Abstract
Gastric cancer (GC) remains the second leading cause of cancer-related death worldwide. Owing to the lack of early diagnostic techniques, GC is often diagnosed at advanced stage and that leading to low survival rate. Growing evidences have been suggesting that circulating microRNAs play an important role in earlier diagnostic of disease. In the present study, we analyze the circulating miRNAs expression in plasma of volunteers with/without GC aiming to identifying early diagnostic biomarkers. Plasma samples were collected form 6 volunteers including 3 early patients with GC and 3 healthy adults. And then miRNAs microarrays were performed to detect the expression profile of miRNAs in these plasma samples. For further validate the results from miRNAs microarray, qRT-PCR was performed. Finally, target genes of miRNAs were predicted by bioinformatic means. Compared to control plasma, 11 up-regulated and 13 down-regulated miRNAs were detected in the plasma from earlier patients with GC (fold change ≥ 2, P < 0.05). Then, 5 differential expression miRNAs (miR-223, miR-19b-2*, miR-194*, miR-141, miR-1233) were chose to confirm by qRT-PCR. The result is nearly consistent with previous data from miRNAs microarray. Finally, 53 target genes of the 5 miRNAs are predicted by bioinformatics. These differential expression miRNAs may be used as biomarker candidates for minimally invasive diagnosis of early patients with GC in the future.
Keywords: Gastric cancer, miRNAs, early diagnosis, biomarkers, plasma
Introduction
Gastric cancer (GC) remains the second leading causes of cancer-related death worldwide. Although diagnostic technique and treatment methods have been improving over the past few decades, the prognosis of GC is still poor. Meanwhile, most of patients are diagnosed at the advanced stage of GC so that missing the best opportunity of treatment [1]. Biomarkers have been hoping to be promising targets for improvement early diagnosis rate. Circulating miRNAs as early diagnostic molecule own minimally invasive characteristic compared to biopsy [2].
miRNAs are highly conserved small non-coding RNA molecules (17~22 nt), which regulate their target genes usually on the post-transcriptional level by binding to complementary sequences on target mRNA transcripts (mRNAs) [3]. They actively participate in the modulation of important cell physiological processes, and they are involved in the pathogenesis of many diseases, including cancer [2]. A better understanding of the role that miRNAs play in these diseases, especially in gastric cancer, could lead to the development of new diagnostic and therapeutic tools.
Since the first description of miRNAs in 1993 [4], nearly 2,000 human miRNA genes have known, and more than 1,000 new ones are continuously discovered [5]. In recent years, miRNAs either directly through binding to the target mRNA or indirectly through repressing nonsense-mediated RNA decay [6,7]. MicroRNAs are not only playing an essential role in cellular processes, but also contributing to many human pathologies including cancer [8]. Subsequently, a multitude of studies about miRNA expression changes in cancer have been reported.
In this article, we investigated the change of differentially expressed miRNAs in plasma of gastric cancer patients through microarray analysis and predicted their target genes with prediction software aiming to identifying novel early diagnostic molecules for GC.
Materials and methods
Plasma samples
Three plasma samples of pathologically diagnosed early gastric cancer patients and 3 normal plasma samples as controls were collected from gastric cancer patients of the First Affiliated Hospital, Anhui Medical University, with informed consent to the study. Pathological diagnosis and classification of all patients were conducted by experienced pathologists according to the 2003 WHO Tumor Classification Standard. Pathological diagnoses of the included patients are shown in Supplementary materials. Informed consent from all patients was obtained and this study was approved by Ethics Committee of the First Affiliated Hospital, Anhui Medical University.
RNA isolation
Peripheral blood samples were collected, mixed and centrifuged in the morning. Total RNA was isolated utilizing mirVanaTM PARISTM Kit (Applied Biosystem, USA), and then swabbed off it and concentrated in vacuum. RNA titer was conducted using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA). RNA concentration and OD (A260/A280) were recorded. Then RNA quantification was conducted by electrophoresis.
MiRNA microarray analysis
After the samples’ dephosphorylation and denaturation, RNA was marked by marker mixture (10 × T4 RNA Ligase Buffer, Cyanine 3-pCp, T4 RNA Ligase). 10 × GE blockers (Agilent, USA) were used to prepare hybrid samples, then, hybridization was conducted utilizing hybrid box (Agilent, USA). After washing the chips, we conducted microarrays scanning by Laser Scanner (Agilent, USA) and Scan Control Software (Agilent, USA). Data analysis was conducted using GenesPring GX 10.0.
Fluorescent quantitative RT-PCR validation
Primer sequences were designed as Table 1. We designed RT primer with stem-loop taking sequence 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC-3’ as a reference, using Primer Premier 5.0 to design forward primer according to cDNA sequences after reverse transcription. At last we added GC at 5’ of PCR forward primer based on Tm calculated by Primer Premier 5.0 to make the temperature of forward primer to be around 60°C. The reverse primer was a general sequence 5’-GTGCAGGGTCCGAGGT-3’. U6 RNAof snRNA was as internal standard of normalization. Then primers were used to synthesize cDNA which would be samples of Real TimePCR.
Table 1.
Primers for real time PCR in this study
| Primers | Sequence (5’-3’) |
|---|---|
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| miR-223-F | GCTGTCAGTTTGTCAAATAC |
| miR-223-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGGGT |
| miR-19b-2*-F | AGTTTTGCAGGTTTGCAT |
| miR-19b-2*-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGAAAT |
| miR-194*-F | CCAGTGGGGCTGCTGTTA |
| miR-194*-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGATA |
| miR-141-F | GCTAACACTGTCTGGTAAAG |
| miR-141-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCATCT |
| miR-1233-F | TGAGCCCTGTCCTCCC |
| miR-1233-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGCGG |
| Universal primer R | GTGCAGGGTCCGAGGT |
Target gene prediction of miRNAs
Targetscans (http://www.targetscan.org/), Pictar (http://pictar.mdc-berlin.de/) and Miranda (http://www.microrna.org/microrna/home.do) were used to analyze miRNA target genes.
Statistical analysis
GenesPring Gx10.0 Software was used to conduct bioinformatic analysis of the data extracted by Feature Extraction Software. After introducing data, percentile shift was used for normalization and logarithmic transformation. Unexpressed probes were filtered out by parameter Flag Value; t-test was used to analysis differentially expressed miRNAs in tumor samples and adjacent normal tissues of cancer; fold change test was used to screen miRNAs whose expression differentiation was more than 2 times. Data analysis was conducted using SPSS 13.0 software, t-test. P < 0.05 was considered statistically significant.
Results
Total RNA concentration and quality
OD260/OD280 ratio ranged among 1.8~2.0 which indicated a high purity of total RNA and it was free from DNA and protein contamination. The agarose gel electrophoresis showed that the density of rRNA 28S band was about 2-fold of 18S band (Figure 1), which demonstrated a good integrity of the extracted total RNA. Quality testing showed that the total RNA samples met the quality requirements.
Figure 1.
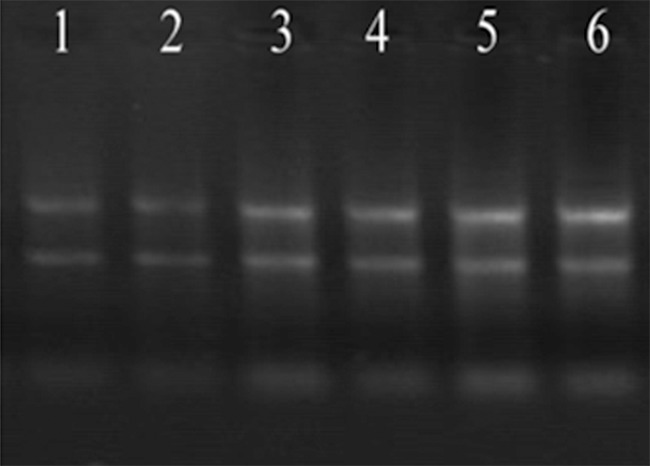
Agarose gel electrophoresis of denatured RNA. 1-3: Control groups 4-6: Gastric cancer.
Expression differences of miRNAs in plasma between gastric cancer patients and healthy human
The results of miRNA microarray showed that 24 differentially expressed of miRNAs were found compared to control groups, among which 11 miRNAs were up-regulated and 13 down-regulated (fold change ≥ 2, p-value < 0.05). The differentially expressed miRNAs are shown in Figure 2, with fold change value and adjusted P-value. These results indicate that these miRNAs may be involved in the development and progress of GC, and can be used as early diagnostic molecule for GC.
Figure 2.
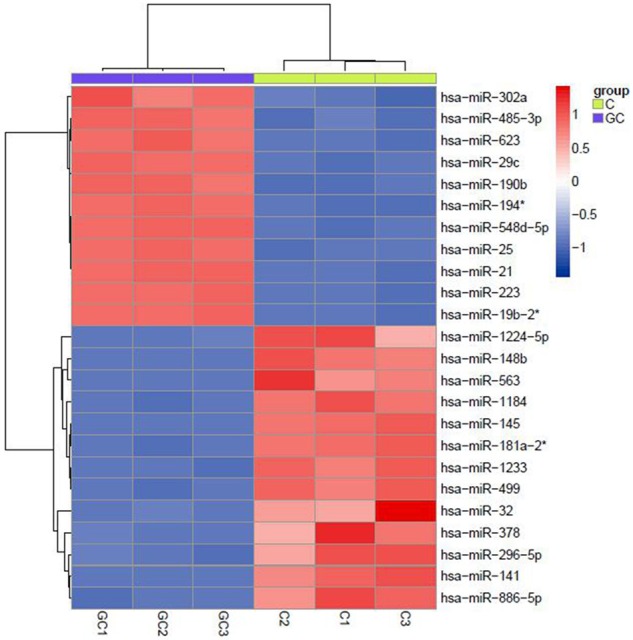
miRNA microarray scanning of plasma samples from gastric cancer patients and normal adults.
Fluorescent quantitative RT-PCR validation
Because of the indirect association between miRNA microarrays and miRNA expression characteristics, disadvantages like specificity and low sensitivity still exist, which may lead to a false positive result. A further fluorescent quantitative RT-PCR experiment was conducted for more accurate results.
Five miRNAs, either over 6 times up-regulated ones (hsa-miR-223, hsa-miR-19b-2* and hsa-miR-194*) or more than 3 times down-regulated ones (hsa-miR-141 and hsa-miR-1233), were the materials of quantitative RT-PCR validation. PCR result showed an up-regulated expression of miR-223, miR-19b-2*, miR-194* in gastric cancer patients’ plasma, and a down-regulated expression of miR-141 and miR-1233 (Figure 3), which demonstrated a consistent result with miRNA microarrays.
Figure 3.
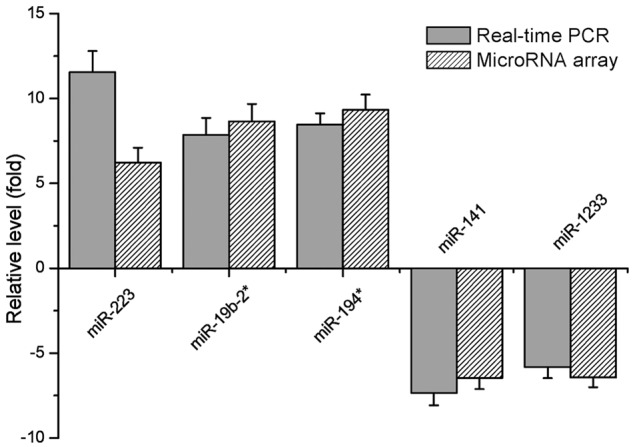
Relative level of differentially expressed miRNAs based on the results of real-time PCR and microRNA array.
Target gene prediction of differentially expressed miRNAs
For further understand the function of the five miRNAs mentioned above, their target genes were predicted by bioinformatics. As shown in Table 2, each miRNA at least match to 5 target genes. Functional analysis shown that these genes are involved in many important life processes such as metastasis.
Table 2.
Target genes predicted with Targetscan, Pictar and Miranda softwares
| miRNA | Fold Change | Target Genes |
|---|---|---|
| hsa-miR-223 | 6.22568749 | ARL6IP2, C13ORF18, FBXO8, HHEX, INPP5B, NFIA, PAPD5, PDE4D, RASA1, RNF34, SCN3A, SP3 |
| hsa-miR-19b-2* | 8.65487785 | DNAJA2, NME7, PCDH10, RHEBL1, SCN4B, ZDHHC7 |
| hsa-miR-194* | 9.33622365 | ARHGAP21, CHD4, C11ORF9, DDEF1, DEPDC1B, HOOK3, HS3ST2, LPHN2, MGAT4A, MEIS2, MID1IP1, NUDC, PDHB, PHF1, RSBN1L, SNX1, YTHDF1 |
| hsa-miR-141 | 0.15487933 | ARPC5, CHD9, CXCL12, EPHA7, IPO5, KLF12, LMO3, MYH10, PRKACB, PLAG1, RANBP6, ZEB2, ZFR |
| hsa-miR-1233 | 0.15558856 | DLGAP4, FMNL2, LRRC57, PDRG1, STIM1 |
Discussion
Presently, a large number of studies discovered that more and more miRNAs are involved in tumorigenesis of gastric cancer and playing crucial roles in its evolution, invasion, metastasis and angiogenesis, etc [9-11]. With deeper and deeper understanding the regulating effects of miRNAs on genetic expression and miRNA differential expression, the potential value of miRNAs in tumor diagnosis, therapy and prognosis evaluation was gradually realized [12,13].
In our present research, differential expression profile of miRNAs in plasma of gastric cancer patients was obtained, which provides useful data for further studying the regulating effects of miRNAs on molecular mechanism of the tumorigenesis and progress of gastric cancer. We found 11 miRNAs were found up-regulated in the plasma of gastric cancer patients, 13 were down-regulated, in which, the expressions of 5 miRNAs were significantly up-regulated (hsa-miR-223, hsa-miR-19b-2* and hsa-miR-194*) or down-regulated (hsa-miR-141 and hsa-miR-1233).
The plasma level of hsa-miR-223 was significantly higher in gastric cancer patients than in healthy controls, promoting gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3 [14], and plasma hsa-miR-223 is a novel potential biomarker for gastric cancer detection as well [15]. Hsa-miR-19b-2* were not detected in osteosarcoma [16] and T cell leukemia [17], but it was found significantly raised in this study, suggesting that hsa-miR-19b-2* might specifically or more highly expressed in gastric cancer. Contrary to our results, it was found by other researchers that hsa-miR-194* and hsa-miR-1233 were totally altered in an opposite way in gastric cancers or other ones: hsa-miR-194* was significantly down-regulated in intestinal-type gastric cancers [18], and hsa-miR-1233 level is distinctly increased in renal cell carcinoma [19]. Consistent with our results, hsa-miR-141 was significantly down-regulated in gastric cancer tissues and serum of ovarian cancer patients [20,21]. These facts demonstrated that the above significantly altered miRNAs in serum may serve as potential biomarkers or prognosis for gastric cancer [15,19,21-23].
Based on the analysis of online prediction, differentially expressed miRNAs were shown to take parts in regulating 74 predicted target genes, and they probably function in many physiological processes. With further analysis, we found there are no genes that are regulated by two or miRNAs in these target genes. When the target genes were analyzed, we noticed that parts of these genes play roles in the differentiation, proliferation, adhesion, apoptosis, and angiogenesis of tumor cells, indicating that these miRNAs are close related to tumor. For instance, KLF12 (target gene of hsa-miR-141) induces cell proliferation, angiogenesis and invasion in gastric cancer cell [24]. The depletion of PDRG1 (target gene of hsa-miR-1233) in colon cancer cells induces a decrease in cell proliferation [25]. RASA1 and PCDH10 (target genes of hsa-miR-223 and hsa-miR-19b-2* respectively) are also tumor suppressor genes [26,27].
Although some of the miRNAs in gastric carcinoma, including hsa-miR-223 [15] and hsa-miR-141 [20], or in other types of carcinomas, have already been reported [28], some significantly altered miRNAs, such as hsa-miR-19b-2*, hsa-miR-194* and hsa-miR-1233, were newly discovered in gastric carcinoma in this study, which may serve as new biomarkers for gastric carcinoma diagnosis in the future. Besides, these abnormally expressed miRNAs might trigger or improve the tumorigenesis and progression of gastric carcinoma [12]. Further studies are still required to validate whether the microRNAs we selected can potentially function as either biomarkers or therapeutic targets and investigate their clinical significance and role in the development of gastric cancer.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NO. 81272743 and No. 81302094), Shanghai Committee of Science and Technology (NO. 13XD1402500 and NO. 13411950902) and Doctoral Innovation Fund Projects from Shanghai Jiao Tong University School of Medicine (BXJ 201219). The author gratefully acknowledges the support of Young Teachers Abroad Visiting Scholar Fellowship Program of Shanghai Education Commission and Shanghai Jiao Tong University K. C. Wong Medical Fellowship Fund.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Espinosa-Parrilla Y, Munoz X, Bonet C, Garcia N, Vencesla A, Yiannakouris N, Naccarati A, Sieri S, Panico S, Huerta JM, Barricarte A, Menendez V, Sanchez-Cantalejo E, Dorronsoro M, Brennan P, Duarte-Salles T, B As Bueno-de-Mesquita H, Weiderpass E, Lund E, Clavel-Chapelon F, Boutron-Ruault MC, Racine A, Numans ME, Tumino R, Canzian F, Campa D, Sund M, Johansson M, Ohlsson B, Lindkvist B, Overvad K, Tjonneland A, Palli D, Travis RC, Khaw KT, Wareham N, Boeing H, Nesi G, Riboli E, Gonzalez CA, Sala N. Genetic association of gastric cancer with miRNA clusters including the cancer-related genes MIR29, MIR25, MIR93 and MIR106: Results from the EPIC-EURGAST study. Int J Cancer. 2014;135:2065–2076. doi: 10.1002/ijc.28850. [DOI] [PubMed] [Google Scholar]
- 3.Leidinger P, Keller A, Meese E. MicroRNAs-Important Molecules in Lung Cancer Research. Front Genet. 2011;2:104. doi: 10.3389/fgene.2011.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Friedlander MR, Lizano E, Houben AJ, Bezdan D, Banez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B, Gonzalez J, Chen KC, LeProust EM, Marti E, Estivill X. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15:R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 7.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 11.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu D, Wang N, Liao X, Huang X, Zhang J, Wang Z, editors. Promising Biomarkers: MicroRNAs at Diagnosis, Therapy and Prognostic Evaluation of Breast Cancer; Proceedings of the 2012 International Conference on Applied Biotechnology (ICAB 2012); Springer; 2014. [Google Scholar]
- 13.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNA Cancer Regulation. Springer; 2013. MicroRNAs in human cancer; pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, Han S, Nie Y, Chen X, Zhao Q, Ding J, Wu K, Daiming F. MiRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 15.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, Xiao B. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namlos HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer ML, Serra M, Burger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: Might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Luo F, Li Q, Xu M, Feng D, Zhang G, Wu W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep. 2011;26:1431–1439. doi: 10.3892/or.2011.1437. [DOI] [PubMed] [Google Scholar]
- 19.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brunagel G, von Ruecker A, Muller SC, Ellinger J. MicroRNAs in renal cell carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T, Si J. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol. 2009;44:556–561. doi: 10.1007/s00535-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, Imoto I, Katai H, Yamaguchi T, Inazawa J, Hirohashi S, Ishikawa Y, Shibata T. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–1867. doi: 10.1002/ijc.24538. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L, Luo X, Shi J, Sun H, Sun Q, Sheikh MS, Huang Y. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol Ther. 2011;11:567–573. doi: 10.4161/cbt.11.6.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J Biol Chem. 2013;288:9508–9518. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Chim JC, Yang M, Ye J, Wong BC, Qiao L. Role of PCDH10 and its hypermethylation in human gastric cancer. Biochim Biophys Acta. 2012;1823:298–305. doi: 10.1016/j.bbamcr.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


