Abstract
Some studies were conducted to investigate the association between signal transducer and activator of transcription 4 (STAT4) polymorphisms and diabetes risk. However, the results were inconsistent. We thus did a meta-analysis. We searched the articles in the PubMed, Embase and CNKI databases (the last search updated on November 2014). Pooled odds ratios (OR) with 95% confidence intervals (CI) were derived from random-effects models or fixed-effects models. Ten case-control studies with 18931 cases and 23833 controls were included in this study. STAT4 rs7574865 polymorphism was significantly associated with diabetes risk (OR = 1.28; 95% CI 1.16-1.42; P < 0.00001). This polymorphism also increased type 1 diabetes risk significantly (OR = 1.27; 95% CI 1.15-1.41; P < 0.00001). In the subgroup analysis by ethnicity, a significant association was found among Asians (OR = 1.33; 95% CI 1.04-1.71; P = 0.02) and Caucasians (OR = 1.24; 95% CI 1.12-1.38; P < 0.0001). In the subgroup analysis by age, both children (OR = 1.28; 95% CI 1.12-1.45; P = 0.0002) and adults (OR = 1.27; 95% CI 1.13-1.42; P < 0.0001) with this polymorphism showed increased diabetes risk. Other STAT4 polymorphisms were not investigated in this meta-analysis due to insufficient data. This meta-analysis indicated that STAT4 rs7574865 polymorphism was associated with diabetes risk.
Keywords: Diabetes, STAT4, meta-analysis, polymorphism, association
Introduction
Diabetes is one of the metabolic diseases. More than 20 million Americans and over 170 million individuals worldwide suffer from diabetes [1]. Type 1 diabetes (T1D) and T2D are two main forms of this disease. Diabetes is thought to develop from an interaction between environmental and genetic factors, indicating that there is a strong genetic basis for this disease [2-4].
Signal transducer and activator of transcription 4 (STAT4) is a central mediator in generating inflammation during protective immune responses and immune-mediated diseases. Because STAT4 is required for the development of fully functional Th1 cells, STAT4-deficient mice are protected from the effects of T-cell-mediated autoimmune diseases. In models of experimental diabetes, STAT4-deficient mice display less disease and decreased parameters of inflammation compared with WT mice [5,6]. Therefore, STAT4 might play an important role in the development of diabetes, especially T1D.
STAT4 was mapped on chromosome 2q32.2-q32.3. Several polymorphisms, such as rs11889341, rs7574865, rs8179673, and rs10181656 have been identified. The most commonly studied functional variant in the STAT4 gene is rs7574865. This polymorphism has been reported to be associated with some diseases, such as rheumatoid arthritis, hepatocellular carcinoma, ulcerative colitis, and pulmonary tuberculosis [7-10]. Some studies also investigated the association between STAT4 polymorphisms and diabetes risk [11-20]. However, the results remained inconclusive. One meta-analysis including three studies tried to determine the association between STAT4 rs7574865 polymorphism and diabetes risk [21]. However, Zheng et al. found that this polymorphism was not associated with T1D [21], which was inconsistent with recent studies [19,20]. Therefore, we performed a meta-analysis to clarify the association between STAT4 polymorphisms and diabetes risk.
Materials and methods
Search for publications
We searched the articles using the search terms “Signal transducer and activator of transcription 4”, “STAT4”, “diabetes” and “polymorphism” in the PubMed, Embase and CNKI databases, and the last search updated on November 2014. Additional studies were identified by a hand search of references of original studies or review articles on the association between STAT4 polymorphisms and diabetes risk. No publication date or language restriction were imposed.
Inclusion and exclusion criteria
The following inclusion criteria were used: (1) the study should have evaluated the association between the STAT4 polymorphisms and diabetes risk; (2) the study should have a case-control design; (3) sufficient data should have been provided in order to calculate odds ratios (OR) and 95% confidence intervals (CI). Studies were excluded if any of the following conditions applied: (1) irrelevant to diabetes or STAT4; (2) abstract or review; (3) non-clinical study; (4) studies were repeated or publications overlapped.
Data extraction
Two investigators independently extracted data and reached consensus on the following characteristics of the selected studies: the first author’s name, year of publication, ethnicity of the study population, diabetes type, ages group, numbers of cases and controls, and STAT4 polymorphisms.
Statistical analysis
OR and 95% CI were employed to evaluate the strength of the association between STAT4 polymorphisms and diabetes risk. Departure from Hardy-Weinberg equilibrium (HWE) in controls was tested by the chi-square test. The Q statistic and the I2 statistic were used to assess the degree of heterogeneity among the studies included in the meta-analysis. If heterogeneity was observed among the studies, the random-effects model was used to estimate the pooled OR (the DerSimonian and Laird method). Otherwise, the fixed-effects model was adopted (the Mantel-Haenszel method). The Galbraith plot was used to find the source of the heterogeneity. Subgroup analyses were carried out by ethnicity, diabetes type, and age group. Cumulative meta-analysis was carried out. Sensitivity analysis was performed through sequentially excluded individual studies to assess the stability of the results. The potential publication bias was examined visually in a funnel plot of log [OR] against its standard error (SE), and the degree of asymmetry was tested using Egger’s test.
All statistical tests were performed using Revman 5.1 software (Nordic Cochrane Center, Copenhagen, Denmark) and STATA 11.0 software (Stata Corporation, College Station, TX, USA). A P value < 0.05 was considered statistically significant, except for tests of heterogeneity where a level of 0.10 was used.
Results
Characteristics of studies
A total of 10 case-control studies with 18931 cases and 23833 controls on the association between STAT4 polymorphisms and diabetes risk were included for this meta-analysis [10-20]. There were 3 studies of Asian population and 7 studies of Caucasian population. One study included T2D patients, while other studies all included T1D patients. Three studies used adults and 7 studies used Children. All studies investigated the association between STAT4 rs7574865 polymorphism and diabetes risk. Two studies reported the results of rs11889341, rs8179673 and rs10181656, one study reported the result of rs3024866. All studies suggested that the distribution of genotypes in the controls was consistent with HWE. The characteristics of each case-control study are presented in Table 1.
Table 1.
Characteristics of the included studies in this meta-analysis
| First author | Year | Ethnicity | Diabetestype | Age group | Case number | Control number | HWE | Polymorphisms |
|---|---|---|---|---|---|---|---|---|
| Lee | 2008 | Asian | T1D | Children | 389 | 152 | Yes | rs11889341, rs7574865, rs8179673, rs10181656 |
| Martinez | 2008 | Caucasian | T1D | Children | 311 | 716 | Yes | rs7574865 |
| Zervou | 2008 | Caucasian | T1D | Children | 101 | 203 | Yes | rs7574865 |
| Fung | 2009 | Caucasian | T1D | Children | 8010 | 9733 | Yes | rs7574865 |
| Kofteridis | 2009 | Caucasian | T2D | Adult | 126 | 344 | Yes | rs7574865 |
| Howson | 2011 | Caucasian | T1D | Adult | 590 | 938 | Yes | rs7574865 |
| Park | 2011 | Asian | T1D | Children | 418 | 1060 | Yes | rs11889341, rs7574865, rs8179673, rs10181656 |
| Plagnol | 2011 | Caucasian | T1D | Children | 8506 | 10118 | Yes | rs7574865 |
| Fourati | 2012 | Caucasian | T1D | Adult | 70 | 162 | Yes | rs7574865 |
| Bi | 2013 | Asian | T1D | Children | 410 | 407 | Yes | rs7574865, rs3024866 |
PCR, polymerase chain reaction; HWE, Hardy-Weinberg equilibrium; NA, not available.
Results of meta-analysis
Because the data of rs11889341, rs8179673, rs10181656, and rs3024866 polymorphisms were limited, only the association between STAT4 rs7574865 polymorphism and diabetes risk was calculated in this meta-analysis. STAT4 rs7574865 polymorphism was significantly associated with diabetes risk (OR = 1.28; 95% CI 1.16-1.42; P < 0.00001), as shown in Figure 1. This polymorphism also increased T1D risk significantly (OR = 1.27; 95% CI 1.15-1.41; P < 0.00001). In the subgroup analysis by ethnicity, a significant association was found among Asians (OR = 1.33; 95% CI 1.04-1.71; P = 0.02) and Caucasians (OR = 1.24; 95% CI 1.12-1.38; P < 0.0001). In the subgroup analysis by age, both children (OR = 1.28; 95% CI 1.12-1.45; P = 0.0002) and adults (OR = 1.27; 95% CI 1.13-1.42; P < 0.0001) with this polymorphism showed increased diabetes risk. All the results are listed in Table 2.
Figure 1.
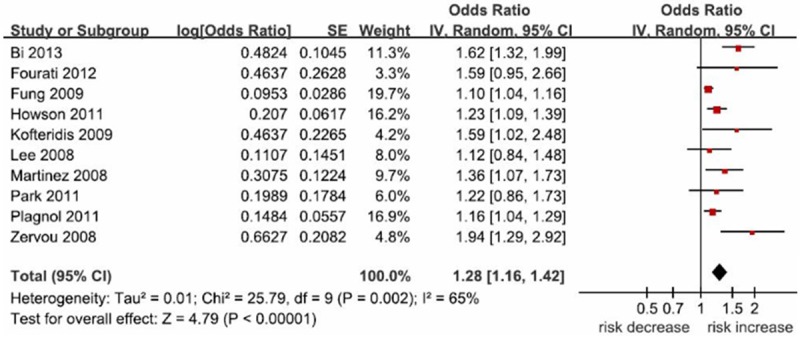
Meta-analysis for the association between STAT4 rs7574865 polymorphism and diabetes risk.
Table 2.
The effect of STAT4 rs7574865 polymorphism on diabetes
| No. of studies | Test of association | Model | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| OR (95% CI) | Z | P Value | χ 2 | P Value | I 2 (%) | |||
| Overall | 10 | 1.28 (1.16-1.42) | 4.79 | < 0.00001 | R | 25.79 | 0.002 | 65 |
| T1D | 9 | 1.27 (1.15-1.41) | 4.55 | < 0.00001 | R | 23.91 | 0.002 | 67 |
| Asian | 3 | 1.33 (1.04-1.71) | 2.25 | 0.02 | R | 4.95 | 0.08 | 60 |
| Caucasian | 7 | 1.24 (1.12-1.38) | 4.13 | < 0.0001 | R | 15.38 | 0.02 | 61 |
| Children | 7 | 1.28 (1.12-1.45) | 3.74 | 0.0002 | R | 21.52 | 0.001 | 72 |
| Adult | 3 | 1.27 (1.13-1.42) | 4.07 | < 0.0001 | F | 1.98 | 0.37 | 0 |
F, fixed-effects model; R, random-effects model.
Cumulative meta-analysis and sensitivity analysis
With regard to the cumulative meta-analysis, the evidence was observed to support a significant association of the STAT4 rs7574865 polymorphism and diabetes risk (Figure 2). As shown in Figure 3, sensitivity analysis did not influence the result excessively by omitting any single study.
Figure 2.
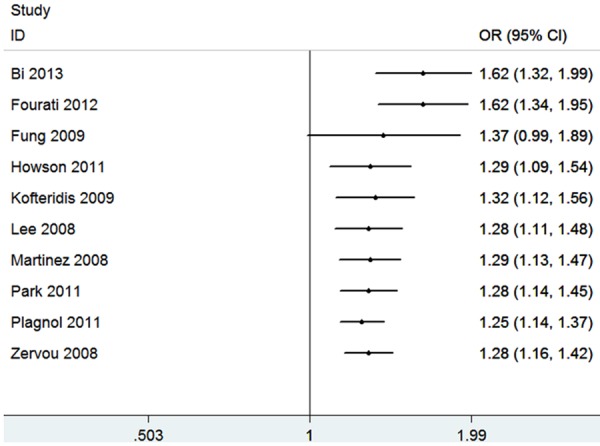
Cumulative meta-analysis of the association between STAT4 rs7574865 polymorphism and diabetes risk.
Figure 3.
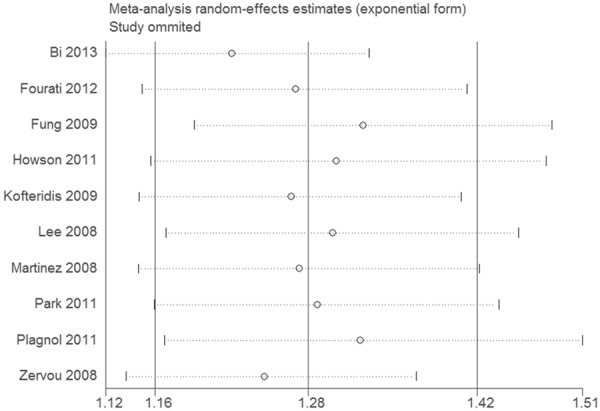
Sensitivity analysis of the association between STAT4 rs7574865 polymorphism and diabetes risk.
Heterogeneity analysis
There was significant heterogeneity (I 2 = 65%, P = 0.002) in the meta-analysis. As shown in Figure 4, three studies were the outliers. After excluding these studies, the heterogeneity effectively decreased and there was no obvious heterogeneity among the remaining studies (I 2 = 0%, P = 0.63). Besides, the result was still statistically significant (OR = 1.22, 95% CI 1.13-1.31, P < 0.00001).
Figure 4.
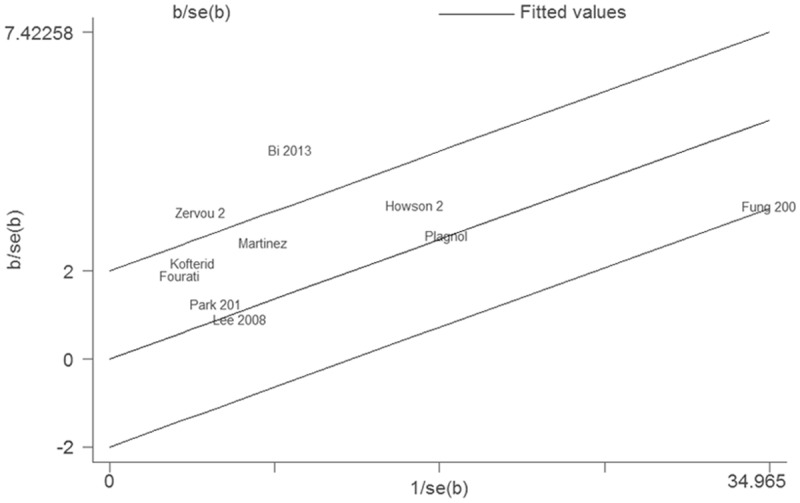
Galbraith plot of STAT4 rs7574865 polymorphism and diabetes risk.
Test of publication bias
The shape of the funnel plot seemed symmetrical (Figure 5). Egger’s test showed no evidence of publication bias (P = 0.122).
Figure 5.
Funnel plot of the associations between STAT4 rs7574865 polymorphism and diabetes risk.
Discussion
This meta-analysis systematically investigated the STAT4 polymorphisms and diabetes risk. Ten case-control studies with a total of 18931 cases and 23833 controls were included. In this meta-analysis, STAT4 rs7574865 polymorphism was significantly associated with diabetes risk, especially T1D. In the subgroup analysis by ethnicity, both Asians and Caucasians with STAT4 rs7574865 polymorphism had an increased diabetes risk. In the subgroup analysis by age, the increased risk of diabetes was detected in both children and adults. Moreover, to investigate the stability of the result, we performed sensitivity analyses. Removal of each study did not alter the result, suggesting the reliability of our result. The cumulative meta-analysis showed a trend of significant association between this polymorphism and the risk of diabetes. Again, this procedure proved that our result was robust. Thus, results from this meta-analysis suggested that STAT4 rs7574865 polymorphism was associated with diabetes risk.
A previous study suggested that selective inhibition of the STAT4 signal transduction pathway might prevent clinical insulin-dependent diabetes mellitus in prediabetic individuals at risk [6]. In addition, Yang et al. indicated that interruption of IL-12-mediated STAT4 activation prevented autoimmune diabetes in NOD mice [22]. The rs7574865 polymorphism is located in the third intron of the STAT4 gene, and its actual functional consequence remains to be identified, given that the connection between diabetes and STAT4 rs7574865 polymorphism is not clear so far. It was possible that rs7574865 polymorphism might influence STAT4 production or phosphorylation procedure. This question should be assessed in the future.
Some limitations in this meta-analysis should be addressed. First, only published studies that were included in the selected electronic databases were identified; it is possible that some relevant published or unpublished studies may have been missed. Second, the effect of gene-gene and gene-environment interactions was not addressed in this meta-analysis, because of limited available data. Third, other STAT4 polymorphisms were not investigated in this meta-analysis due to insufficient data.
This meta-analysis suggested that STAT4 rs7574865 polymorphism was a risk factor of diabetes.
Disclosure of conflict of interest
None.
References
- 1.Dall T, Nikolov P, Hogan P. Economic costs of diabetes in the US in 2002. Diabetes Care. 2003;26:917–932. doi: 10.2337/diacare.26.3.917. [DOI] [PubMed] [Google Scholar]
- 2.Yan ST, Li CL, Tian H, Li J, Pei Y, Liu Y, Gong YP, Fang FS, Sun BR. Association of calpain-10 rs2975760 polymorphism with type 2 diabetes mellitus: a meta-analysis. Int J Clin Exp Med. 2014;7:3800–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Xuan M, Wang Y, Wang W, Yang J, Li Y, Zhang X. Association of LRP5 gene polymorphism with type 2 diabetes mellitus and osteoporosis in postmenopausal women. Int J Clin Exp Med. 2014;7:247–54. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao L, Huang P. Plasminogen activator inhibitor-1 4G/5G polymorphism is associated with type 2 diabetes risk. Int J Clin Exp Med. 2013;6:632–40. [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Chen M, Ellett JD, Fialkow LB, Carter JD, McDuffie M, Nadler JL. Autoimmune diabetes is blocked in Stat4-deficient mice. J Autoimmun. 2004;22:191–200. doi: 10.1016/j.jaut.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Holz A, Bot A, Coon B, Wolfe T, Grusby MJ, von Herrath MG. Disruption of the STAT4 signaling pathway protects from autoimmune diabetes while retaining antiviral immune competence. J Immunol. 1999;163:5374–82. [PubMed] [Google Scholar]
- 7.Jiang X, Zhou Z, Zhang Y, Yang H, Ren K. An updated meta-analysis of the signal transducer and activator of transcription 4 (STAT4) rs7574865 G/T polymorphism and rheumatoid arthritis risk in an Asian population. Scand J Rheumatol. 2014;43:477–80. doi: 10.3109/03009742.2014.918174. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X, Jiang K, Liang B, Huang X. STAT4 Gene Polymorphism and Risk of Chronic Hepatitis B-Induced Hepatocellular Carcinoma. Cell Biochem Biophys. 2015;71:353–7. doi: 10.1007/s12013-014-0205-0. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Dai WQ, Wang F, He L, Zhou YQ, Lu J, Xu XF, Guo CY. Association of STAT4 gene rs7574865G > T polymorphism with ulcerative colitis risk: evidence from 1532 cases and 3786 controls. Arch Med Sci. 2014;10:419–24. doi: 10.5114/aoms.2014.43735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabri A, Grant AV, Cosker K, El Azbaoui S, Abid A, Abderrahmani Rhorfi I, Souhi H, Janah H, Alaoui-Tahiri K, Gharbaoui Y, Benkirane M, Orlova M, Boland A, Deswarte C, Migaud M, Bustamante J, Schurr E, Boisson-Dupuis S, Casanova JL, Abel L, El Baghdadi J. Association study of genes controlling IL-12-dependent IFN-γ immunity: STAT4 alleles increase risk of pulmonary tuberculosis in Morocco. J Infect Dis. 2014;210:611–8. doi: 10.1093/infdis/jiu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HS, Park H, Yang S, Kim D, Park Y. STAT4 polymorphism is associated with early-onset type 1 diabetes, but not with late-onset type 1 diabetes. Ann N Y Acad Sci. 2008;1150:93–8. doi: 10.1196/annals.1447.013. [DOI] [PubMed] [Google Scholar]
- 12.Martínez A, Varadé J, Márquez A, Cénit MC, Espino L, Perdigones N, Santiago JL, Fernández-Arquero M, de la Calle H, Arroyo R, Mendoza JL, Fernández-Gutiérrez B, de la Concha EG, Urcelay E. Association of the STAT4 gene with increased susceptibility for some immune-mediated diseases. Arthritis Rheum. 2008;58:2598–602. doi: 10.1002/art.23792. [DOI] [PubMed] [Google Scholar]
- 13.Zervou MI, Mamoulakis D, Panierakis C, Boumpas DT, Goulielmos GN. STAT4: a risk factor for type 1 diabetes? Hum Immunol. 2008;69:647–50. doi: 10.1016/j.humimm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, Wicker LS, Todd JA. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10:188–91. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 15.Kofteridis D, Krasoudaki E, Kavousanaki M, Zervou MI, Panierakis C, Boumpas DT, Goulielmos GN. STAT4 is not associated with type 2 diabetes in the genetically homogeneous population of Crete. Genet Test Mol Biomarkers. 2009;13:281–4. doi: 10.1089/gtmb.2008.0128. [DOI] [PubMed] [Google Scholar]
- 16.Howson JM, Rosinger S, Smyth DJ, Boehm BO ADBW-END Study Group. Todd JA. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60:2645–53. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park Y, Lee HS, Park Y, Min D, Yang S, Kim D, Cho B. Evidence for the role of STAT4 as a general autoimmunity locus in the Korean population. Diabetes Metab Res Rev. 2011;27:867–71. doi: 10.1002/dmrr.1263. [DOI] [PubMed] [Google Scholar]
- 18.Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, Wallace C, Stevens H, Jackson L, Simmonds MJ Type 1 Diabetes Genetics Consortium. Bingley PJ, Gough SC, Todd JA. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourati H, Bouzid D, Abida O, Kharrat N, Mnif F, Haddouk S, Fesel C, Costa J, Ben Ayed M, Abid M, Rebai A, Penha-Gonçalves C, Masmoudi H. Non-HLA autoimmunity genetic factors contributing to Autoimmune Polyglandular Syndrome type II in Tunisian patients. Hum Immunol. 2012;73:740–6. doi: 10.1016/j.humimm.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Bi C, Li B, Cheng Z, Hu Y, Fang Z, Zhai A. Association study of STAT4 polymorphisms and type 1 diabetes in Northeastern Chinese Han population. Tissue Antigens. 2013;81:137–40. doi: 10.1111/tan.12057. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J, Yin J, Huang R, Petersen F, Yu X. Meta-analysis reveals an association of STAT4 polymorphisms with systemic autoimmune disorders and anti-dsDNA antibody. Hum Immunol. 2013;74:986–92. doi: 10.1016/j.humimm.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Chen M, Fialkow LB, Ellett JD, Wu R, Nadler JL. Inhibition of STAT4 activation by lisofylline is associated with the protection of autoimmune diabetes. Ann N Y Acad Sci. 2003;1005:409–11. doi: 10.1196/annals.1288.069. [DOI] [PubMed] [Google Scholar]


