Abstract
Objective: To investigate the expression of mir-16 in lung adenocarcinoma cancer line and to observe the effect of mir-16 on the biological behaviors of human lung adenocarcinoma cancer A549 cell. Methods the expression of mir-16 in A549 cells was examined by quantitative real-time (qRT)-PCR. mir-16 minics was chemically synthesized and transfected into A549 cells by Lipofectamine 2000. The cell cycle and apoptosis changes were assayed by flow cytometry, the cell proliferation was measured by MTS assay. The wild-type and mutant wip1 3’-UTR luciferase reporter rectors were constructed. The relative activity of renila luciferase was detected to confirm the binding site of mir-16 on wip1 mRNA. Results, the expression of mir-16 is reduced in A549 cell compared with the normal bronchial epithelial cell. Transfection of mir-16 minics significantly suppressed the luciferase reporter containing wild type not mutant wip1 3’-UTR. Furthermore enforced expression of mir-16 lead to reduced A549 cell proliferation and promote apoptosis. Conclusion Therapeutic strategies to resume miRNA-16 expression may be benefit to patients with NSCLC in the feature.
Keywords: Mir-16, lung adenocarcinoma, apoptosis
Introduction
microRNA (miRNA) are a class of small non-coding RNA molecules, composed of 18-25 nucleotides in length, as a regulation of gene expression at the posttranscriptional level, Different miRNAs are in the form of oncogenes or tumor suppressor genes, respectively, which play an important role in occurrence and development of malignancy [1-3]. mir-16 was first discovered in chronic granulocytic leukemia, and considered it is a tumor suppressor gene [4]. However, the mir-16 of lung cancer is proto-oncogene or tumor suppressor gene, and the mechanism of action remains unclear [5,6]. In this study, we first detect the expression of Mir-16 in lung adenocarcinoma cell. On this basis, we transiently transfect mir-16 into A549 cell and observe the effect of mir-16 on proliferation, cell cycle and apoptosis in lung adenocarcinoma cells, in order to explore new targets in the pathogenesis and treatment of lung adenocarcinoma.
Materials and methods
Cell lines and primary reagent
Human A549 lung adenocarcinoma cells were purchased from the Animal Laboratory of Sun Yat-sen University, normal human bronchial epithelial cells HBE were purchased from Hunan yongtai Biotechnology Co., Ltd; 1640 culture and fetal calf serum were purchased from hyclone; Opti-MEM culture, PBS, pancreatin were purchased from Gibco; Trizol, Lipofectamine™ RNAiMAX Transfection reagents were purchased from invitrogen; M-MLV, Rnasin inhibitor, psiCHECK-2, DpnI enzyme were purchased from Promega; SYBR Green PCR Master Mix were purchased from TOYOBO; PrimeSTAR HS, DNA-Polymerse, T4 DNA Ligase, DL2000, 1 kbp DNA Ladder Marker, NotI, XhoI, TaKaRa TaqTM were purchased from TaKaRa; OneStep RT-PCR kits were the products of Qiagen; Dual-Luciferase Reporter Assay System was the products of Promega; DNA purification kit and plasmid prep kit were purchased from DONGSHENG BIOTECH; High purity plasmid mini prep kit were purchased from G-SHUN; Annexin V-FITC apoptosis assays kit and cell cycle kit were purchased from Keyge; Primers was synthesized by Sangon; Gene sequencing was completed by Beijing genomics institute; mir-16 fragment was synthesized by Guangzhou Rui Bo Biological Technology Co., Ltd.
Cell culture
A549 cells were seeded at low glucose DMEM culture medium containing 10% FBS, cultured in 5% CO2 saturated humidity incubator at 37°C, inverted, observed under the microscope, passaged 2 or 3 times weekly, cells within 50 generations are selected to transfection.
Group of experiments
Non-transfected A549 cell was a blank control group, transfected empty vector A549 cell was a negative control group, Transfected mir-16 A549 cell was a specific transfection group, and normal bronchial epithelial cells group.
Transfection
24 hours before transfection, A549 cells in the logarithmic phase were inoculated in 6-well plates in the form of 5 × 104 cells/well, respectively. When the degree of cell fusion reached 30%-50%, dissolved 20 um mir-16 mother liquors into 100 μL Opti-MEM culture medium as A liquid; dissolved 2 μL Lipofectamine™ RNAiMAX reagent into 100 μL Opti-MEM culture medium as B liquid. A liquid and B liquid were mixed, and let stand for 20 minutes. And then add mixed liquor to cell culture plate with 300 μL control medium without antibiotic. 4 hours after transfection, changed to 1640 medium.
Detect the expression of mir-16 in transfected cells by quantitative PCR
48 hours after transfection, collected cells of each group, respectively. Total RNA was extracted by Trizol reagent, U 6 as internal control performed RT-PCR amplification reaction, the volume was 20 μL, amplification system: 2 × PCR premix 10 μL, primers 0.8 μL, cDNA 1 μL, H2O make up the volume of 20 μL. Reaction conditions: 95°C: 5 min; 95°C: 15 s; 65°C: 15 s; 72°C: 32 s; 40 cycles. Each sample set three wells, separated by 2% agarose gel electrophoresis and visualized by UVP gel imaging system.
Detect effects of mir-16on cell proliferation by MTS
Non-transfected cell, transfected mir-16 A549 cell and cells of empty vector were digested and resuspended, respectively.adjust the cell concentration of 1 × 105/ml, added into 96-well plates, each well 100 μL (1 × 104 per well ). At 24 h, 48 h, 72 h, 96 h, adding MTS reagent (CellTiter 96® AQueous One Solution Cell Proliferation Assay) into corresponding culture wells, respectively, 20 μL per well. Incubate 3 hours in 5% CO2 environment at 37°C. The absorbance in 490 nm was Detect the apoptosis with flow cytometry.
48 h after transfection cell were digested with trypsin and made into cell suspension, collected the cell by centrifuge, washed with PBS twice and then mixed 4 × 105 cells with 500 μL binding buffer, 5 μL Annexin V-FITC and 5 μL PI staining solution, stained at dark for 15 min at room temperature, detected the apoptosis with flow cytometry by standard procedure.
Detect the cell cycle with flow cytometry
48 h after transfection, cell were digested with trypsin and made into cell suspension, washed with pre-cooling PBS twice, fixed with 70% pre-cooling ethanol overnight, collected the cell by centrifuge, washed with 1 ml PBS, added 100 μg/mL RNase A incubated at 37°C for 30 min, added 500 μL PI incubated at 4°C for 30 min at dark, 3 × 104 cells for cell cycle analysis with flow cytometry by standard procedure.
Luciferase reporter gene detection
According to microRNA target prediction software (Targetscan), it showed that mir-16 can bind to 3’-UTR of wip1 mRNA. In vitro synthetize the PCR primer with restriction enzyme digestion sites, inserted the product to the psi-CHECK-2 vector downstream of renilla luciferase reporter gene, constructed the expression vector of 3’-UTR of wild type wip1, mutate the vector according to the binding sites of mir-16 and 3’-UTR of wip1, and constructed the mutant 3’-UTR of wip1 expression vector. Added 5 µL ligased product to the 50 µL DH5α competent cell, plate the cell on the LB plate containing Amp (100 μg/mL), picking monoclone for PCR determination, and extracted the positive clone plasmid for XhoI and NotI digestion, detect by 1% agarose with EB, take picture with gel imaging system. Contransfected mir-16 fragment mimics with plasmid constructed above, for blank group transfected plasmid alone. After 48 hours transfection, detected the activity of renilla luciferase reporter gene with Dual-Luciferase Assay Kit, the firefly luciferase act as internal reference.
Statistical methods
The results were performed in 3 replications. Statistical differences of measurement data were compared with ANOVA method, P<0.05, with statistical difference.
Results
Detect the expression of mir-16 in A549 cells, normal bronchial epithelial cells and in transfected cells
The differential expression of mir-16 in transfected mir-16 A549 cell, transfected empty vector A549 cell, non-transfected A549 cell and normal bronchial epithelial cells were detected by (qRT) -PCR. The results showed that the expression of mir-16 in A549 cells was low than in normal bronchial epithelial cells, the expression of mir-16 in transfected A549 cells showed significantly increase, significantly higher than transfected empty vector A549 cell and non-transfected A549 cells, suggesting that the expression of mir-16 in lung adenocarcinoma cells and normal bronchial epithelial cells decreased, and expression of mir-16 in transfected mir-16 A549 cell increased, indicating that mir-16 was successfully transfected into A549 cells (See Figure 1).
Figure 1.
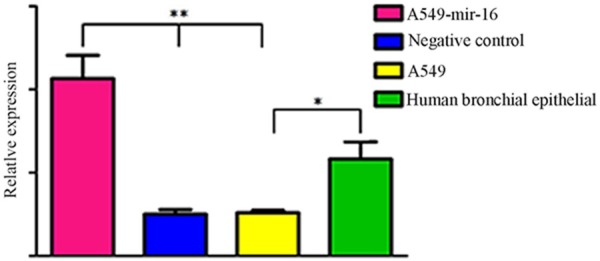
The expression of mir-16. Real-time RT-PCR revealed that the expression level of mir-16 is decreased in A549 cells compared to the Normal human bronchial epithelial cells (*P<0.05). After transfection, the expression level of mir-16 in the A549 cells is higher than other cells (**P<0.05), values represent means from three separate experiments.
Effects of mir-16 on cell proliferation
After the cells were cultured 24, 48 h, 72 h, 96 h, detect cell proliferation. The results showed that cell proliferation of transfected mir-16 A549 cells was significantly slowed down after the cells were cultured 96 h (See Figure 2).
Figure 2.
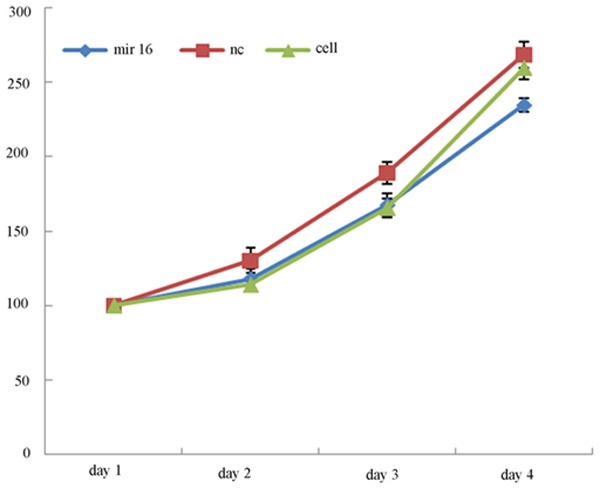
Effect of mir-16 on growth of A549 cells. MTS growth curves indicated that the proliferation rate of A549 cells transfected with mir-16 is significantly decreased compare to the other cells (values represent means from there separate experiments, P<0.05).
Effects of mir-16 on cell apoptosis
Cells were stained by Annexin V-FITC, flow cytometry were used to detect different apoptosis of three groups, respectively, verified the effects of mir-16 on A549 cell apoptosis. The results showed that early apoptosis of transfected mir-16 A549 cells was significantly increased than the blank control group and non-transfected cells group (See Figure 3A, 3B).
Figure 3.
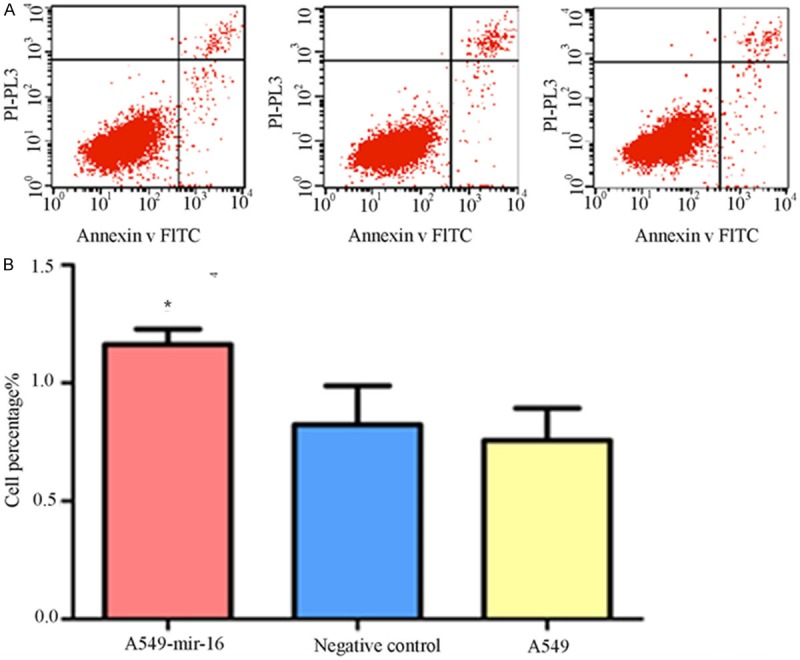
Effect of mir-16 on cell apoptosis. A. Flow cytometry pictures of cell apoptosis (lower right quadrant, one of the three separate results was showed); B. Percentage of three groups in early stage apoptosis. Flow cytometry showed that the percentage of mir-16 transfected A549 cells in early stage apoptosis is higher than that of other two groups (values represent means from three separate experiments, *P<0.05).
Effects of mir-16 on cell cycle
Flow cytometry were used to detect the effects of mir-16 on cell cycle, the results showed that G1 phase of transfected mir-16 A549 cell was significantly prolong than NC group and cell group, S phase was significantly shortened, cell growth delay (Figure 4A, 4B).
Figure 4.
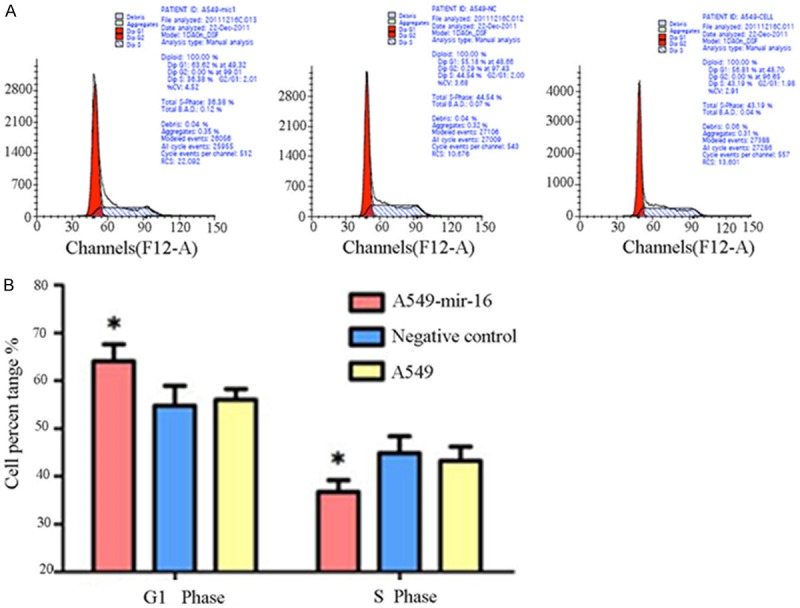
Effect of mir-16 on cell cycle of A549 cells. A. Flow cytometry pictures of cell cycle (one of the three separate results was showed). B. Flow cytometry results showed that the percentage of A549 cells transfected with mir-16 in the G0-G1 phase is higher than the other two groups, which paralleled with decrease in the S phase (values represent means from three separate experiments, *P<0.05).
Luciferase reporter gene detection
Prediction software suggested wip1 was one of target gene of mir-16 (see Figure 5). Construct the vector contain 3’UTR of wip1 gene. At first, amplified 3’UTR of wip1 gene by PCR, purified the products by gel purification, and ligased to the vector, the restriction enzymes digestion and sequencing showing that the vector was successfully constructed (see Figure 6), and then the same law for 3’UTR same gene mutation. Two kinds of luminescent signal of luciferase were detected by luciferase reporter gene. the results show that compared to mutant wip1 exist in the co-transfected, mir-16 and wild-type wip1 exist in the co-transfected experimental group can significantly reduced the activity of luciferase, the fluorescence signal was significantly reduced, indicating that mir-16 can be combined with 3’UTR of the wild-type wip1 gene, thereby inhibiting the activity of luciferase, but not in combination with 3’UTR of the mutant wip1 gene, and therefore considered wip1 may be a target gene of mir-16. NC group and the blank control group were not significantly different (See Figure 7).
Figure 5.

wip1 is one of the target gene of mir-16. Predicted mir-16 sequence and putative recognition sites with 3’-UTR of wip1.
Figure 6.
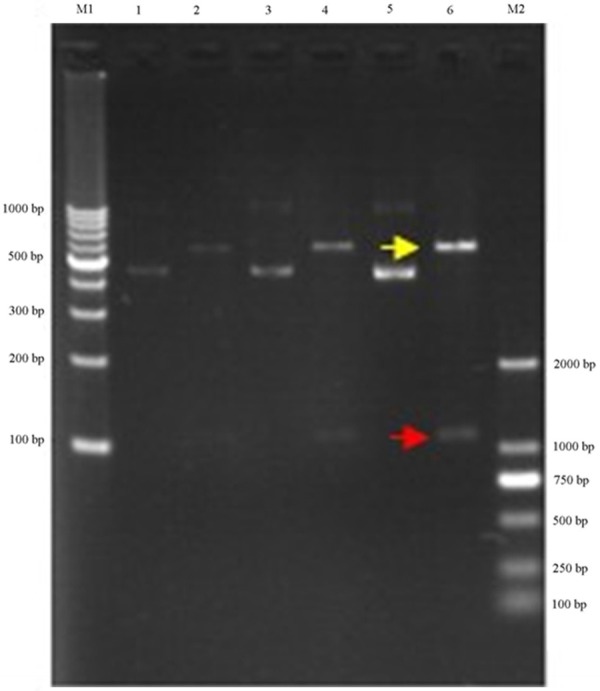
Construct renila luciferase reporter. Clone of wip1 3’-UTR protein containing miRNA-16 target site and not containing the mir-16 target site (mutant wip1 3’-UTR) into a renila luciferase reporter construct.
Figure 7.
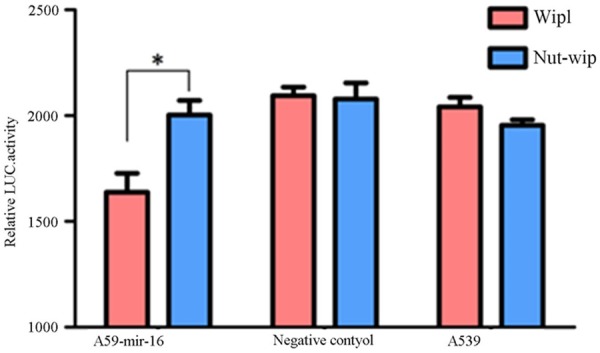
Luciferase activity using wild-type and mutated wip1-3’-UTR constructs. Luciferase activity of A549 cells after transfection with mir-16, negative control or no transfected mir-16 and then also co-transfected with vector containing either WT-wip1-3’-UTR or MUT-wip1 3’-UTR. A significant reduction of the luciferase activity was observed in A549 transfected with mir-16 and WT-wip1-3’-UTR than with MUT-wip1 3’-UTR. There were no significant differences in other two groups (values represent means from three separate experiments, *P<0.05).
Discussion
Currently, the study found about 40-45 species of the expression of miRNA are abnormal in lung cancer, some of which plays an important role in the occurrence and development of lung cancer , the most famous is the Let-7 family [7]. mir-16 was first discovered in chronic granulocytic leukemia, mir-16 plays the role of cancer suppressor gene in colorectal cancer, pancreatic cancer, glioma and so on. But the mechanism of mir-16 remains unclear in lung cancer.
Abnormal cell proliferation and apoptosis plays an important role in cell growth and the occurrence of tumours. Compared to normal cells, death and apoptosis of malignant cell had defects, and apoptosis resistance of tumor cells often caused tumor cells to grow out of control. Most chemotherapeutic agents kill tumor cells by inducing apoptosis, malignant cells escape apoptosis through some mechanisms [8,9]. At present, inducing apoptosis of tumor cells by genetic technology is therapeutical hot spot, it can enhance the sensitivity of tumor cells to chemotherapeutic drugs and reduce drug resistance to chemotherapy. What is being explored here is whether or not miRNA-16 can play an role in inhibiting tumor cell proliferation and promoting apoptosis in lung adenocarcinoma. Studied the function of mir-16 by using transient transfection of microRNA, made the expression of mir-16 up-regulate by transfecting mir-16 into A549 cell. The results showed the up-regulation of miRNA-16 can significantly inhibit cell growth and promote early apoptosis in lung adenocarcinoma cells, indicating miRAN-16 had a function of cancer suppressor gene, so increasing the expression level of miRAN-16 can be as a new strategy of treatment of lung adenocarcinoma.
In order to understand the mechanism of miRNA-16 may affect the biological characteristics of cancer cells, we used dual-luciferase reporter gene system. It is the most effective verification methods, with high sensitivity and specificity. We found that miRNA-16 can significantly reduce luciferase activity of the reporter plasmid of wild-type wip1 gene 3’-UTR, and luciferase activity of the reporter plasmid of the mutant gene 3’-UTR wip1 did not significant function, thus validated miRNA-16 can be combined directly with 3’-UTR of wip1 gene, so wip1 may be a target genes of miRNA-16.
wip1 expressed in most tumor types, and played an important role in tumor formation. It has proved Wip1 can promote tumor transformation and enhance tumor biological aggressiveness through the silence target genes [10,11], but the carcinogenic mechanism of Wip1 is not yet explained clearly.
In conclusion, this study carried out basic research on biological function of mir-16, also found that there is a certain relationship between the proto-oncogene wip1 and mir-16, then whether or no the mir-16 play a role of tumor suppressor genes through its inhibition of wip1, and future research will be done in the near future.
Disclosure of conflict of interest
None.
References
- 1.Calin GA, Dumitru CD, Shimizu M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 3.Volinia S, Calin GA, Liu CG. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Cimmino A, Fabbri M. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y, Okubo T, Rawlins E, Hogan BL. Epithelial progenitor cells of the embryonic lung and the role of microRNAs in their proliferation. Proc Am Thorac Soc. 2008;5:300–304. doi: 10.1513/pats.200710-162DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cory S, Adams CS. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 9.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 10.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace AJ Jr, Appella E. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Yang Y, Peng Y. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]


