Abstract
With a meta-analysis and narrative review, we evaluated the clinical and prognostic role of all CD44 family proteins in gastric cancer (GC). Literatures published up to August 2014 were searched on PubMed. Among the 37 eligible studies (6606 patients), 34 were included in meta-analysis, and 10 were subjected to narrative review. With meta-analysis, standard CD44 (CD44s) was demonstrated to predict reduced overall survival (OS) (HR = 1.93, 95% CI: 1.58-2.34, PHR = 0.0222) and disease free survival (HR = 3.13, 95% CI: 1.02-9.68, PHR = 0.0469), advanced N-stage (RR = 1.12, 95% CI: 1.04-1.21, PRR = 0.0019), and distant metastasis (RR = 2.14, 95% CI: 1.46-3.14, PRR < 0.0001) of GC. CD44 variant 6 (CD44v6) in GC might influence OS (5 studies; HR = 1.27, 95% CI: 0.75-2.14, PHR = 0.3783; 4 studies; HR = 1.52, 95% CI: 1.09-2.14, PHR = 0.0139), while significantly associated with N-stage (RR = 1.23, 95% CI: 1.03-1.48, PRR = 0.0240), M-stage (RR = 2.54, 95% CI: 1.08-6.00, PRR = 0.0333), TNM-stage (RR = 1.72, 95% CI: 1.18-2.50, PRR = 0.0045), Lauren type (RR = 0.67, 95% CI: 0.50-0.91, PRR = 0.0106), lymphatic invasion (RR = 1.13, 95% CI: 1.04-1.23, PRR = 0.0057), and liver metastasis (RR = 3.20, 95% CI: 1.94-5.27, PRR < 0.0001) of the disease. Moreover, a narrative review was performed for CD44 isoforms, such as v3, v5, v7, v8-10, and v9, in GC. In conclusion, CD44s and CD44v6 as evaluated by immunohistochemistry, respectively, predicts the prognosis and disease severity of GC.
Keywords: CD44s, CD44 variants, gastric cancer, meta-analysis, narrative review, prognosis
Introduction
Gastric cancer (GC) is a major public health issue, as the fourth most common and the second most deadly human malignancy worldwide [1,2]. Although great advances have been made for the diagnosis and therapy of the disease, the clinical outcome of patients is still poor [3-5]. Increasing evidence suggests that cancer stem cells (CSCs) within GC show the potential for the initiation and progression of cancer, such as inducing heterogeneity, metastasis, and therapeutic resistance of GC, and thus resulting in the poor prognosis of patients [6-8]. Furthermore, special biomarkers account for the particular property of CSCs [9-11]. Amongst the several stem cell surface markers of GC, the class I transmembrane glycoprotein CD44 family represents the novel and most robust surface marker for GC stem cells [9,11].
CD44 family includes standard CD44 (CD44s) that expressed ubiquitously and CD44 splicing variants (CD44v) with specific distributions in keratinocytes (CD44v3-v10), epithelial cells (CD44v8-v10), and activated lymphocytes and macrophages (CD44v6) [12,13]. Functionally, CD44 was initially identified as the receptor for the extracellular matrix component, hyaluronic acid, and involved in multiple physiological and pathological processes, like cancer development, angiogenesis, cell adhesion, wound healing and inflammation [14,15]. Later studies suggested CD44 to be an important stem cell marker for multiple solid tumors including GC [9,16]. Following the identification of various CD44 isoforms, the studies for CD44 became more broad and complex. Currently, it is revealed that CD44 family proteins mediate a variety of biological processes, such as epithelial-mesenchymal transition (EMT), DNA repair, over expression of ABC transporters, chemo-radioresistance and invasiveness, in GC cells [5,10,17-19].
Although all of the aforementioned molecular functions of CD44 lead to the development and progress of cancer, the clinical studies evaluating the validity of CD44 as a therapeutic or diagnostic target in human GC are diversified, and the findings are also controversial. Mayer, B. et al first found that CD44 expression in GC independently predicted poor survival of patients [20], which were confirmed by some later studies [4,6,9,12,16,21]. However, there were still some other studies showing the insignificant association between the presence of CD44 in GC and the poor clinical outcome of patients [22,23]. Moreover, the reports on the clinical and prognostic role of CD44 variants in GC are also inconsistent [23-25]. To reveal the current research status, clarify the controversial issues and present some potential clues for the future research directions, we performed the meta-analysis and narrative review for the association of CD44 family proteins with the prognosis and clinicopathologic features in GC.
Methods
Publication search
In the PubMed database, publications were identified with the following search terms: “Stomach Neoplasms” or “gastric cancer” [Title/Abstract] or “gastric carcinoma” [Title/Abstract] or “gastric cancers” [Title/Abstract] or “gastric cancer*” [Title/Abstract] or “gastric carcinomas” [Title/Abstract] or “gastric carcinoma*” [Title/Abstract] or “gastric adenocarcinoma” [Title/Abstract] or “gastric adenocarcinomas” [Title/Abstract] or “gastric adenocarcinoma*” [Title/Abstract] or “stomach cancer” [Title/Abstract] or “stomach cancers” [Title/Abstract] or “stomach cancer*” [Title/Abstract] or “stomach carcinoma” [Title/Abstract] or “stomach carcinomas” [Title/Abstract] or “stomach carcinoma*” [Title/Abstract] or “stomach adenocarcinoma” [Title/Abstract] or “stomach adenocarcinomas” [Title/Abstract] or “stomach adenocarcinoma*” [Title/Abstract] or “gastric neoplasm” [Title/Abstract] or “gastric neoplasms” [Title/Abstract] or “gastric neoplasm*” [Title/Abstract] or “stomach neoplasm” [Title/Abstract] or “stomach neoplasms” [Title/Abstract] or “stomach neoplasm*” [Title/Abstract] or “cancer of the stomach” [Title/Abstract] or “cancer of stomach” [Title/Abstract], AND “Antigens, CD44” or “Hyaluronan-Binding Protein” [Title/Abstract] or “Hyaluronan Binding Protein” [Title/Abstract] or “CD44 Antigen” [Title/Abstract] or “Hyaluronan Receptor” [Title/Abstract] or “Hyaluronan Receptors” [Title/Abstract] or “Hyaluronic Acid Binding Protein” [Title/Abstract] or “CD44 Antigens” [Title/Abstract] or “MC56 protein” [Title/Abstract] or “homing-associated cell adhesion molecule” [Title/Abstract] or “MC56 drug-sensitivity marker protein” [Title/Abstract] or “HCAM protein” [Title/Abstract] or “CD44*” [Title/Abstract]. Articles included in the present analysis were published from 1991 through August 2014. The articles that detected CD44 by immunohistochemistry (IHC) method were included in the current study. To identify relevant articles, title/abstract scanning and full-text browsing were sequentially performed.
Inclusion criteria
The inclusion criteria for literatures were: (1) studies published as original research in English regardless of publication time; (2) studies presenting sufficient data for evaluating the impact of the expression of CD44 and its variants on the clinicopathological outcome in GC; (3) studies dealing with primary GC samples removed by surgery (not metastatic GC or GC adjacent tissue) and confirmed pathologically; (4) studies that is the newest or the most completed amidst duplicated reports on the same cohorts at different time, as checked out by references manager software EndNote (X7 version). Letters, case reports, reviews, conference abstracts and researches using animal or cell lines, and studies unrelated to our analysis were excluded.
Data extraction
With the standardized principle and tool for data extraction, two reviewers independently abstracted the data. The disagreements were resolved by consensus after referring to the original reports. Nonspecific-defined CD44 in the previous reports was regarded as CD44s, and CD44 isoforms were named as the original reports. The following information were collected from the eligible publications: name of first author, publication year, patients’ country, number of patients analyzed, research technique used, cut off value of CD44 family proteins, clinicopathological variables including cancer location, differentiation, Lauren type, lymphatic invasion, vascular invasion, liver metastasis, peritoneal metastasis, perineural metastasis, depth of invasion (T-stage), lymph node metastasis (N-stage), distant metastasis (M-stage), and TNM-stage. For ease of analysis, CD44 expression was categorized as high/positive and low/negative, and the following clinicopathological variables were combined into dichotomous categories: cardia and non-cardia location, well/moderate (WD/MD) and poor/undifferentiated (PD/UN) differentiation, intestinal and diffuse type, negative (-) and positive (+) lymphatic/vascular invasion, negative (-) and positive (+) liver/peritoneal/perineural metastasis, T1-2 and T3-4 stage, N0 and N1-3 stage, M0 and M1 stage, as well as I-II and III-IV TNM-stage. Hazard ratio (HR) and 95% confidence interval (CI) from univariate analysis was preferably taken if both univariate and multivariate analysis were reported. Calculation method was applied to extract HR and 95% CI where HR was not reported [26]. In those studies with only Kaplan-Meier (K-M) curve available, survival curves were read by Engauge Digitizer version 4.1 (downloaded from http://sourceforge.net), and HR, 95% CI, the significance as well as the orientation (favor protective or hazardous) were extracted from original publications as described by Parmar et al [27].
For the above categories, data reported not less than 3 times were meta-analyzed, and the others were subjected to narrative review. Additionally, data not shown in the primary articles were designated as “N/A (not available)” in our study. We did not request additional or unreported information of the primary studies. We also did not evaluate the studies with quality score, considering the quality score system in meta-analysis of observational studies is still controversial.
Statistical methods
R/meta software (R 3.0.2) was utilized to perform the statistical analysis. Unless specifically indicated, all statistic tests were two tailed with P < 0.05 as statistically significant. Heterogeneity of publications was calculated with the Chi-square-based Q statistic and inconsistency index (I2) statistic (P < 0.10 and I2 > 50% indicated substantial heterogeneity). A fixed-effect model was used if homogeneity was present, and a random-effect model was used if heterogeneity was demonstrated. Log HRs were used to make the forest plot in the survival analysis using R software, and 95% CI not overlap 0 was considered significant. Pooled HR and 95% CI were obtained from log HR by calculation, and a HR > 1 implied that CD44 high/positive expression predicted worse survival of patients. Risk ratio (RR) and 95% CI were utilized for the analysis of dichotomous data. It was considered as statistically significant if the 95% CI for RR did not overlap 1. To assess the stability of the results, we conducted sensitivity analysis, which means to delete one at a time to check the influence of the individual data set on the pooled RR (or HR). Egger’s regression tests were performed to evaluate the publication bias.
Results
Description of studies
Figure 1 showed the detailed search steps. A total of 293 studies were retrieved with the search strategy described above. After title/abstract scanning, 81 studies were considered relevant and further evaluated by reviewing full text in detail. Of these publications, 44 were excluded: 15 were not immunohistochemical research for CD44, and 29 were with non-extractable data for analysis. Finally, there were totally 37 eligible studies involving 38 cohorts. Thirty-four studies including 35 observational cohorts (totally 5450 patients, ranging from 28 to 729 patients per cohort) were included in meta-analysis, and 7 of them were also subjected to narrative review. The other 3 studies were merely analyzed by narrative review. Table 1 showed the major characteristics of the eligible studies.
Figure 1.
Flowchart for the selection of studies according to the predefined inclusion criteria in meta-analysis.
Table 1.
Main characteristics of the eligible studies
| Author | Year | Country | Cut off | Stage | Cancer | Cohort | Sample | Detection | Gene | Survival | HR Extraction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cao, L. | 2014 | China | 0 | N/A | GC | 203 | FFPE | WTS-IHC | CD44 | OS | Calculation |
| Qiu, Y. | 2014 | China | 4 score | I-IV | GC | 243 | FFPE | TMA-IHC | CD44 | N/A | |
| Hirata, K. | 2013 | Japan | 3.55% | N/A | EGC | 65 | FFPE | WTS-IHC | CD44v9 | DFS | Report-mul |
| Chen, S. | 2013 | China | 65% | I-IV | GC | 152 | FFPE | WTS-IHC | CD44 | OS | Report-mul |
| Jung, W. Y. | 2013 | Korea | N/A | I-IV | GC | 430 | FFPE | TMA-IHC | CD44 | N/A | |
| Doventas, A. | 2012 | Turkey | 0 | I-IV | GC | 48 | FFPE | WTS-IHC | CD44 | OS | SC |
| Wakamatsu, Y. | 2012 | Japan | 10% | I-IV | GC | 96 | FFPE | WTS-IHC | CD44 | CRS | Report-uni |
| Fanelli, M. F. | 2012 | Brazil | 0 | I-IV | GC | 137 | FFPE | TMA-IHC | CD44v6 | OS | Calculation |
| Liang, Yi-Zhi | 2012 | China | 5% | N/A | GC | 59 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Ryu, H. S. | 2012 | Korea | 5% | I-IV | GC | 276 | FFPE | TMA-IHC | CD44 | N/A | |
| Wang, T. | 2011 | Singapore | 5 score | I-IV | GC | 106 | FFPE | TMA-IHC | CD44 | OS | SC |
| Dhingra, S. | 2011 | United States | N/A | I-IV | GC | 138 | FFPE | WTS-IHC | CD44 | N/A | |
| Kim, J. Y. | 2009 | Korea | 10% | I-IV | GC | 210 | FFPE | TMA-IHC | CD44 | OS | SC |
| Okayama, H. | 2009 | Japan | 5% | I-III | GC | 135 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Songun, I. | 2007 | Netherlands | 25% | N/A | R0 GC | 286 | FFPE | WTS-IHC | CD44v6 | OS | Calculation |
| Kim, M. A. | 2005 | Korea | 10% | I-IV | GC | 729 | FFPE | TMA-IHC | CD44 | N/A | |
| Chen, X. Y. | 2005 | China | 0 | I-IV | GC | 28 | FFPE | TMA-IHC | CD44v6 | N/A | |
| Chen, J. Q. | 2004 | China | 10% | I-IV | D2/D3 GC | 43 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Polkowski, W. P. | 2004 | Poland | 10% | II-IV | Cardia GC | 49 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Joo, M. | 2003 | Korea | 10% | I-IV | GC | 99 | FFPE | WTS-IHC | CD44/CD44v6 | N/A | |
| Yamaguchi, A. | 2002 | Japan | 0 | I-IV | AGC | 201 | FFPE | WTS-IHC | CD44v6 | OS | SC |
| Xin, Y. | 2001 | Ireland | 5% | I-IV | GC | 155 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Li, H. | 2000 | China | N/A | N/A | GC | 74 | Frozen | WTS-IHC | CD44v5/v6/v7/v8-10 | N/A | |
| Yoo, C. H. | 1999 | Korea | 5% | II/IIIA | GC | 261 | FFPE | WTS-IHC | CD44 | OS | Report-mul |
| Saito, H. | 1998 | Japan | 5% | I-IV | Diffuse GC | 46 | FFPE | WTS-IHC | CD44v6 | OS | SC |
| Saito, H. | 1998 | Japan | 5% | I-IV | Intestinal GC | 71 | FFPE | WTS-IHC | CD44v6 | OS | SC |
| Kurozumi, K. | 1998 | Japan | 30% | I-III | GC | 98 | FFPE | WTS-IHC | CD44v6 | N/A | |
| Isozaki, H. | 1998 | Japan | 10% | I-IV | GC | 108 | FFPE | WTS-IHC | CD44 | N/A | |
| Yasui, W. | 1998 | Japan | 5% | I-IV | GC | 1074 | FFPE | WTS-IHC | CD44v9 | N/A | |
| Chong, J. M. | 1997 | Japan | N/A | N/A | GC | 104 | FFPE | WTS-IHC | CD44v6/v3-5 | N/A | |
| Muller, W. | 1997 | Germany | 5% | N/A | GC | 418 | FFPE | WTS-IHC | CD44v5 | OS | Calculation |
| Ura, H. | 1996 | Japan | 10% | N/A | GC | 110 | FFPE | WTS-IHC | CD44v6/v3 | N/A | |
| Hong, R. L. | 1995 | China | 0 | N/A | GC | 103 | Frozen | WTS-IHC | CD44/CD44v6 | OS/DFS | SC |
| Mirecka, J. | 1995 | Poland. | N/A | N/A | GC | 112/105 | FFPE | WTS-IHC | CD44v5/v6 | N/A | |
| Yamaguchi, A. | 1995 | Japan | 25% | I-IV | GC | 194 | FFPE | WTS-IHC | CD44v8-10 | N/A | |
| Harn, H. J. | 1995 | China | N/A | N/A | GC | 49 | FFPE | WTS-IHC | CD44v5/v6 | N/A | |
| Dammrich, J. | 1995 | Germany | N/A | N/A | GC | 42 | Frozen | WTS-IHC | CD44v6 | N/A | |
| Mayer, B. | 1993 | Germany | N/A | N/A | GC | 31 | FFPE | WTS-IHC | CD44 | CRS/DFS | Calculation |
GC: gastric cancer; EGC: early gastric cancer; AGC: advanced gastric cancer; N/A: not available; FFPE: formalin-fixed, paraffin-embedded; WTS: whole tissue section; TMA: tissue microarray; IHC: immunohistochemistry; OS: overall survival; CRS: cancer related survival; DFS: disease free survival; HR: hazard ratio; SC: survival curve; mul: multivariate analysis; uni: univariate analysis.
Meta-analysis for CD44s expression in GC
The correlation between CD44s and OS of GC patients was illustrated in Figure 2. For the systematic evaluation of 9 eligible studies (1210 patients), the pooled HRs were got directly (3 studies) or extracted (6 studies) with the aforementioned methods (Table 1). In a fixed-effect model (I2 = 18.4%, P = 0.2792), the presence of CD44s highly indicated reduced OS (pooled HR = 1.93, 95% CI: 1.58-2.34, transformed from log HR and its 95% CI indicated in Figure 2 and Table 2; PHR = 0.0222). Moreover, CD44s expression tended to associate with poorer disease free survival (DFS) in GC (random effect; pooled HR = 3.13, 95% CI: 1.02-9.68, transformed from log HR and its 95% CI indicated in Figure 3 and Table 2; PHR = 0.0469). Thirteen studies (2336 patients) evaluated the correlation of CD44s expression with lymph node metastasis of cancer (Table 2). The pooled RR was 1.12 (95% CI: 1.04-1.21, PRR = 0.0019, fixed-effect), and there was no significant heterogeneity (I2 = 00.0%, P = 0.5891), indicating that presence of CD44s was highly related with advanced N-stage of GC. Furthermore, CD44s expression was significantly associated with distant metastasis (Table 2, 4 studies, 426 patients; pooled RR = 2.14, 95% CI: 1.46-3.14, PRR < 0.0001, fixed-effect) with no obvious heterogeneity observed (I2 = 00.0%, P = 0.6093). As shown in Table 2, there was no statistically significant association between CD44s expression and other parameters such as cancer location, T-stage, TNM-stage, differentiation, Lauren type, lymphatic invasion, vascular invasion, and perineural metastasis.
Figure 2.
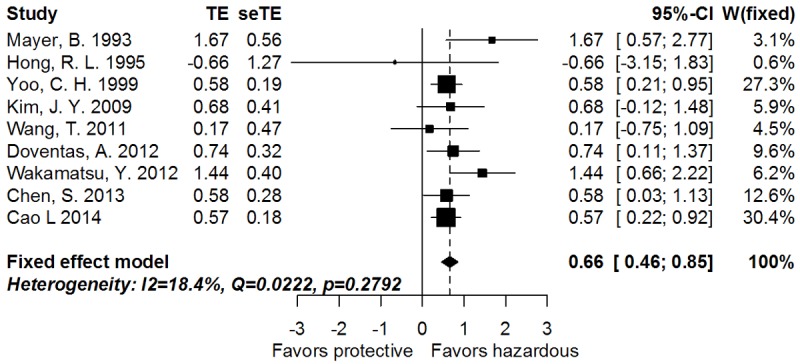
Forrest plot of log hazard ratio for the correlation between CD44s expression and overall survival.
Table 2.
Meta-analysis of CD44s in gastric cancer
| Stratification | Studies (N) | Patients (N) | Model | Log HR/RR (95% CI) | PHR/PRR | P | I2 (%) | P bias |
|---|---|---|---|---|---|---|---|---|
| Overall survival | 9 | 1210 | Fixed | 0.66 (0.46-0.85) | 0.0222 | 0.2792 | 18.4 | 0.6264 |
| Disease free survival | 3 | 261 | Random | 1.14 (0.02-2.27) | 0.0469 | 0.0168 | 75.5 | 0.6582 |
| Location of cancer | 7 | 1646 | Fixed | 0.95 (0.89-1.01) | 0.1049 | 0.0791 | 47.0 | 0.7651 |
| T-stage | 11 | 1927 | Random | 0.97 (0.85-1.11) | 0.6909 | 0.0008 | 66.7 | 0.0297 |
| N-stage | 13 | 2336 | Fixed | 1.12 (1.04-1.21) | 0.0019 | 0.5891 | 00.0 | 0.9071 |
| M-stage | 4 | 426 | Fixed | 2.14 (1.46-3.14) | < 0.0001 | 0.6093 | 00.0 | 0.5477 |
| TNM-stage | 10 | 2103 | Fixed | 1.09 (0.99-1.20) | 0.0854 | 0.0536 | 46.1 | 0.3935 |
| Lymphatic invasion | 7 | 1105 | Fixed | 1.09 (0.97-1.22) | 0.1529 | 0.7111 | 00.0 | 0.5177 |
| Vascular invasion | 4 | 693 | Fixed | 1.07 (0.85-1.33) | 0.5677 | 0.9133 | 00.0 | 0.6898 |
| Degree of differentiation | 9 | 1479 | Random | 1.12 (0.97-1.29) | 0.1122 | 0.0144 | 58.1 | 0.6852 |
| Lauren type | 7 | 852 | Random | 0.96 (0.71-1.30) | 0.7923 | 0.0070 | 66.1 | 0.9145 |
| Perineural metastasis | 3 | 598 | Fixed | 0.98 (0.78-1.25) | 0.8972 | 0.6098 | 00.0 | 0.5374 |
HR: hazard ratio; RR: risk ratio; N: number of studies or patients; CI: confidence interval; PHR: significance of HR; PRR: significance of RR; P: significance for heterogeneity of publications; P bias: significance for publication bias.
Figure 3.
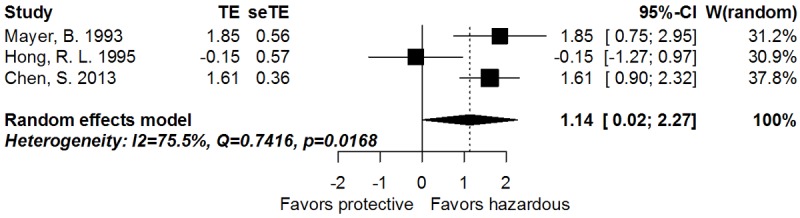
Forrest plot of log hazard ratio for the correlation between CD44s expression and Disease free survival.
Meta-analysis for CD44v expression in GC
Five studies including 6 cohorts evaluated the impact of CD44 variant 6 (CD44v6) on OS of GC. The pooled HR was 1.27 (95% CI: 0.75-2.14, transformed from log HR and its 95% CI indicated in Figure 4A and Table 3; PHR = 0.3783, random effect) and the heterogeneity was significant (I2 = 71.00%, P = 0.0041). However, CD44v6 could predict poorer OS while the study by Songun was omitted according to the sensitivity assay (pooled HR: 1.52, 95% CI: 1.09-2.14, transformed from log HR and its 95% CI indicated in Figure 4B and Table 3; PHR = 0.0139, I2 = 17.4%, P = 0.3035, fixed-effect). In the further analysis for clinicopathological variables (Table 3), CD44v6 was significantly correlated with N-stage (14 studies, 1716 patients; pooled RR = 1.23, 95% CI: 1.03-1.48, PRR = 0.0240, I2 = 67.1%, P = 0.0002, random-effect), M-stage (3 studies, 299 patients; pooled RR = 2.54, 95% CI: 1.08-6.00, PRR = 0.0333, I2 = 00.0%, P = 0.7393, fixed-effect), TNM-stage (3 studies, 277 patients; pooled RR = 1.72, 95% CI: 1.18-2.50, PRR = 0.0045, I2 = 32.7%, P = 0.2250, fixed-effect), Lauren type (8 studies, 1166 patients; pooled RR = 0.67, 95% CI: 0.50-0.91, PRR = 0.0106, I2 = 75.5%, P = 0.0002, random-effect), lymphatic invasion (8 studies, 1270 patients; pooled RR = 1.13, 95% CI: 1.04-1.23, PRR = 0.0057, I2 = 38.3%, P = 0.1242, fixed-effect), and liver metastasis (4 studies, 509 patients; pooled RR = 3.20, 95% CI: 1.94-5.27, PRR < 0.0001, I2 = 00.0%, P = 0.8192, fixed-effect), whereas exhibited no impact on cancer location, T-stage, differentiation, vascular invasion, or peritoneal metastasis. Additionally, the expression of CD44 variant 5 (CD44v5) had no relationship with GC differentiation in the systematic review of 4 studies (652 patients; pooled RR = 0.93, 95% CI: 0.66-1.31, PRR = 0.6852, I2 = 79.4%, P = 0.0022, random-effect; Table S1).
Figure 4.
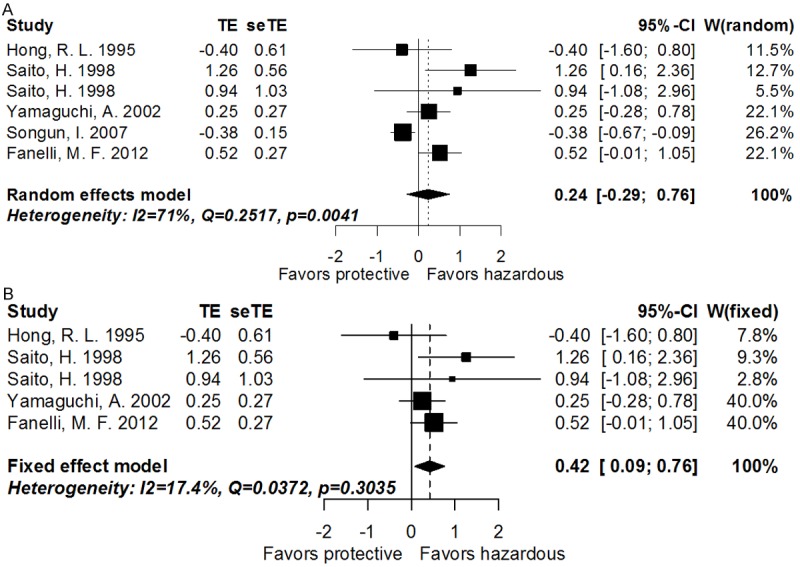
A. Forrest plot of log hazard ratio for the correlation between CD44v6 expression and overall survival. B. Forrest plot of log hazard ratio for the correlation between CD44v6 expression and overall survival after omitting the study by Dr. Songun, I.
Table 3.
Meta-analysis of CD44v6 in gastric cancer
| Stratification | Studies (N) | Patients (N) | Model | Log HR/RR (95% CI) | PHR/PRR | P | I2 (%) | P bias |
|---|---|---|---|---|---|---|---|---|
| Overall survival | 6 | 844 | Random | 0.24 (-0.29-0.76) | 0.3783 | 0.0041 | 71.0 | 0.1738 |
| *Subgroup overall survival | 5 | 558 | Fixed | 0.42 (0.09-0.76) | 0.0139 | 0.3035 | 17.4 | 0.7620 |
| Location of cancer | 3 | 248 | Fixed | 0.99 (0.88-1.12) | 0.8813 | 0.3534 | 3.9 | 0.3638 |
| T-stage | 10 | 1295 | Random | 1.18 (0.97-1.44) | 0.1002 | 0.0261 | 52.4 | 0.5069 |
| N-stage | 14 | 1716 | Random | 1.23 (1.03-1.48) | 0.0240 | 0.0002 | 67.1 | 0.2888 |
| M-stage | 3 | 299 | Fixed | 2.54 (1.08-6.00) | 0.0333 | 0.7393 | 00.0 | 0.9129 |
| TNM-stage | 3 | 277 | Fixed | 1.72 (1.18-2.50) | 0.0045 | 0.2265 | 32.7 | 0.2250 |
| Lymphatic invasion | 8 | 1270 | Fixed | 1.13 (1.04-1.23) | 0.0057 | 0.1242 | 38.3 | 0.3916 |
| Vascular invasion | 7 | 1172 | Random | 1.08 (0.90-1.30) | 0.3839 | 0.0479 | 52.8 | 0.7268 |
| Peritoneal metastasis | 3 | 361 | Fixed | 0.85 (0.58-1.24) | 0.3994 | 0.5619 | 00.0 | 0.1358 |
| Liver metastasis | 4 | 509 | Fixed | 3.20 (1.94-5.27) | < 0.0001 | 0.8192 | 00.0 | 0.6360 |
| Degree of differentiation | 8 | 895 | Random | 0.98 (0.76-1.26) | 0.8661 | 0.0004 | 73.8 | 0.4538 |
| Lauren type | 8 | 1166 | Random | 0.67 (0.50-0.91) | 0.0106 | 0.0002 | 75.5 | 0.2197 |
Subgroup assay for overall survival after omitting the study by Songun.I, et cl.
HR: Hazard ratio; RR: risk ratio; N: number of studies or patients; CI: confidence interval; PHR: significance of HR; PRR: significance of RR; P: significance for heterogeneity of publications; P bias: significance for publication bias.
Egger’s test and sensitivity assay
Egger’s test was performed to evaluate the publication bias of the eligible studies (Tables 2, 3, S1). The results indicated no publication bias for most subgroup assays, except for the depth of invasion for the CD44 subgroup (P bias = 0.0297). In addition, most sensitivity analysis did not reveal significant variation on the pooled RR (or HR) after omitting any single study at a given time, except that for OS and distant metastasis for the CD44v6 analysis, and DFS for the CD44s analysis (Table S2).
Narrative review for CD44 family proteins in GC
As shown in Table S3, CD44v5 phenotype was demonstrated to indicate poorer OS of GC by Dr. Muller, W [28] (HR = 1.36, 95% CI: 1.00-1.84, PHR = 0.049), whereas neither CD44v5 (HR = 0.97, 95% CI: 0.74-1.28, PHR = 0.840) nor CD44 variant 9 (CD44v9; HR = 1, 95% CI: 0.76-1.31, PHR = 0.230) could predict OS as shown by Dr. Songun, I [25]. Moreover, DFS of GC was significantly related with CD44v9 [29] (HR = 21.8, 95% CI: 5.71-83.1, PHR < 0.001), but not CD44v6 [23] (HR = 0.77, 95% CI: 0.28-2.12, PHR = 0.610). Table S4 summarizes the review of clinicopathological parameters. Dr. Muller, W. reported that CD44v5 significantly predicted Lauren classification (Preport = 0.001), differentiation (Preport = 0.001), T-stage (Preport = 0.001), vascular invasion (Preport = 0.004), and lymphatic invasion (Preport = 0.001) [28]. However, Dr. Mirecka, J. failed to demonstrate the association of CD44v5 with T-stage and differentiation of GC [30]. CD44 variant 8-10 (CD44v8-10) was associated with vascular invasion (Preport = 0.032) and liver metastasis (Preport = 0.015) [31], but not T-stage, N-stage, TNM-stage, lymphatic invasion, peritoneal invasion [31] or differentiation [32]. In addition, CD44s significantly predicted hepatic metastasis (Preport = 0.0002) [33] and tumor relapse (Preport = 0.025) [4]; CD44 variant 3-5 (CD44v3-5) was related with lymphatic invasion (Preport < 0.050) and N-stage (Preport < 0.050) [34]; CD44 variant 7 (CD44v7) could predict poorly differentiation (Preport < 0.010) [32]; and CD44 variant 9 (CD44v9) was correlated with T-stage (Preport < 0.010), N-stage (Preport < 0.010), TNM-stage (Preport < 0.050), and differentiation (Preport < 0.010) [35], but not location of GC [29]. Moreover, CD44 variant 3 (CD44v3) had no relationship with either liver metastasis or N-stage in GC [36].
Discussion
Since the indicative significance of CD44 family phenotype in GC is controversial and the related studies are diversified, a quantitative meta-analysis and comprehensive narrative review of the previous studies is warranted. Currently, CD44s is shown to associate with poor OS by combining 1210 patients from 9 studies, and with reduced DFS in 261 patients from 3 studies (Figures 1, 2 and 3; Table 2). Moreover, CD44s expression significantly predicts lymph node and distant metastasis of GC (Table 2). As for CD44 splicing variants, CD44v6 shows distinct impact on OS in the meta-analysis of 5 (6 cohorts; 844 patients) and 4 studies (5 cohorts; 558 patients) (Figure 4; Table 3), whereas statistically relates with Lauren type, lymphatic invasion, liver metastasis, and N-, M- and TNM- stage of GC. CD44v3-5, v5, v7, v8-10, and v9 show the potential to influence disease severity and outcome of GC in the narrative review (Tables S3, S4). As far as we know, this is the first study to simultaneously assess the predictive value of standard CD44 and CD44 variants for the clinicopathological characteristics and survival status of GC.
As the principal cell surface receptor for hyaluronic acid, CD44 family exerts important functions in cell survival, proliferation, motility, and extracellular matrix adherence and degradation [11,13,14]. Some researchers have reported the prognostic value of CD44 family for GC and the results show that CD44 could be used as a novel marker for the characteristics and management of GC [4,20]. In our study, there was no association between CD44s expression and cancer location, differentiation, Lauren type, vascular invasion, lymphatic invasion, depth of invasion, TNM-stage, or perineural metastasis. On the contrary, our meta-analysis demonstrated that CD44s phenotype was positively correlated with lymph node and distant metastasis, and poorer outcome of GC patients (Table 2; Figures 2, 3), suggesting the potential value of this marker for clinical applications.
As for CD44 variants, CD44v6 in GC could be meta-analyzed in the current study. Previously, the only one meta-analysis about CD44 family proteins in GC was about the clinical and survival validity of CD44v6 by Dr. Chen Jing [37]. Although both reports included 5 studies for the analysis of OS and CD44v6, great differences were shown as follows. First, unlike the previous report that focused on Asian cohorts, the current study did not set limitation for ethnology, because there is no evidence so far to show that CD44 family proteins exert ethnology-related functions. Second, the previous report included the Korean study by Dr. Eom, D. W. and indicated the data for analysis was reported in the text of the original report [37]. However, we could not find the necessary data in the original article, thus excluded the article for further analysis. Third, the 2 Chinese of totally 5 Asian studies included by the previous report were published in Chinese, while the current analysis excluded the publications in Chinese and the included Chinese study was published in English. Fourth, the previous report demonstrated CD44v6 to be the predictor of higher risk of death (767 patients) [37]. Presently, the correlation between CD44v6 and OS (Figure 4; Table 3) shifted from insignificance (844 patients, significant heterogeneity) to significance (558 patients, no heterogeneity) after omitting the heterogeneous study that used relatively higher cutoff (25%) comparing with the others (0-5%, Table 1). Furthermore, we failed to get the significant relationship shown by Dr. Chen [37] between CD44v6 and T-stage, vascular invasion, and histological differentiation; while found the influence of CD44v6 on Lauren type, lymphatic invasion, and liver metastasis (Table 3). The current report included the data from the whole world including those from Asian and published more recently, and updated the previous report about the prognostic and clinical pathological role of CD44v6 in GC. Moreover, the difference between the current and the previous report suggested the necessity of further research about the topic.
To interpret the results of the meta-analysis, certain limitations in the present meta-analysis should be concerned. First, publication bias should be taken into account, although most of our obtained statistical results are insignificant. The power of detecting publication bias could be reduced by the small number of eligible studies. Additionally, we excluded some studies from our analysis, for reasons of language restriction, non-extractable or insufficient survival data [5,38,39]. The missing data, especially those reported “negative” or more conservative correlation of CD44 with prognosis [5,38,39], might affect the significance of CD44 phenotype as an indicator of disease severity and patients outcome. Second, the size of included studies for analysis is not large (248-2336 patients), the patient populations are not uniform, and the length of follow-up varies. Yoo et al. focused on patients with stage II and IIIA GC [21], and the median follow-up duration ranged from 9.5 to 137 months [22,23]. All these might influence the significance of the clinical outcome in the current analysis. Third, the survival data are achieved directly, calculated from the available data, or extracted from the K-M curves in the articles (Table 1). The latter two methods are less reliable than direct analysis of primary data [26]. Fourth, the methods for CD44 evaluation, such as the cutoff scores, are not unified between the studies (Table 1), and could affect the conclusion.
In summary, we firstly evaluate the prognostic and clinical pathological role of all CD44 family proteins in GC with systematic or narrative review, and update the previous report for CD44v6 in GC. CD44s and CD44v6 might indicate reduced survival and the potential of metastasis and invasion of GC. The detection of CD44 family by IHC would be helpful to predict the severity of disease, and the development of treatment strategy against subset of CD44 proteins could be novel therapeutic choice in clinical settings. Given the variety of molecular function and clinical validity of CD44 family proteins in GC, further large-sample studies are required.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81201801 and No. 81302023), and the Fund of the Education Department of Liaoning Province (No. L2012278).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Resende C, Thiel A, Machado JC, Ristimaki A. Gastric cancer: basic aspects. Helicobacter. 2011;16(Suppl 1):38–44. doi: 10.1111/j.1523-5378.2011.00879.x. [DOI] [PubMed] [Google Scholar]
- 3.Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Hou JH, Feng XY, Zhang XS, Zhou ZW, Yun JP, Chen YB, Cai MY. Clinicopathologic significance of putative stem cell marker, CD44 and CD133, in human gastric carcinoma. J Surg Oncol. 2013;107:799–806. doi: 10.1002/jso.23337. [DOI] [PubMed] [Google Scholar]
- 5.Ryu HS, Park do J, Kim HH, Kim WH, Lee HS. Combination of epithelial-mesenchymal transition and cancer stem cell-like phenotypes has independent prognostic value in gastric cancer. Hum Pathol. 2012;43:520–528. doi: 10.1016/j.humpath.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang T, Ong CW, Shi J, Srivastava S, Yan B, Cheng CL, Yong WP, Chan SL, Yeoh KG, Iacopetta B, Salto-Tellez M. Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer. 2011;105:658–665. doi: 10.1038/bjc.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, Los M. Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Ishigami S, Ueno S, Arigami T, Uchikado Y, Setoyama T, Arima H, Kita Y, Kurahara H, Okumura H, Matsumoto M, Kijima Y, Natsugoe S. Prognostic impact of CD133 expression in gastric carcinoma. Anticancer Res. 2010;30:2453–2457. [PubMed] [Google Scholar]
- 9.Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y, Uraoka N, Anami K, Sentani K, Oue N, Yasui W. Expression of cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and lymph node metastasis of gastric cancer. Pathol Int. 2012;62:112–119. doi: 10.1111/j.1440-1827.2011.02760.x. [DOI] [PubMed] [Google Scholar]
- 10.Dhingra S, Feng W, Brown RE, Zhou Z, Khoury T, Zhang R, Tan D. Clinicopathologic significance of putative stem cell markers, CD44 and nestin, in gastric adenocarcinoma. Int J Clin Exp Pathol. 2011;4:733–741. [PMC free article] [PubMed] [Google Scholar]
- 11.Li K, Dan Z, Nie YQ. Gastric cancer stem cells in gastric carcinogenesis, progression, prevention and treatment. World J Gastroenterol. 2014;20:5420–5426. doi: 10.3748/wjg.v20.i18.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doventas A, Bilici A, Demirell F, Ersoy G, Turna H, Doventas Y. Prognostic significance of CD44 and c-erb-B2 protein overexpression in patients with gastric cancer. Hepatogastroenterology. 2012;59:2196–2201. doi: 10.5754/hge10498. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh HF, Yu JC, Ho LI, Chiu SC, Harn HJ. Molecular studies into the role of CD44 variants in metastasis in gastric cancer. Mol Pathol. 1999;52:25–28. doi: 10.1136/mp.52.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang BI, Li Y, Graham DY, Cen P. The Role of CD44 in the Pathogenesis, Diagnosis, and Therapy of Gastric Cancer. Gut Liver. 2011;5:397–405. doi: 10.5009/gnl.2011.5.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004;95:930–935. doi: 10.1111/j.1349-7006.2004.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Yao D, Liu W, Wang N, Lv H, He N, Shi B, Hou P, Ji M. Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci. 2012;13:4714–4726. doi: 10.3390/ijms13044714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, Gordon SA, Shimada Y, Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, Masuko T, Shimizu T, Ishikawa T, Kai K, Takahashi E, Imamura Y, Baba Y, Ohmura M, Suematsu M, Baba H, Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida GJ, Saya H. Inversed relationship between CD44 variant and c-Myc due to oxidative stress-induced canonical Wnt activation. Biochem Biophys Res Commun. 2014;443:622–627. doi: 10.1016/j.bbrc.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Mayer B, Jauch KW, Gunthert U, Figdor CG, Schildberg FW, Funke I, Johnson JP. De-novo expression of CD44 and survival in gastric cancer. Lancet. 1993;342:1019–1022. doi: 10.1016/0140-6736(93)92879-x. [DOI] [PubMed] [Google Scholar]
- 21.Yoo CH, Noh SH, Kim H, Lee HY, Min JS. Prognostic significance of CD44 and nm23 expression in patients with stage II and stage IIIA gastric carcinoma. J Surg Oncol. 1999;71:22–28. doi: 10.1002/(sici)1096-9098(199905)71:1<22::aid-jso5>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 22.Kim JY, Bae BN, Kim KS, Shin E, Park K. Osteopontin, CD44, and NFkappaB expression in gastric adenocarcinoma. Cancer Res Treat. 2009;41:29–35. doi: 10.4143/crt.2009.41.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong RL, Lee WJ, Shun CT, Chu JS, Chen YC. Expression of CD44 and its clinical implication in diffuse-type and intestinal-type gastric adenocarcinomas. Oncology. 1995;52:334–339. doi: 10.1159/000227485. [DOI] [PubMed] [Google Scholar]
- 24.Saito H, Tsujitani S, Katano K, Ikeguchi M, Maeta M, Kaibara N. Serum concentration of CD44 variant 6 and its relation to prognosis in patients with gastric carcinoma. Cancer. 1998;83:1094–1101. [PubMed] [Google Scholar]
- 25.Songun I, Litvinov SV, van de Velde CJ, Pals ST, Hermans J, van Krieken JH. Loss of Ep-CAM (CO17-1A) expression predicts survival in patients with gastric cancer. Br J Cancer. 2005;92:1767–1772. doi: 10.1038/sj.bjc.6602519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Muller W, Schneiders A, Heider KH, Meier S, Hommel G, Gabbert HE. Expression and prognostic value of the CD44 splicing variants v5 and v6 in gastric cancer. J Pathol. 1997;183:222–227. doi: 10.1002/(SICI)1096-9896(199710)183:2<222::AID-PATH923>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 29.Hirata K, Suzuki H, Imaeda H, Matsuzaki J, Tsugawa H, Nagano O, Asakura K, Saya H, Hibi T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer. 2013;109:379–386. doi: 10.1038/bjc.2013.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirecka J, Marx D, Schauer A. Immunohistochemical localization of CD44 variants 5 and 6 in human gastric mucosa and gastric cancer. Anticancer Res. 1995;15:1459–1465. [PubMed] [Google Scholar]
- 31.Yamaguchi A, Saito M, Gio T, Iida A, Takeuchi K, Hirose K, Nakagawara G, Urano T, Furukawa K, Shiku H. Expression of CD44 variant exons 8-10 in gastric cancer. Jpn J Cancer Res. 1995;86:1166–1171. doi: 10.1111/j.1349-7006.1995.tb03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Guo L, Li JW, Liu N, Qi R, Liu J. Expression of hyaluronan receptors CD44 and RHAMM in stomach cancers: relevance with tumor progression. Int J Oncol. 2000;17:927–932. [PubMed] [Google Scholar]
- 33.Isozaki H, Ohyama T, Mabuchi H. Expression of cell adhesion molecule CD44 and sialyl Lewis A in gastric carcinoma and colorectal carcinoma in association with hepatic metastasis. Int J Oncol. 1998;13:935–942. doi: 10.3892/ijo.13.5.935. [DOI] [PubMed] [Google Scholar]
- 34.Chong JM, Fukayama M, Hayashi Y, Funata N, Takizawa T, Koike M, Muraoka M, Kikuchi-Yanoshita R, Miyaki M, Mizuno S. Expression of CD44 variants in gastric carcinoma with or without Epstein-Barr virus. Int J Cancer. 1997;74:450–454. doi: 10.1002/(sici)1097-0215(19970822)74:4<450::aid-ijc16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 35.Yasui W, Kudo Y, Naka K, Fujimoto J, Ue T, Yokozaki H, Tahara E. Expression of CD44 containing variant exon 9 (CD44v9) in gastric adenomas and adenocarcinomas: relation to the proliferation and progression. Int J Oncol. 1998;12:1253–1258. doi: 10.3892/ijo.12.6.1253. [DOI] [PubMed] [Google Scholar]
- 36.Ura H, Denno R, Hirata K. Correlation between nm23 protein and several cell adhesion molecules in human gastric carcinoma. Jpn J Cancer Res. 1996;87:512–517. doi: 10.1111/j.1349-7006.1996.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Li T, Liu Q, Jiao H, Yang W, Liu X, Huo Z. Clinical and prognostic significance of HIF-1alpha, PTEN, CD44v6, and survivin for gastric cancer: a meta-analysis. PLoS One. 2014;9:e91842. doi: 10.1371/journal.pone.0091842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setala L, Lipponen P, Tammi R, Tammi M, Eskelinen M, Alhava E, Kosma VM. Expression of CD44 and its variant isoform v3 has no prognostic value in gastric cancer. Histopathology. 2001;38:13–20. doi: 10.1046/j.1365-2559.2001.01038.x. [DOI] [PubMed] [Google Scholar]
- 39.Xie JW, Huang CM, Zheng CH, Li P, Wang JB, Lin JX, Lu J. [Expression tumor stem cell surface marker CD44 in gastric cancer and its significance] . Zhonghua Wei Chang Wai Ke Za Zhi. 2013;16:1107–1112. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


