Abstract
Background: Intracellular pathogen resistance 1 (Ipr1) has been found in macrophages and plays a pivotal role in fighting against Mycobacterium tuberculosis (Mtb) infection. This study is designed to evaluate the effect of Iprl on the expression of macrophage genes related to the anti-infection of Mtb. Design or methods: In the experimental and control groups, the macrophages were infected with Mycobacterium H37Ra, and then the related immune genes between two groups were detected using microarray assay. Real-time quantitative PCR was applied to detect the differences in the expression of three up-regulated genes detected by microarray assay and to verify the reliability of microarray assay. Results: The expression of Iprl up-regulated 11 genes related to macrophage anti-immunity involved TLRs signaling pathway including TLR2 and TLR4, Irak1, Traf7, Ifngr1 and Tnfrsfla. No significant difference was found in terms of the molecular expression involved in regulation of the adaptive immune response, such as IL-1 and IL-12. The results of real-time PCR were consistent with the findings of microarray assay. Conclusions: The expression of Iprl gene probably promotes macrophage activity and enhances the ability of macrophages to fight against Mtb infection. The underlying mechanism may be achieved by up-regulating the expression levels of innate immunity genes, especially TLR2/TLR4 and signal transduction molecules, which is determined using microarray assay. All these findings offer the basis for subsequent study of the mechanisms of Ipr1 gene in host innate immunity against Mtb infection.
Keywords: Iprl, mycobacterium tuberculosis, macrophages, innate immunity, gene expression
Introduction
Tuberculosis is a major global disease threatening public health. Albeit substantial progress has been made, tuberculosis is still one of the most severe infectious with a high morbidity and mortality worldwide [1]. Tuberculosis is caused by Mycobacteria tuberculosis, which belongs to the family of Mycobacteriaceae and the order of Actinomycetales. Among the pathogenic species of the M. tuberculosis (Mtb) complex, Mtb acts as the most prevalent and vital pathogen causing human infectious diseases [2]. Many studies have shown that innate immune response plays an essential role in the Mtb infection. As a vital part of the first host defense against Mtb infection, macrophage is involved in the course of innate immune response by performing certain important functions [3,4]. The activated macrophages could secrete several cytokines that bind to the signaling receptors of other cells and thereby instruct those cells to respond in ways that contribute to host defense. Macrophages are activated to perform their functions by recognizing a variety of microbial molecules and host molecules produced in response to infections. These activated molecules further bind to specific signaling receptors located on the surface or inside the macrophages. Toll-like receptor is one category of these signaling receptors, which plays a central role in innate immunity [5]. In 2005, Pan et al found an intracellular pathogen resistance gene 1 (Ipr1), a macrophage-mediated intracellular pathogen resistance gene in murine macrophages, plays a role in regulating the response of macrophages towards Mtb in vitro, mainly by suppressing the replication of Mtb and regulating cell death pathways of infected macrophages (apoptosis or necrosis) [6,13-15]. The scientists from our laboratory have successfully constructed eukaryotic expression gene pEGFP-Iprl and verified that Iprl gene product is mainly expressed in the nucleus [7,16,17].
In this study, a correlation between gene expression and the innate immune mechanisms of macrophage anti-mycobacterial infections was determined using microarray assay and the mechanisms underlying Iprl gene in resistant tuberculosis of macrophages in innate immunity were clarified.
Materials and methods
Plasmids, cells and strains
Plasmid pEGFP-Iprl was constructed and preserved by Laboratory of Pathogenic Biology, Binzhou Medical University. Plasmid pEGFP-C and cell strain RAW264.7 were preserved by our laboratory the fore-mentioned [7]. Attenuated strain of Mtb H37Ra was provided by Beijing Institutes for Food and Drug Control (strain No. 93020).
Main reagents
RPMl l640 powdered cell culture media and 1:125 trypsins were purchased from Gibco Company, USA. The 6- and 24-well cell culture plates were purchased from Costar Company, USA. Fetal bovine serum was purchased from Hangzhou Sijiqing Biological Engineering Materials Co., Ltd., China. Eukaryotic cell transfection reagent Lipofectamine 2000 was provided by Invitrogen Corporation. M7H9 and M7H10 dry medium and OADC enrichment were purchased from Difco Company. Innate and adaptive immune response gene chip (Oligo Innate & Adaptive Immune Responses Microarray) originating in SuperArray Company of USA was purchased from Shanghai Kangcheng Company.
Screening of cell line RAW264.7 stably expressing Ipr1 gene
RAW264.7 cells were transfected with plasmid pEGFP-Ipr1 and the control vector pEGFP-C1, respectively. RAW264.7 cells stably expressing Iprl and green fluorescent protein were screened using G418 [8]. The obtained cells were assigned into the experimental and control groups.
Culture of Mtb H37Ra and preparation of bacterial suspension
Fresh Mtb H37Ra colony in logarithmic growth phase was selected from modified L-J culture medium, fully grinded by 0.05% Twain saline, then inoculated in M9H9 liquid culture medium for 3-week at 37°C, sealed and centrifuged and the supernatant was discarded. The cells were irrigated repeatedly by 0.9% saline and precipitated for 3 times every, 10 minutes for each. The supernatant was collected and the concentration of bacteria was adjusted to 1 × 107/ml using RPM 1640 serum-free culture medium.
Mtb infection of macrophages and detection of bacterial quantity by phagocytosis
In the experimental and control groups, the cells screened by G418 were cultured in RPMI 1640 medium (containing 10% fetal bovine serum, 600 μg/ml G418) at 37°C in a 5% CO2 saturated humidity condition. When the cells were in good condition, the cells were treated with 0.25% trypsin and then harvested, transferred into a 6-well plate at a cell density of 1 × 106 cells for each and continuously cultured in RPM1640 medium containing 10% fetal bovine serum and 600 μg/ml G418. Meanwhile, 6-well plate was prepared using conventional methods for subsequent acid-fast staining. Treated cells were further cultured in RPM1640 medium containing 10% fetal bovine serum (without G418 or antibiotics) until cell adhesion. PMA was simultaneously supplemented to a final concentration of 100 nmol/L. The suspension of Mtb H37Ra prepared in the previous step was added to the 6 well plates at a bacteria/cell ratio of 1:10 in both groups. Cultured cells were cultured at 37°C and 5% CO2 conditions for 6 hours. The bacteria free from phagocytosis by phagocytes were discarded after washing for three times with PBS containing 1% fetal bovine serum, and further cultured using complete RPM1640 culture medium without antibiotics (the time point was counted as 0 h for ease of description). The obtained cells were washed with PBS containing 1% fetal bovine serum every 6 h for 3 times and complete RPM1640 culture medium without antibiotics was supplemented. After 24 h, the cells were collected from both groups for acid-fast staining to observe the situation of phagocytosis.
Microarray assay
The total RNA of cells in each group was extracted after 96 h-bacterial infection. The purity and concentration of the total RNA samples were determined by agarose gel electrophoresis and the accuracy of OD value was verified by ultraviolet spectrophotometer. The synthesis of cDNA and synthesis, amplification and purification of linear biotin-labeled cRNA were performed strictly according to the manufacturers’ instructions. Microarray hybridization was performed according to microarray detection manual of Oligo Innate & Adaptive Immune Responses Microarray. The outcomes of microarray hybridization were detected by chemiluminescence detection kit and subject to X ray film exposure. The image was scanned on the film by Microtek 120TF scanner and converted to grayscale TIFF format image files. The original data was completely analyzed using GEArray Expression Analysis Suite (SABiosciences). The percentage of the expression level of testing gene among the expression level of the housekeeping gene was calculated and the differences in expression levels of specific genes were then compared between the control and the experimental groups.
Real-time PCR detection
Three up-regulated genes were selected for real-time PCR detection to verify the reliability of the results of microarray assay. The primers of three detected genes and internal reference GAPDH were designed using primer design software Primer 5 as follows: GAPDH F 5’AAGAAGGTGGTGAAGCAGGC3’R 5’TCCACCACCCTGTTGCTGTA3’; Ifngrl F 5’CTTTGACGAGCACTGAGGA3’R 5’CCAGGAACCCGAATACACC3’; Tlr2F 5’TGTCGTTCAAGGAGGTGCG 3’R 5’TCCAGAAGAGCCAAAGAGC3’; Tlr4 F 5’GAGAATCTGGTGGCTGTGG3’R 5’TTCCCTGAAAGGCTTGGTC3’. Bacterial culture and cell infection were conducted using the methods above. RNA extraction and synthesis of cDNA were performed by real-time PCR the same with microarray assay. Slope dilutions of DNA template were constructed for standard curves. cDNA templates that expressed each detected gene and housekeeping gene were selected and subject to carry real-time PCR. PCR product and 100 bp DNA Ladder were subject to 2% agarose gel electrophoresis and ethidium bromide staining to confirm whether the real-time PCR product was a monospecific amplified band. PCR product was diluted by 10-fold and the concentration was adjusted to 1. The gradient concentration was 1 × 10-1, 1 × 10-2, 1 × 10-3, 1 × 10-4, 1 × 10-5, 1 × 10-6, and 1 × 10-7. Several gradient dilutions of DNA template and all samples of GAPDH, Ifngrl, Tlr2 and Tlr4 cDNA were subject to real-time PCR and the fluorescence was collected at 86°C, 85°C, 81°C and 85°C. Based upon the gradient dilutions of DNA standard curve, the concentration of target and the housekeeping genes of each sample were directly generated by the machine. The corrected relative content was calculated as the concentration of target gene divided by that of the housekeeping gene.
Results
Screening of RAW264.7 stably expressing Iprl
RAW264.7 cells transfected with plasmid pEGFP-Ipr1 and empty plasmid pEGFP-C1 were obtained after G418 selection pressure. Positive clones with resistance were acquired. Cells stably expressing Iprl in the experimental group were identified by real-time PCR. Inverted fluorescence microscope revealed that fluorescence was mainly found in the nucleus of the experimental group (Figure 1A) and fluorescence could be seen in the cytoplasm of the control group (Figure 1B).
Figure 1.
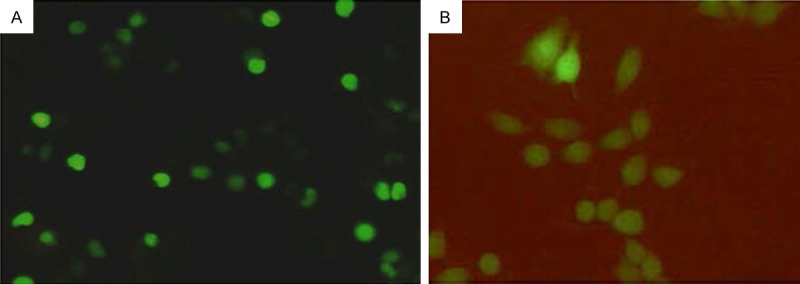
Exogenously expressed Ipr1 is located in the nucleus. Fluorescence microscopy image of RAW264.7 cell transfected with pEGFP-Ipr1 (A) or pEGFP-C1 (B) (×250).
RAW 264.7 cells infected with Mtb H37Ra
After 24 h infection with H37Ra, acid-fast staining was performed to observe the situation of bacterial phagocytosis. Observational results revealed that approximately 70% of cells engulfed at least one acid-fast bacillus, 20 acid-fast bacillus for each cell on average, indicating that RAW 264.7 cells were successfully infected with Mtb H37Ra, as shown in Figure 2.
Figure 2.
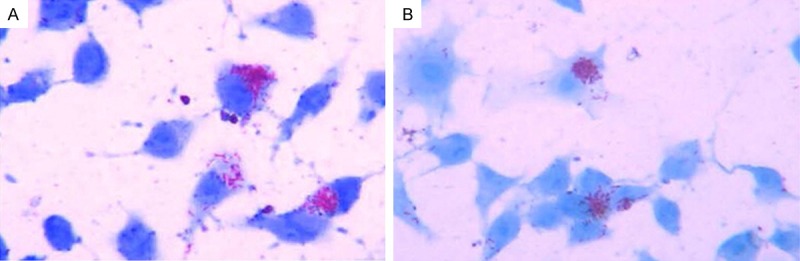
RAW264.7 cells infected with H37Ra for 24 h in the experimental group (A) and control group (B) (×1000).
Extraction of total RNA and quality verification
In both groups, total RNA was extracted from the RAW 264.7 cells and subject to agarose gel electrophoresis observations. The band pattern of total RNA 28s and 18s rRNA was clear and bright, and the brightness of RNA 28s band doubled that of 18s rRNA (Figure 3). OD values were measured by UV absorption. OD260/OD280 values in the experimental and control groups were 2 and 1.98, suggesting that the total RNA extracted with a high purity can be used for subsequent experiments. The concentrations of total RNA in the experimental and control groups were 1765.56 ng/μL and 1486.74 ng/μL.
Figure 3.
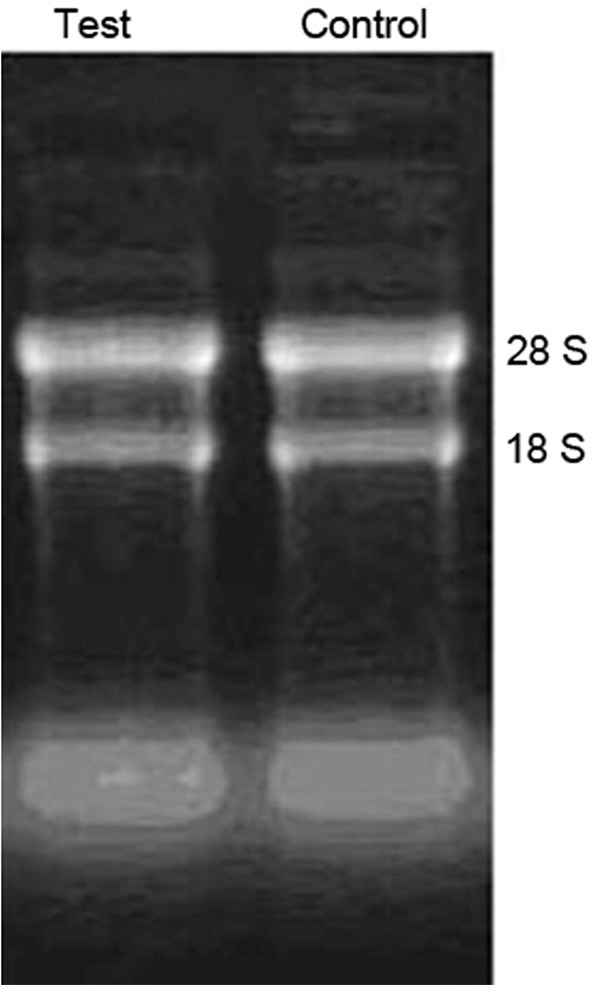
Comparison of total RNA between the experimental and control groups.
Gene chip hybridization chemiluminescence detection
In both groups, linear biotin-labeled cRNA samples and the Mice Oligo Innate & Adaptive Immune Responses Microarray Chip were hybridized, detected by chemiluminescence and then subject to X-film imaging, as shown in Figure 4A and 4B. A complete analysis of microarray data was conducted by the GEArray Expression Analysis Suite software to analyze the differences in expression levels of specific genes between the experimental and control groups. The results showed that 11 genes in the experimental group were up-regulated more than 2 times of that in the control group including TLRs and its signaling pathway genes (TLR2, TLR4, Irakl, and Traf6), IFN-γ receptor genes [Ifngrl (IFN-7R1)], TNF receptor-associated genes [Tnfrsfla (TNFRl)], cellular recognition and intercellular interactions associated genes (Lbp, Fcgrl, and Colccl2) and inflammation-related genes (Cebpe and Treml), as illustrated in Table 1. Up-regulation of these genes was mainly manifested as innate immune response function of macrophages, whereas no significant difference was found in the expression of genes and the cytokines associated with the adaptive immune response of macrophages to regulate T lymphocyte, such as IL-1, IL-12, and TNF-γ.
Figure 4.

cDNA expression of cells infected with H37Ra for 96 h in the experimental group (A) and control group (B).
Table 1.
Genes twice up-regulated in the experimental group than those in the control group
| Location | Gene sequence number | Gene | Gray scale of gene expression (standard value) | ||
|---|---|---|---|---|---|
|
| |||||
| Experimental group | Control group | Experimental group/control group | |||
| 14 | NM_207131 | Cebpe | 0.1935 | 0.0931 | 2.0772* |
| 19 | NM_130449 | Colec12 | 0.6088 | 0.2781 | 2.1877* |
| 37 | NM_010186 | Fcgr1 | 0.3058 | 0.0895 | 3.4167* |
| 45 | NM_010511 | Ifngr1 | 0.4478 | 0.2060 | 2.1741* |
| 68 | NM_008363 | Irak1 | 0.0997 | 0.0271 | 3.6697* |
| 73 | NM_008489 | Lbp | 0.2219 | 0.0827 | 2.6806* |
| 103 | NM_011905 | Tlr2 | 0.1284 | 0.0526 | 2.4394* |
| 105 | NM_021297 | Tlr4 | 0.1757 | 0.0519 | 3.381* |
| 111 | NM_011609 | Tnfrsf1a | 0.1456 | 0.0693 | 2.1012* |
| 113 | NM_009424 | Traf6 | 0.1638 | 0.0699 | 2.3427* |
| 114 | NM_021406 | Trem1 | 0.1761 | 0.0626 | 2.8138* |
defines statistical significance between the experimental and control groups.
Abbreviations: Cebpe (CCAAT/enhancer Binding Protein, C/EBP-ε), Colec12 (Collectin sub-family member 12/CL-P1), Fcgr1 (IgG Fc Receptor), Ifngr1 (IFN-γR1), Irak1 (IL-1R associated protein kinase), Lbp (Lipopolysaccharide binding protein), Tlr2 (Toll-like receptor 2), Tlr4 (Toll-like receptor 4), Tnfrsf1a (TNFR1), Traf6 (Tnf receptor-associated factor 6), Trem1 (Triggering receptor expressed on myeloid cells 1).
Real-time PCR
Up-regulated expression of Ifngrl, Tlr2 and TIr4 by microarray assay was verified by real-time PCR. Initial copy number of measurement template was expressed by Ct values. The smaller Ct value was, the larger initial copy number was, and vice versa. Standard and melting curves of each target gene were drawn to ensure the accuracy and specificity of the amplification results. Linear regression coefficient of standard curve was R2 > 0.99 and amplification efficiency ranged from 0.83 to 1.0. The results showed that the expression of internal reference was similar while the expression of Ifngrl, Tlr2 and Tlr4 was different between two samples (Table 2). The expression levels of Ifngrl, Tlr2 and Tlr4 in the experimental group was 3.05, 2.27 and 3.40 times of that in the control group, respectively, which was consistent with the results of micro-array (Table 3 and Figure 5), indicating that the results of gene micro-array were reliable.
Table 2.
Relative expression levels of Ifngr1, Tlr2 and Tlr4 between the experimental and control groups
| Gene | Experimental group | Control group | Experimental group/control group |
|---|---|---|---|
| GAPDH | 9.85 × 10-3 | 10.6 × 10-3 | 0.93 |
| Ifngr1 | 1.77 × 10-3 | 6.25 × 10-4 | 2.83* |
| Tlr2 | 2.36 × 10-3 | 1.12 × 10-3 | 2.11* |
| Tlr4 | 2.95 × 10-4 | 9.36 × 10-5 | 3.15* |
| Ifngr1/GAPDH | 0.18 | 0.059 | 3.05* |
| Tlr2/GAPDH | 0.24 | 0.106 | 2.27* |
| Tlr4/GAPDH | 0.03 | 0.00883 | 3. 40* |
defines statistical significance between the experimental and control groups.
Table 3.
Verification of the results of microarray assay by real-time PCR
| Gene | Microarray assay (fold) | Real-time PCR (fold) |
|---|---|---|
| Ifngr1 | 2.17 | 3.05 |
| Tlr2 | 2.43 | 2.27 |
| Tlr4 | 3.38 | 3.40 |
Figure 5.
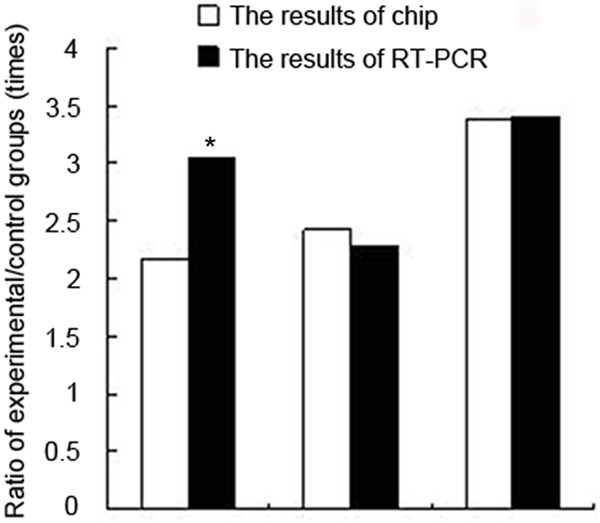
Verification of the results of microarray assay by real-time PCR, *defines statistical significance.
Discussion
Recent studies found that Iprl gene is associated with susceptibility to tuberculosis in mice, but the exact mechanism remains elusive. Present studies showed that expression of Iprl gene could enhance the ability of macrophage to anti-intracellular pathogens, such as Mtb. However, these studies are merely limited to transgenic animals and cells in vitro. In this study, micro-array assay was used to detect the effects of Iprl expression on anti-mycobacterial associated immune gene expression of macrophage and whether the expression of Iprl interrelated with known mechanisms of the anti-mycobacterial infection of macrophage.
Oligo Innate & Adaptive Immune Responses Microarray contains 113 gene loci, which have been known to participate in the immune response of anti-microbial infections. These gene loci included IL-1R/TLRs members and related genes, pathogen recognition-related genes, inflammatory response genes, apoptosis-related genes, NF-κB signaling pathway-related genes and cytokines, interleukins, chemokines and corresponding ligand genes. The expression of immune response genes of different transcription levels was detected using microarray assay.
In this study, the differences in expression levels of immunity-related genes were detected using microarray assay after the macrophages were infected with H37Ra for 96 h in both groups. The microarray results showed that Iprl expression mainly increased the expression of genes involving with anti-bacterial infection immune response of macrophages, especially innate immunity-related genes of macrophages were up-regulated significantly. In the upregulated genes, macrophages against intracellular pathogens, such as, the genes involved in TLRs signaling pathways (TLR2, TLR4, Irak1, and Traf6), IFN-γ receptor gene (Ifngrl) and TNF-γ receptor gene (Tnfrsfla) were validated.
The genes of TLR2, TLR4, Irakl, and Traf6 are involved in TLRs signal transduction. A large number of studies have shown that TLRs signaling pathway mediating macrophage resistance to infection of Mtb plays a pivotal role in the process of anti-tuberculosis. More research has been conducted on TLR2 and TLR4 associated macrophages anti-infection at present. Ingredients or secretory component of Mtb contains a series of ligand interacted with TLR2 and TLR4, such as LAM (lipoarabinomannan), 19kD lipoprotein, 38kD sugar lipoprotein, HSP70, etc. NF-κB is activated by MyD88 dependent and non-dependent pathway (TLR4) after TLR2 or TLR4 combined with the corresponding ligand, and a variety of inflammatory cytokine are released and APC cell surface costimulatory molecules such as CD80, CD86 are increased. It plays an important role in pathogen destruction of anti-infection innate immunity in the early stage, regulating adaptive immune responses in the late stage and the process of the formation of granulomas [9]. In TLR2/TLR4 MyD88-dependent pathway mediated macrophage activation process, MyD88 inside the macrophages as an adaptor protein form complex with the intracellular domain of TLR2/TLR4, and then recruit and activate IRAK-4, IRAK-1 and TRAF-6, and further activate the downstream cascade and activate NF-κB [10], ultimately promoting macrophage activation and exerting intracellular bactericidal immune function. In this process, the participation and activation of IRAK-l and TRAF-6 molecule has an important significance in TLRs signal transduction. The regulation effect of Iprl upon the expression of the gene mentioned above strongly suggests that Iprl expression may be associated with transcription and expression of TLR related genes, and participate in macrophage-mediated immune responses in anti-Mtb.
Interferon-γ (IFN-γ) that a major Th1-type cytokines secreted by T cells and NK cells plays a very important role in Macrophages against infection of Mtb. Its function of anti-Mtb reactive infection was executed mainly by stimulating the host macrophages to produce nitric oxide synthase (NOS2) [11]. Tumor necrosis factor-γ (TNF-γ) that a proinflammatory cytokine secreted by mononuclear macrophages activated macrophages to kill replicating Mtb cells synergy with IFN-γ, and closely related to the Mtb induced apoptosis in infected macrophages [12]. This study found that Iprl expression up-regulated the gene expression of macrophage IFN-γRl. It can be speculated that the antimicrobial action of Iprl expression also possible increased the sensitivity of macrophages stimulus for IFN-γ, and the up-regulation of TNF R1 gene also suggest Iprl expression may be involved in promoting the TNF-a mediated macrophage cell killing Mtb and apoptosis. However, NOS2 expression in the experimental and control groups did not significantly differ. In the experimental group, the differences in gene expression of TNF-γ and NF-κB apoptosis-related genes were not detected. It may have been caused by the experimental design that only macrophages alone without any other stimulating factors were involved during the study.
The results showed that the expression of Iprl up-regulated a series of gene expression antibacterial immune responses of macrophages, especially anti-microbial innate immune response related gene expression of macrophages, included TLR2/TLR4 gene and signaling pathways related genes Irakl and Traf6. As well as macrophage activation and killing intracellular pathogens related cytokine receptors: Ifngrl and Tnfrsfla. However, no significant difference was found in gene expression of IL-1, TNF-γ and IL-12 involved in macrophages regulates the adaptive immune response. It may be speculated Iprl mainly achieved by enhanced innate immune response of Macrophage to destruct and phagocytic Mtb, especially the role of TLRs activation enhances the body’s ability to resist infection of Mtb. Further experiments will be necessary to confirm how to interact between Iprl and TLRs activation mechanism, and where the action sites of Iprl expression products are located.
Disclosure of conflict of interest
None.
References
- 1.Raviglione M, Marais B, Floyd K, Lönnroth K, Getahun H, Migliori GB, Harries AD, Nunn P, Lienhardt C, Graham S, Chakaya J, Weyer K, Cole S, Kaufmann SH, Zumla A. Scaling up interventions to achieve global tuberculosis control: progress and new developments. Lancet. 2012;379:1902–1913. doi: 10.1016/S0140-6736(12)60727-2. [DOI] [PubMed] [Google Scholar]
- 2.Harrison’s infectious diseases. McGraw-Hill Medical. 2010:596–617. [Google Scholar]
- 3.Sundaramurthy V, Pieters J. Interactions of pathogenic mycobacteria with host macrophages. Microbes Infect. 2007;9:1671–1679. doi: 10.1016/j.micinf.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Fremond CM, Yeremeev V, Nicolle DM, Jacobs M, Quesniaux VF, Ryffel B. Fatal Mycobacterium tuberculosis infection despite adaptive immune response in the absence of MyD88. J Clin Invest. 2004;114:405–412. doi: 10.1172/JCI21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbas AK, Lichtman AH, Pillai S. Cellular and molecular immunology. Philadelphia: Saunders; 2012. pp. 17–18. [Google Scholar]
- 6.Pan H, Yan BS, Rojas M, Shebzukhov YV, Zhou H, Kobzik L, Higgins DE, Daly MJ, Bloom BR, Kramnik I. Gene mediates innate immunity to Tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li N, Zhu DY, Tie RX, Yu WW. Construction of a fusion gene expression vector containing mouse Ipr1 and EGFP and its expression in murine macrophage. Cell Mol Neurobiol. 2008;24:231–237. [PubMed] [Google Scholar]
- 8.Shi CG, Zhang H, Xi L, Wang XW, Wang LM, Zhao Y. Establishment of a stable cell line expressing the fusion protein of Mycobacterium tuberculosis Hsp65 and IL-2. Chinese Journal of Zoonoses, ISTIC. 2008:24. [Google Scholar]
- 9.Quesniaux V, Fremond C, Jacobs M, Parida S, Nicolle D, Yeremeev V, Bihl F, Erard F, Botha T, Drennan M, Soler MN, Le Bert M, Schnyder B, Ryffel B. Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect. 2004;6:946–959. doi: 10.1016/j.micinf.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Mohan C. Toll-like receptor signaling pathways-therapeutic opportunities. Mediators Inflamm. 2010;2010:781235. doi: 10.1155/2010/781235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng CG, Kaviratne M, Rothfuchs AG, Cheever A, Hieny S, Young HA, Wynn TA, Sher A. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J Immunol. 2006;77:7086–7093. doi: 10.4049/jimmunol.177.10.7086. [DOI] [PubMed] [Google Scholar]
- 12.Arcila ML, Sánchez MD, Ortiz B, Luis FB, Luis FG, Mauricio R. Activation of apoptosis, but not necrosis, during Mycobacterium tuberculosis infection correlated with decreased bacterial growth: role of TNF-alpha, IL-10, caspases and phospholipase A2. Cell Immunol. 2007;249:80–93. doi: 10.1016/j.cellimm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Kramnik I. Genetic dissection of host resistance to Mycobacterium tuberculosis: the sst1 locus and the Ipr1 gene. Curr Top Microbiol Immunol. 2008;321:123–148. doi: 10.1007/978-3-540-75203-5_6. [DOI] [PubMed] [Google Scholar]
- 14.He XN, Su F, Lou ZZ, Jia WZ, Song YL, Chang HY, Wu YH, Lan J, He XY, Zhang Y. Ipr1 gene mediates RAW 264.7 macrophage cell line resistance to Mycobacterium bovis. Scand J Immunol. 2011;74:438–444. doi: 10.1111/j.1365-3083.2011.02596.x. [DOI] [PubMed] [Google Scholar]
- 15.Cai L, Pan H, Trzciński K, Thompson CM, Wu Q, Kramnik I. MYBBP1A: a new Ipr1’s binding protein in mice. Mol Biol Rep. 2010;37:3863–3868. doi: 10.1007/s11033-010-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Niyonsaba F, Ushio H, Okuda D, Nagaoka I, Ikeda S, Okumura K, Ogawa H. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J Dermatol Sci. 2005;40:123–132. doi: 10.1016/j.jdermsci.2005.03.014. [DOI] [PubMed] [Google Scholar]


