Abstract
Objective: This study aims to identify the target gene of hsa-miR-195 and to research the molecular mechanism of hsa-miR-195 which is through its target genes in the colorectal cancer invasion and metastasis. Methods: We used biological informatics (RNAhybrid and Target Scan analysis database) to predict the target genes of hsa-miR-195. Collected colon cancer tissues from clinical colorectal cancer patients by surgical removal of the carcinoma and control tissues, and researched the expression of Bcl-2 in tissues by immunohistochemical. Next, Real-time PCR was used to research the expression of hsa-miR-195 in Caco-2 and NCM460 cell line. hsa -miR-195 Mimics was transient transfered to Caco-2 cells, western blot was used to analysis the expression changes of Bcl-2. To analysis the possibility that hsa-miR-195 can affect the invasive ability of tumor cells by Bcl-2, we transferred hsa-miR-195 Mimics and Bcl-2 expression plasmid, and used the cell invasion experiment to discusses hsa-miR-195 effect on the ability of tumor cell invasion. Results: the immunohistochemical results showed that, the semi-quantitative parameters for the Bcl-2: control by 0.89 ± 0.51, 6 colon cancer by 31 ± 0.79. The expression of has-miR-195 in Caco-2 is 0.39 ± 1.5 while the value in control is2.01 ± 0.2, **P < 0.01. Conclusion: In colorectal cancer, has-miR-195 can promote cell apoptosis and inhibit the invasion and metastasis by inhibiting the expression of Bcl-2.
Keywords: MicroRNA-195, colorectal cancer, Bcl-2, Caco-2, NCM460
Introduction
MicroRNAs (miRNAs) is a kind of endogenous small RNA with length of about 17-25 nucleotides. miRNAs degraded mRNA or blocked its translation mainly by pairing with mRNA base of its target genes, it had a variety of roles in cells. Each miRNA can have multiple target genes and several miRNAs can work together to regulate the same gene. Recent studies showed that miRNAs played important regulating roles in the occurrence and development of tumor [1-3].
Colorectal cancer is a common malignant tumor of digestive system and its morbidity and mortality rate were only lower than that of primary liver cancer, gastric cancer and esophageal cancer in the digestive system malignant tumor. The occurrence of colorectal cancer is a complex process, which including the activation of oncogene and inactivation of tumor suppressor genes caused by genetic and environmental changes. miRNA exerted its role through regulating the activity of oncogene and tumor suppressor genes. Studies showed the abnormal expressions of MicroRNA-195 in many tumors. It was up-regulated in chronic lymphocytic leukemia and breast cancer while down-regulated in hepatocellular carcinoma, gastric cancer and bladder cancer [4-6]. These results suggested that MicroRNA-195 played different roles in different tumors.
It is not clear for the roles of MicroRNA-195 in colon cancer. Therefore, we explored the molecular mechanism of invasion and metastasis of microRNA-195 in colorectal carcinoma.
Materials and methods
General data
A total of 12 resected specimens of patients with colorectal cancer in China-Japan Union hospital of Jilin University were collected in this study. These patients were operated and hospital treatment during 2010 ~ 2013. At the same time, a total of 9 corresponding normal operational specimens which are the adjacent tissue of the cancer were collected as controls. All specimens are tested by two pathologists according to WHO diagnostic criteria for the histopathologic diagnosis. There were 7 males and 5 females patients. Ages ranged from 50 to 68 years (mean of 61.3 years). All subjects signed an informed consent form. This study was approved by the Ethics Committee of the China-Japan Union Hospital of Jilin University.
Cell culture
Caco-2 and NCM460 cells were obtained from ATCC (American Type Culture Collection). The cells were grown in DMEM supplemented with 15% fetal bovine serum. They were cultured at 37°C with 5% CO2.
Immunohistochemical staining
The Clinical samples were cut into small pieces (< 5 mm) and fixed with 10% neutral formalin for 24 h, dewatered with gradient alcohol and embedded tissue with automatic paraffin embedding instrument. Then they were cut into 3 ~ 5 µm slices. The tissue sections were de-waxed, hydrated and incubated them in H2O2 for 20 min in order to block the endogenous peroxidase. Microwave repaired the antigen for 5 min in citrate buffer solution and repeated it. Washed them with distilled water and PBS for 5 min respectively and repeated it. Drop-adding the 1st antibody dilution (1:100) and incubated at 37°C in water bath for 2 h, then washed them with PBS every time for 2 min, a total of 3 times; Drop-adding the 2nd antibody and incubated at 37°C in water bath for 2 h, washed them with PBS every time for 2 min, a total of 3 times. Developed with the DAB solution and observed them under microscope, flushed them completely, hematoxylin counterstained them, washed with water, dehydration, transparency, mount and observed under microscope. The data were analyzed with Image-ProPlus software. Integrated option density (IOD) of interesting area (AOI) was measured and density mean (IOD/AOI) were calculated as the semi quantitative parameters of Bcl-2 gene expression levels.
RNA extraction and RT-PCR
Total RNA of cells was extracted using RNeasy Mini Kit according to the manufacturer’s protocol. 1 μg total RNA was subjected to reverse transcription using reverse transcription kit. Real-time PCR were performed using TaqMan probe method. TaqMan probe Sequence of has-miR-195: CCAAUAUUGGCUGUGCUGCUCC. U6 was used as an internal control for normalization of RNA quantity and quality differences in all samples. Quantifications of target genes mRNA was performed using the 2−ΔΔCt method [7]. ΔCT = Ct(has-miR-195) - Ct(U6); Δ(ΔCT) = ΔCT - ΔCT (contro1).
Construction and transfection of recombinant plasmid
The Bcl-2 gene was ligated with eukaryotic expression plasmid pcDNA3.1 to construct the recombinant plasmid. The recombinant plasmid was transfected in cells using lipofectamine 2000 kit according to the manual. The transfection of hsa-miR-195 Mimics and Negative scramble control (NC) RNA were performed using Lipofectamin RNAi MAX according to the manual.
Transwell in vitro invasion assays
The Matrigel was diluted with pre cooling serum-free DMEM to the concentration of 1 mg/ml and 100 μl of it was added in the bottom center of upper chamber of 8 micron TrHanswells and incubated at 37°C and 200 μl DMEM was added in each well. The cells were placed in the upper chamber and DMEM with 10% FBS was added in the lower chambers. They were incubated at 37°C in 5% CO2. The upper chamber was removed after incubation and cleansed the filter side of the upper chamber with a cotton swab. The filter was fixed with 4% formaldehyde for 10 min and stained with 0.1% crystal violet. Gently cut the filter from the chamber and count the cells that have migrated through the filter pores from the underside of the filter in 4 high-power fields per insert, and average values afterwards. For each migration condition, three identical replicates should be performed.
Western blotting
The cells were digested with trypsin and total proteins were extracted and their concentration was determined using Protein Assay Kit. The proteins were separated by SDS polyacrylamide gel electrophoresis. The separated proteins were electrotransferred to the PVDF membrane. The membrane containing the proteins was used for immunoblotting with required antibodies. The protein bands were scanned and quantified as a ratio to GAPDH control using Quantity one 4.62 software.
Statistical analyses
The data were analyzed using SPSS 11.5 soft ware. The results are expressed as mean ± SD. One-way ANOVA was used to evaluate the differences among multi-groups and t-test was used to evaluate the differences between two groups. A value of P < 0.05 and P < 0.01 was taken to denote statistical significance.
Results
The predicted target genes of hsa-miR-195
The interaction between miRNAs and its target genes has certain regularity. It can be predicted by programming. We analyzed and predicted the target genes of hsa-miR-195 by searching RNAhybrid and TargetScan database, the results were shown in Table 1. The results suggested that the sequence of hsa-miR-195 5’-UAGCAGCA-3’ can combine with the mRNA sequence 3’UTR 5’-UGCUGCUA-3’ of Bcl-2 gene.
Table 1.
The predicted target genes of hsa-miR-195
| microRNA family | Target gene | Estimated false discovery rate | Target gene name | Target gene description |
|---|---|---|---|---|
| miR-15/16/195 | FGF2 | 0.00096 | fibroblast growth factor 2 | FGF family |
| CCND2 | 0.0017 | cyclin D2 | Conservative cell cycle protein family | |
| CCND1 | 0.0018 | cyclin D1 | Conservative cell cycle protein family | |
| CCNE1 | 0.0038 | cyclin E1 isoform 1 | Conservative cell cycle protein family | |
| DCAMKL1 | 0.0039 | doublecortin and CaM kinase-like 1 | NA | |
| ARL2 | 0.0042 | ADP-ribosylation factor-like 2 | GTP-binding RAS super family | |
| PAPPA | 0.006 | pregnancy-associated plasma protein A | Metalloproteinase secretion of insulin like growth factor binding protein | |
| KCNJ2 | 0.0074 | potassium inwardly-rectifying channel J2 | Potassium ion channel protein | |
| SLC12A2 | 0.011 | solute carrier family 12 | NA | |
| LUZP1 | 0.02 | leucine zipper protein 1 | leucine zipper | |
| PAFAH1B1 | 0.021 | platelet-activating factor acetylhydrolase, | Miller-Dieker lissencephaly syndrome associated protein | |
| CPD | 0.023 | carboxypeptidase D precursor | NA | |
| WEE1 | 0.028 | wee1 tyrosine kinase | Tyrosine kinase | |
| KIF1B | 0.028 | kinesin family member 1B isoform alpha | NA | |
| DEDD | 0.091 | death effector domain-containing protein | DED containing protein. | |
| BCL2 | 0.091 | B-cell lymphoma protein 2 beta isoform | The mitochondrial outer membrane protein, prevent apoptosis | |
| NEBL | 0.091 | nebulette non-muscle isoform | NA | |
| PRDM4 | 0.091 | PR domain containing 4 | PR-domain protein family | |
| RECK | 0.092 | RECK protein precursor | Cysteine rich protein |
Immunohistochemical staining results
The expression levels of Bcl-2 in cancer tissues were detected by immunohistochemical staining method and the positive cells were stained brown (Figure 1; Table 2). The density mean (IOD/AOI) were calculated using Image-ProPlus (IPP) software. The density mean of Bcl-2 in adjacent cancer normal tissues was 1.22 ± 0.63 and that of cancer tissues was 5.45 ± 1.49.
Figure 1.
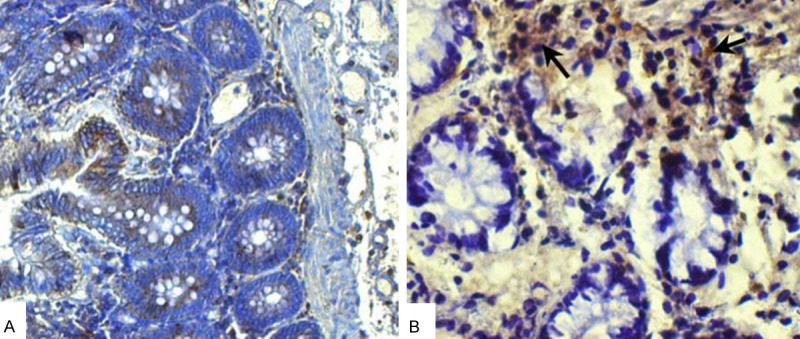
Immunohistochemical staining results. A: Control; B: Cancer tissues.
Table 2.
The expression of Bcl-2 in colorectal cancer tissues
| Slicea | Normal specimens | Resected colorectal specimens | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Random fieldb | ||||||
|
| ||||||
| 1 | 2 | 3 | 1 | 2 | 3 | |
| 1 | 1.70 | 0.50 | 0.46 | 5.70 | 6.13 | 7.09 |
| 2 | 0.72 | 0.69 | 1.14 | 3.90 | 3.02 | 5.43 |
| 3 | 2.50 | 1.82 | 0.70 | 4.92 | 4.83 | 6.20 |
| 4 | 0.52 | 1.05 | 0.99 | 8.57 | 7.72 | 5.43 |
| 5 | 1.57 | 2.03 | 1.88 | 3.57 | 5.29 | 3.99 |
Selected 5 slices;
Selected 3 random field of each slice.
RT-PCR results
As shown in Figure 2, the normalized expression of has-miR-195 in Caco-2 cells was 0.39 ± 1.5 while that of NCM460 cells was 2.01 ± 0.2. The differences between them were significant (P < 0.01).
Figure 2.
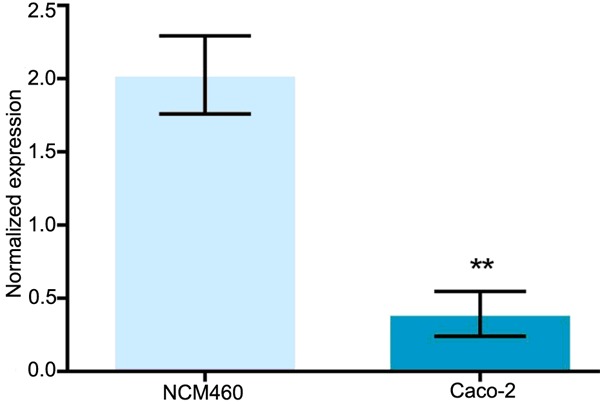
Real-time PCR results of hsa-miR-195 expression. **P < 0.01.
Western blotting results
The expression levels of Bcl-2 and hsa-miR-195 in cells transfected with hsa-miR-195 Mimics were detected by Western blotting method. As shown in Figure 3A, the expression levels of hsa-miR-195 increased after transfection (P < 0.01), while as shown in Figure 3B and 3C, the expression levels of Bcl-2 decreased after transfection (P < 0.01).
Figure 3.
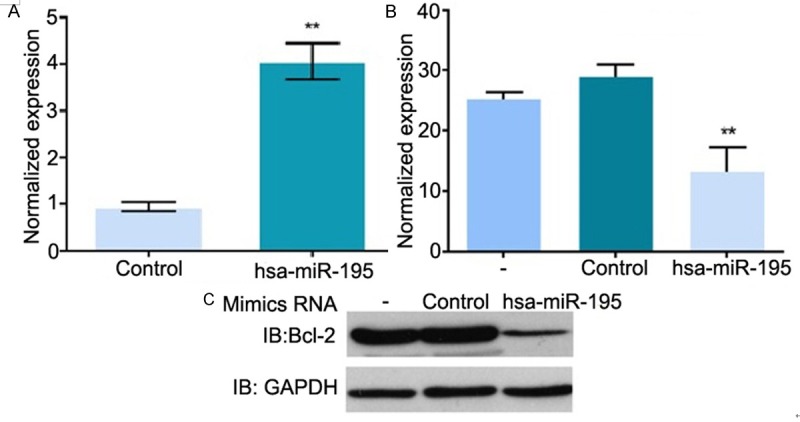
Western blotting results. A: Normalized expression of hsa-miR-195; B: Normalized expression of Bcl-2; C: Western blotting picture. Control: Transfected with negative scramble control (NC) RNA, -: Empty plasmid; hsa-miR-195: Transfected with hsa-miR-195 Mimics.
Results of invasion assays
As shown in Figure 4 and Table 3, the invasion ability of Caco-2 cells increased after overexpression of Bcl-2. On the contrary, the invasion ability of Caco-2 cells was inhibited after transfected with hsa-miR-195.
Figure 4.
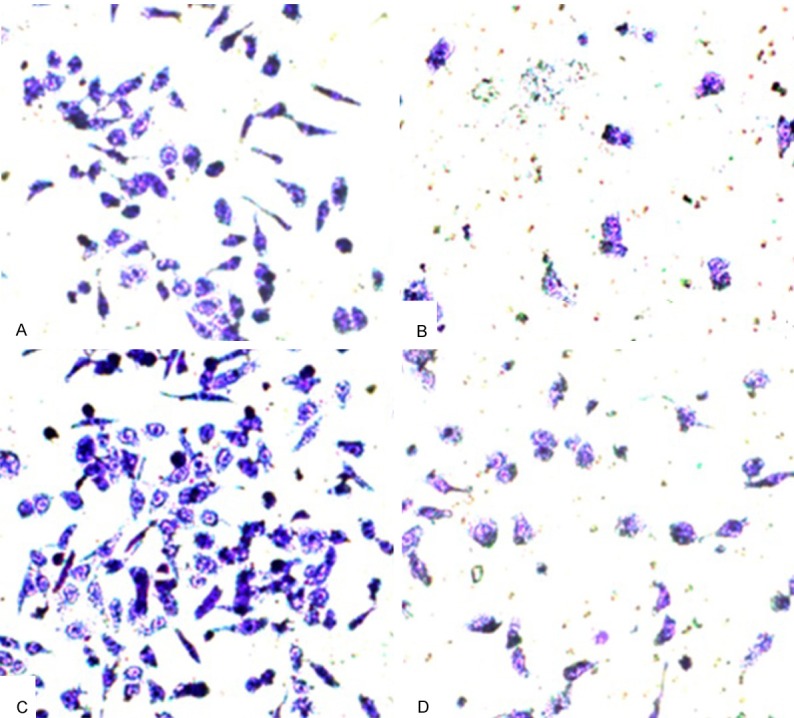
Transwell in vitro invasion assays results. A: hsa-miR-195 Mimics (-) and Bcl-2 recombinant plasmid (-); B: hsa-miR-195 Mimics (+) and Bcl-2 recombinant plasmid (-); C: hsa-miR-195 Mimics (-) and Bcl-2 recombinant plasmid (+); D: hsa-miR-195 Mimics (+) and Bcl-2 recombinant plasmid (+).
Table 3.
The results of transwell in vitro invasion assays (Mean ± SD)
| Hsa-miR-195 Mimics - | Hsa-miR-195 Mimics + | |
|---|---|---|
| Bcl-2 plasmid - | 35 ± 7 | 16 ± 2* |
| Bcl-2 plasmid + | 52 ± 3** | 23 ± 5 |
Compared with control group,
P < 0.01;
P < 0.05.
Discussion
MicroRNAs inhibit or block the translation of mRNA mainly through the completely or partially combination with nucleotide in 3’ end untranslated region of its target gene mRNA to regulate the genes expression. MicroRNAs and its target gene or other MicroRNAs can form a regulatory network to be widely involved in cellular proliferation and apoptosis. The abnormal expression of MicroRNAs plays a similar biological effect as oncogene or tumor suppressor gene. It was found that the abnormal expression of microRNA-15/16 occurred in chronic lymphatic leukemia and the abnormal expression of microRNA -155 was closely related to the incidence of lung cancer [9,10]. These results suggested that microRNAs could be potential target biomarkers for cancer treatment. Recent study showed that the abnormal expression of microRNAs was one important reason for the occurrence of colorectal cancer [11].
MicroRNA-195 was first found to be the gene of rat and it was confirmed to exist in human lately. Has-miR-195 is located on human chromosome 17p13.1 and expressed abnormally in many cancers [12,13]. We investigated the target gene of has-miR-195 by using prediction methods of bioinformatics or literature [3]. In many predicting results we found that the sequence 5’-UAGCAGCA-3’ of miR-195 could combine with the sequence 3’UTR 5’-UGCUGCUA-3’ of Bcl-2 gene mRNA thus inhibiting the transcription and translation of Bcl-2 gene. Bcl-2 is an important proto-oncogene inhibiting apoptosis and its abnormal expression has close relationship with the occurrence and development of many diseases and is more closely with tumor [14]. Therefore, we took the important target gene Bcl-2 of has-miR-195 as the research object in this study to explore the biological effects of has-miR-195 in the development of colorectal cancer.
In this study we found that the expression of Bcl-2 gene was up-regulated in colorectal cancer tissues (P < 0.01) which confirmed that Bcl-2 protein played an important role in the pathogenesis of colorectal cancer. RT-PCR results showed that the expression levels of hsa-miR-195 was lower in Caco-2 cells than that of NCM460 cells (P < 0.01), which blocked the regulation ability of hsa-miR-195 on Bcl-2 gene. Western blotting results also confirmed that the expression levels of Bcl-2 was lower in Caco-2 cells transfected with hsa-miR-195 Mimics than that of Caco-2 cells transfected with Negative scramble control (NC) RNA (P < 0.01). Results of Transwell invasion assay showed that the transfection of hsa-miR-195 Mimics could reduce the invasive ability of Caco-2 cells. When hsa-miR-195 and Bcl-2 were over expressed simultaneously, the invasive ability of Caco-2 cells was stronger that of only transfected with hsa-miR-195 Mimics. These results suggested that Bcl-2 protein was involved in regulation of tumor invasion and metastasis.
There are many different research perspectives in the molecular mechanism that Bcl-2 protein can promote the proliferation and invasion of tumor cells. Kumar found that Bcl-2 can promote the invasion ability of tumor cells by IL-8 mediation [15]. Other studies showed that Bcl-2 can promote the metastasis of prostate cancer [16], lung cancer [17] and glioma [18]. Hsa-miR-195 reducing the invasion ability of Caco-2 cells was at least partly associated with the down-regulation of Bcl-2 protein [19,20]. The regulatory effect of MicroRNAs is not single [21] and Hsa-miR-195 also played more than one kind of biological effects in the occurrence of colorectal cancer. Hsa-miR-195 was significantly down-regulated in adriamycin resistant cell lines of colon cancer HT29 cells, the cytotoxicity of adriamycin was inhibited significantly after silencing hsa-miR-195, which was closely related with the inhibition of BCL2L2 expression by hsa-miR-195 [3,22-25].
In a word, Hsa-miR-195 could inhibit the proliferation and migration of tumor cells by regulating theH expression of Bcl-2 protein but the biological effects of hsa-miR-195 are not single, it is coordinated process with multiple molecules in multiple levels. So it is very important to further research the molecular mechanisms of hsa-miR-195 for explaining the occurrence of tumor and providing new drug target of anticancer drugs.
Disclosure of conflict of interest
None.
References
- 1.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner A, Mayr C, Bach D, Illig R, Plaetzer K, Berr F, Pichler M, Neureiter D, Kiesslich T. MicroRNAs associated with the efficacy of photodynamic therapy in biliary tract cancer cell lines. Int J Mol Sci. 2014;15:20134–20157. doi: 10.3390/ijms151120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 4.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 5.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 6.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919–927. doi: 10.1007/s12032-011-9880-5. [DOI] [PubMed] [Google Scholar]
- 13.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 14.Zhao XD, He YY, Gao J, Zhao C, Zhang LL, Tian JY, Chen HL. High Expression of Bcl-2 Protein Predicts Favorable Outcome in Non-small Cell Lung Cancer: Evidence from a Systematic Review and Meta-analysis. Asian Pac J Cancer Prev. 2014;15:8861–8869. doi: 10.7314/apjcp.2014.15.20.8861. [DOI] [PubMed] [Google Scholar]
- 15.Kumar P, Ning Y, Polverini PJ. Endothelial cells expressing Bcl-2 promotes tumor metastasis by enhancing tumor angiogenesis, blood vessel leakiness and tumor invasion. Lab Invest. 2008;88:740–749. doi: 10.1038/labinvest.2008.46. [DOI] [PubMed] [Google Scholar]
- 16.Bruckheimer EM, Brisbay S, Johnson DJ, Gingrich JR, Greenberg N, McDonnell TJ. Bcl-2 accelerates multistep prostate carcinogenesis in vivo. Oncogene. 2000;19:5251–5258. doi: 10.1038/sj.onc.1203881. [DOI] [PubMed] [Google Scholar]
- 17.Choi J, Choi K, Benveniste EN, Rho SB, Hong YS, Lee JH, Kim J, Park K. Bcl-2 promotes invasion and lung metastasis by inducing matrix metalloproteinase-2. Cancer Res. 2005;65:5554–5560. doi: 10.1158/0008-5472.CAN-04-4570. [DOI] [PubMed] [Google Scholar]
- 18.Wick W, Wild-Bode C, Frank B, Weller M. BCL-2-induced glioma cell invasiveness depends on furin-like proteases. J Neurochem. 2004;91:1275–1283. doi: 10.1111/j.1471-4159.2004.02806.x. [DOI] [PubMed] [Google Scholar]
- 19.Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W, Jiang X, Zhang C, Qu J. MicroRNA-195 Chemosensitizes Colon Cancer Cells to the Chemotherapeutic Drug Doxorubicin by Targeting the First Binding Site of BCL2L2 mRNA. J Cell Physiol. 2015;230:535–545. doi: 10.1002/jcp.24366. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Li M, Chang S, Wang L, Song T, Gao L, Hu L, Li Z, Liu L, Yao J, Huang C. MicroRNA-195 acts as a tumor suppressor by directly targeting Wnt3a in HepG2 hepatocellular carcinoma cells. Mol Med Rep. 2014;10:2643–2648. doi: 10.3892/mmr.2014.2526. [DOI] [PubMed] [Google Scholar]
- 21.Harraz MM, Xu JC, Guiberson N, Dawson TM, Dawson VL. MiR-223 regulates the differentiation of immature neurons. Mol Cell Ther. 2014;2:18. doi: 10.1186/2052-8426-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 25.Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, Weng Y. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep. 2013;30:877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]


