOI] [PubMed] [Google Scholar]
- Dupuis P. The beta-tubulin genes of Paramecium are interrupted by two 27 bp introns. EMBO J. 1992 Oct;11(10):3713–3719. doi: 10.1002/j.1460-2075.1992.tb05456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. The exon theory of genes. Cold Spring Harb Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Go M., Nosaka M. Protein architecture and the origin of introns. Cold Spring Harb Symp Quant Biol. 1987;52:915–924. doi: 10.1101/sqb.1987.052.01.100. [DOI] [PubMed] [Google Scholar]
- Le Blancq S. M., Swinkels B. W., Gibson W. C., Borst P. Evidence for gene conversion between the phosphoglycerate kinase genes of Trypanosoma brucei. J Mol Biol. 1988 Apr 5;200(3):439–447. doi: 10.1016/0022-2836(88)90534-7. [DOI] [PubMed] [Google Scholar]
- Ohno S. Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2421–2425. doi: 10.1073/pnas.81.8.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Original domain for the serum albumin family arose from repeated sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7657–7661. doi: 10.1073/pnas.78.12.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. W., Feng D. F., Doolittle R. F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992 Dec;17(12):489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Methods Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Swinkels B. W., Evers R., Borst P. The topogenic signal of the glycosomal (microbody) phosphoglycerate kinase of Crithidia fasciculata resides in a carboxy-terminal extension. EMBO J. 1988 Apr;7(4):1159–1165. doi: 10.1002/j.1460-2075.1988.tb02926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T. C., Goldstein J. L., Brown M. S., Russell D. W. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985 May 17;228(4701):815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohra G. B., Golding G. B., Tsao N., Pearlman R. E. A phylogenetic analysis based on the gene encoding phosphoglycerate kinase. J Mol Evol. 1992 May;34(5):383–395. doi: 10.1007/BF00162995. [DOI] [PubMed] [Google Scholar]




