Abstract
The polymorphisms in Fas/FasL system were proposed to be associated with susceptibility to leukemia, but recent studies reported controversial findings. Hence, we performed a meta-analysis to assess the association between Fas gene polymorphisms and susceptibility to leukemia. We carried out a literature search in PubMed, Embase, Web of Science and CNKI databases for studies on the associations between Fas/FasL gene polymorphisms and susceptibility to leukemia. The associations were assessed by odds ratio (OR) together with its 95% confidence intervals (CIs). 7 literatures and 14 studies with a total of 8787 subjects were eventually included into our meta-analysis. Overall, there was no association between Fas/FasL polymorphisms and susceptibility to leukemia. In subgroup analysis by ethnicity, there was also no association between Fas/FasL polymorphisms and susceptibility to leukemia in Asians and Caucasians. In addition, there was also a significant association between Fas-1377G/A polymorphism and susceptibility to leukemia in ALL patients, the A allele seemed to be a protective factor in ALL risk. In summary, more studies with large sample size are needed to provide further evidence for association between Fas/FasL polymorphisms and susceptibility to leukemia.
Keywords: Leukemia, polymorphism, meta-analysis
Introduction
Leukemia is one of the most common malignant tumors, which make up 30.4% of all hematological malignancies and are one of the most common hematological malignancies according to the information collected by (SEER) Cancer Statistics Review 1975-2006. The change of apoptosis induced by gene mutation could result in leukemia [1,2]. Fas also known as CD95/TNFSF6/APO-1 belongs to the subgroup of the tumor necrosis factor receptor (TNF-R) family and is one of the important molecules contributed to apoptosis pathway. The human Fas gene located on chromosome 10q 24.1 [3], which contains nine exons and eight introns [4]. Fas-induced apoptosis in the control of the immune system and its critical function as a guardian against autoimmune disease and certain lymphoid malignancies [5]. And the Fas could cooperate with Fas ligand (FasL) to trigger programmed cell death [6]. Previous studies have reported two functional single nucleotide polymorphisms (SNPs) in the promoter region of Fas gene [7,8] and one single nucleotide polymorphism in FasL gene. One of these polymorphisms is A to G substitution at position -670 in the enhancer region which changes the activators of transcription 1 (STAT1) transcription factor-binding site of Fas gene. The other polymorphism is G to A base change at position -1377 situated between two putative silencer regions which alters SP-1 transcription factor-binding site [8,9]. In addition, the change of T to C base at position-844 of FasL gene also has been suggested to alter the expression of FasL gene [10]. In current studies, the association between Fas/FasL gene polymorphisms and susceptibility to cancers including lung cancer [11], gastric cancer [12], breast cancer [13], cervical cancer [14], prostate cancer [15] and leukemia [8] has been showed. Currently, there are many case-control studies published to evaluate the association between Fas gene polymorphisms between susceptibility to leukemia, but these studies reported controversial findings [8,10,16,17]. In addition, there were several meta-analyses on association between Fas/FasL polymorphisms and cancer risk recently [18-20]. However, these meta-analyses did not include the studies on leukemia completely and there is no meta-analysis on association between Fas/FasL polymorphisms and leukemia risk. So, we performed a meta-analysis to assess the association between Fas/FasL polymorphisms and susceptibility to leukemia.
Methods
Search strategy
We carried out a literature search in PubMed, Embase, Web of Science and CNKI databases for studies on the associations between Fas/FasL polymorphisms and susceptibility to leukemia. The search strategy was based on the combination of following key words (“Fas”, “FasL”, “CD95”, “TNFSF6”, “APO-1”, “rs2234767”, “rs1800682” or “rs763110”) and (“polymorphism(s)”, “variants”, “genotype”) and (“leukemia” or “leukaemia” or “leucocythaemia”). There was no language limitation. The last search was updated on April 2014. All searched studies were retrieved, and their references were also checked for other relevant publications. If more than one cancer type was reported in one study, the data for each type was extracted separately. If data or data subsets were published in more than one article, only the publication with the largest sample size was included.
Inclusion criteria
The following criteria were used for the inclusion of eligible articles for our meta-analysis: (1) Studies that assessed the association between the Fas/FasL polymorphisms and risk of leukemia; (2) Studies with a case-control study design; (3) Studies with detailed genotype frequencies for cases and controls or studies provided sufficient data to calculate genotype frequencies; (4) Genetic testing method is reasonable. The exclusion criteria were as follows: (1) Studies without control population; (2) Studies without available genotype frequency for the Fas or FasL polymorphisms; (3) Studies that contained overlapping data; (4) Studies not in Hardy-Weinberg equilibrium (HWE).
Data extraction
Two investigators independently extracted data using a standardized extract form. Disagreement was solved by discussion between the two investigators. If these two investigators could not reach a consensus, another investigator was consulted to resolve the dispute. The following information was extracted from each publication: the first author, year of publication, country of origin, ethnicity of participants and genotyping methods, total number of cases and controls, and source of controls, whether or not the genotype distributions among controls were in accordance with Hardy-Weinberg equilibrium (HWE). Ethnicity of participants was categorized as Caucasians, Asians and South Latinas. The leukemia types were further categorized as acute lymphocytic leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myelocytic leukemia (AML), chronic myeloid leukemia (CML) and other leukemia types.
Statistical analysis
The pooled OR and its 95% CI were used to assess the strength of the associations. The significance of the pooled OR was determined by Z test, and a P value of less than 0.05 was considered significant. We examined the associations between Fas polymorphisms and susceptibility to leukemia under four different models including the allele model, the dominant model, the recessive model and the homozygous model. The heterogeneity between the studies was assessed by the χ2 test-based Q statistic method [21] and I2 statistic which provides values between 0 and 100% with a greater degree of heterogeneity (I2 = 0-25%: no heterogeneity; I2 = 25-50%: moderate heterogeneity; I2 = 50-75%: large heterogeneity; I2 = 75-100%: extreme heterogeneity) [22]. A P value of < 0.10 and I2 > 50% indicated evidence of significant heterogeneity. The combined OR was calculated by the fixed-effects model (Mantel-Haenszel) [23] in the absence of heterogeneity; otherwise, the random-effects model (the DerSimonian and Laird method) [24] was used to calculate the pooled OR. The departure of the SNP from expected frequencies under HWE was assessed in controls using the Pearson χ2 test (P < 0.05 was considered significant). Subgroup analysis by ethnicity was further performed. Sensitivity analysis was performed to assess influence of each study on our pooled results. Publication bias was observed with the funnel plot using the standard error of logOR (An asymmetric plot and suggests a possible publication bias) and Egger’s test (P < 0.05 was considered representative of statistically significant publication bias) [25]. All the statistical tests were performed with Stata 12.0.
Results
Study characteristics
According to the inclusion criteria defined for the studies available for this meta-analysis, 7 publications with a total of 14 studies were finally included into the meta-analysis [8,10,16,17,26-28]. There were 6 studies with a total of 1870 cases and 2788 controls on the association between Fas-670A/G polymorphism and susceptibility to leukemia [8,10,16,17,26,27]. And there were 6 studies with a total of 2338 cases and 3563 controls on the association between Fas-1377G/A polymorphism and susceptibility to leukemia [8,10,16,17,27,28]. In addition, there were 2 studies with a total of 953 cases and 1377 controls on association between FasL-844T/C polymorphism and susceptibility to leukemia. The alleles within control groups of all studies are in Hardy-Weinberg equilibrium, one study only provides the frequency of alleles [28]. The summary characteristics of studies are listed in Table 1.
Table 1.
Characteristics of studies on the association between Fas/FasL polymorphisms and leukemia risk
| Study | Ethnicity | Countries | Type of cases | Type of controls | Sample size Case/Control | Genotype frequencies Case/Control | Allele frequencies Case/Control | HWE | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fas-670A/G (rs1800682) | AA | AG | GG | A | G | ||||||
| Valibeigi, 2013 | Caucasian | Iran | ALL | PB | 142/117 | 44/47 | 77/57 | 21/13 | 165/151 | 119/83 | 0.487 |
| Prajitha, 2013 | Asian | India | CML | PB | 290/300 | 90/83 | 147/155 | 53/62 | 327/321 | 253/279 | 0.506 |
| Tong, 2012 | Asian | China | ALL | PB | 361/519 | 157/198 | 159/255 | 45/66 | 473/651 | 249/387 | 0.249 |
| Kim, 2010 | Asian | Korea | AML | PB | 592/858 | 168/251 | 307/421 | 117/186 | 643/923 | 541/793 | 0.704 |
| Farre, 2008 | Latinas | Brazil | ATL | HB | 31/60 | 12/10 | 8/31 | 11/29 | 32/51 | 30/89 | 0.714 |
| Sibley, 2003 | Caucasian | UK | AML | PB | 454/934 | 129/280 | 228/449 | 97/205 | 486/1009 | 422/859 | 0.324 |
| Fas-1377G/A (rs2234767) | GG | GA | AA | G | A | ||||||
| Valibeigi, 2013 | Caucasian | Iran | ALL | PB | 142/117 | 117/94 | 21/17 | 4/6 | 255/205 | 29/29 | 0.487 |
| Prajitha, 2013 | Asian | India | CML | PB | 290/300 | 176/190 | 106/100 | 8/10 | 458/480 | 122/120 | 0.506 |
| Tong, 2012 | Asian | China | ALL | PB | 361/519 | 177/212 | 139/225 | 45/82 | 493/649 | 229/389 | 0.249 |
| Kim, 2010 | Asian | Korea | AML | PB | 592/858 | 195/286 | 303/427 | 94/145 | 693/999 | 491/717 | 0.704 |
| Rollinson, 2004 | Caucasian | England | AML | PB | 482/838 | NA | NA | NA | 752/1525 | 212/151 | NA |
| Sibley, 2003 | Caucasian | UK | AML | PB | 471/931 | 319/726 | 136/186 | 16/19 | 774/1638 | 168/224 | 0.324 |
| FasL-844T/C (rs763110) | TT | TC | CC | T | C | ||||||
| Tong, 2012 | Asian | China | ALL | PB | 361/519 | 192/132 | 107/276 | 62/111 | 491/540 | 231/498 | 0.249 |
| Kim, 2010 | Asian | Korea | AML | PB | 592/858 | 52/75 | 236/321 | 302/462 | 340/471 | 840/1245 | 0.704 |
PB, population based; NA, not available; HWE, Hardy-Weinberg Equilibrium.
The summary results of the meta-analysis on the association between Fas/FasL polymorphisms and susceptibility to leukemia are shown in Table 2. Overall, there was no association between Fas-670A/G polymorphism and susceptibility to leukemia (G vs A: OR = 0.94, 95% CI = 0.80-1.09; GG vs AA: OR = 0.93, 95% CI = 0.78-1.11; GG/AG vs AA: OR = 1.01, 95% CI = 0.89-1.14; GG vs AG/AA: OR = 0.96, 95% CI = 0.89-1.05). In addition, there was also no association between Fas-1377G/A or FasL-844T/C polymorphisms and susceptibility to leukemia (AA vs GA/GG: OR = 0.91, 95% CI = 0.74-1.12; AA vs GG: OR = 0.89, 95% CI = 0.71-1.12; AA/GA vs GG: OR = 1.06, 95% CI = 0.77-1.45; A vs G: OR = 1.21, 95% CI = 0.82-1.81; CC vs TC/TT: OR = 0.86, 95% CI = 0.72-1.03; CC vs TT: OR = 0.60, 95% CI = 0.25-1.45; CC/TC vs TT: OR = 0.54, 95% CI = 0.17-1.75; C vs T: OR = 0.69, 95% CI = 0.38, 1.25). Subgroup analysis by ethnicity suggested that there was no association between Fas/FasL polymorphisms and leukemia risk under all four genetic models in Asians or Caucasians (Table 2). In the subgroup analysis by leukemia type, we observed that Fas-1377G/A polymorphism was associated with leukemia risk in ALL patients under all models except the recessive model (Table 2).
Table 2.
Meta-analysis of the association between Fas/FasL polymorphisms and susceptibility to leukemia
| Study groups | OR (95% CI), I2 (%) | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Recessive model | Homozygous model | Dominant model | Allele model | |||||
| Fas-670A/G | GG vs AG/AA | GG vs AA | GG/AG vs AA | G vs A | ||||
| Overall (6) | 0.94 (0.80, 1.09) | 0.0 | 0.93 (0.78, 1.11) | 28.7 | 1.01 (0.89, 1.14) | 47.4 | 0.96 (0.89, 1.05) | 38.6 |
| Caucasian (2) | 1.01 (0.78, 1.30) | 0.0 | 1.10 (0.82, 1.48) | 27.5 | 1.15 (0.92, 1.43) | 20.9 | 1.06 (0.92, 1.23) | 37.4 |
| Asian (3) | 0.90 (0.74, 1.09) | 0.0 | 0.88 (0.71, 1.10) | 0.0 | 0.92 (0.78, 1.07) | 13.4 | 0.93 (0.84, 1.04) | 0.0 |
| AML (2) | 0.93 (0.77, 1.12) | 0.0 | 0.98 (0.79, 1.22) | 0.0 | 1.06 (0.90, 1.25) | 0.0 | 1.00 (0.90, 1.11) | 0.0 |
| ALL (3) | 1.06 (0.75, 1.51) | 0.0 | 1.12 (0.58, 2.17) | 55.2 | 1.05 (0.57, 1.93) | 77.4 | 1.05 (0.72, 1.53) | 71.9 |
| Fas-1377G/A | AA vs GA/GG | AA vs GG | AA/GA vs GG | A vs G | ||||
| Overall (6) | 0.91 (0.74, 1.12) | 13.8 | 0.89 (0.71, 1.12) | 48.2 | 1.06 (0.77, 1.45) | 81.8 | 1.21 (0.82, 1.81) | 94.4 |
| Caucasian (3) | 0.99 (0.76, 1.27) | 42.4 | 1.10 (0.62, 1.96) | 55.0 | 1.20 (0.79, 1.80) | 80.3 | 1.41 (0.82, 2.42) | 95.3% |
| Asian (3) | 0.78 (0.54, 1.11) | 0.0 | 0.69 (0.47, 1.00) | 0.0 | 0.89 (0.57, 1.37) | 75.7 | 0.89 (0.66, 1.22) | 69.1 |
| AML (3) | 1.15 (0.66, 2.03) | 61.0 | 1.26 (0.64, 2.48) | 70.3 | 1.31 (0.80, 2.14) | 88.7 | 1.64 (0.88, 3.04) | 96.7 |
| ALL (2) | 0.75 (0.51, 1.09) | 0.0 | 0.64 (0.43, 0.96) | 0.0 | 0.74 (0.58, 0.95) | 0.0 | 0.78 (0.64, 0.94) | 0.0 |
| FasL-844T/C | CC vs TC/TT | CC vs TT | CC/TC vs TT | C vs T | ||||
| Overall (2) | 0.86 (0.72, 1.03) | 0.0 | 0.60 (0.25, 1.45) | 90.6 | 0.54 (0.17, 1.75) | 96.0 | 0.69 (0.38, 1.25) | 95.3 |
The results that are in bold type show statistical significance. OR, odds ratio; CI, confidence interval.
Sensitivity analysis
To examine the stability and reliability of our meta-analysis results, we performed sensitivity analysis by sequentially removing the single studies one at time. In this meta-analysis, the results of sensitivity analysis showed that no single study influenced the recalculated ORs and 95% CIs quantitatively, suggesting robustness and reliability of our results.
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures in the analyses of FAS-670A/G and FAS-1377G/A polymorphisms. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry (Figure 4). Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not show any evidence of publication bias (All P > 0.05, Table 3). The publication bias cannot be analyzed in the analysis of Fas-844T/C polymorphism due to the limit of study number.
Figure 4.
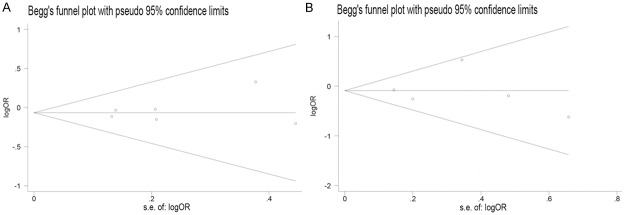
Funnel plots between Fas polymorphisms and leukemia risk. A. GG vs AG/AA (Fas-670); B. AA vs GA/GG (CC vs TC/TT).
Table 3.
Egger’s test for publication bias
| Variables | Models | P | 95% CI |
|---|---|---|---|
| Fas-670A/G | |||
| GG vs AG/AA | 0.510 | (-1.31, 2.22) | |
| GG vs AA | 0.537 | (-4.70, 2.86) | |
| GG/AG vs AA | 0.206 | (-1.79, 6.04) | |
| G vs A | 0.527 | (-5.12, 3.07) | |
| Fas-1377G/A | |||
| AA vs GA/GG | 0.991 | (-3.69, 3.72) | |
| AA vs GG | 0.926 | (-4.94, 5.26) | |
| AA/GA vs GG | 0.767 | (-14.50, 11.83) | |
| A vs G | 0.851 | (-15.54, 17.95) |
Discussion
Leukemia risks were affected by various environmental and genetic factors. Genetic polymorphisms in Fas/FasL gene were also proposed to be associated with susceptibility to leukemia, but recent studies reported controversial findings. We performed a meta-analysis to assess the association between Fas/FasL polymorphisms and susceptibility to leukemia. Our meta-analysis is the first meta-analysis on the association between Fas/FasL polymorphisms and leukemia risk. However, the findings from the overall analyses did not support the associations of Fas/FasL polymorphisms with leukemia (Table 2).
In the meta-analysis for the association between Fas-670A/G and leukemia risk, no association was observed under all models (Figure 1) and there was also no significant heterogeneity. From the subgroup analysis by leukemia type, we found that the between-study heterogeneity was significant among ALL subgroup under all models except the recessive model, suggesting that the studies in ALL and leukemia type might be the source of heterogeneity (Table 2). In the meta-analysis for the association between Fas-1377G/A and leukemia risk, no association was observed (Figure 2) and there was significant heterogeneity in allele model (A vs G) and dominant model (GA/AA vs GG) and there was no significant heterogeneity in the recessive model (AA vs GA/GG) and the homozygous model (AA vs GG). From the subgroup analysis by ethnicity, we found that the between-study heterogeneity in Asians and Caucasians was still significant, suggesting that the ethnicities might be the source of heterogeneity (Table 2). However, from the subgroup analysis by leukemia type, we observed significant association in ALL subgroup and no heterogeneity was observed in ALL subgroup. These results suggested A allele of Fas-1377G/A may be a protective factor to ALL risk (Table 2). As for the FasL-844T/C polymorphism, only two studies meet the Inclusion criteria and no significant association was observed (Figure 3). Thus more studies on FasL-844T/C polymorphism were required in the future.
Figure 1.
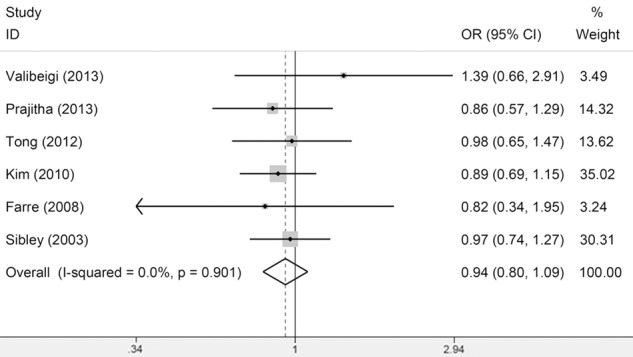
Meta-Analysis of association between Fas-670 polymorphism and susceptibility to leukemia (GG vs AG/AA).
Figure 2.
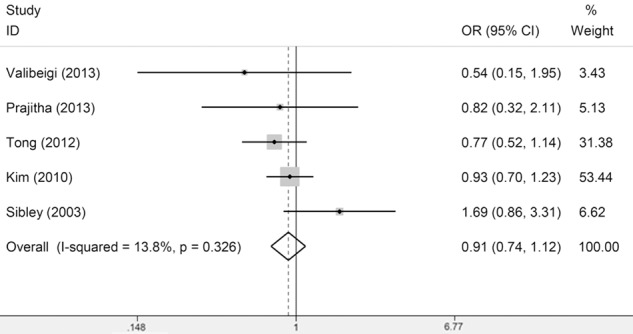
Meta-Analysis of association between Fas-1377 polymorphism and susceptibility to leukemia (AA vs GA/GG).
Figure 3.
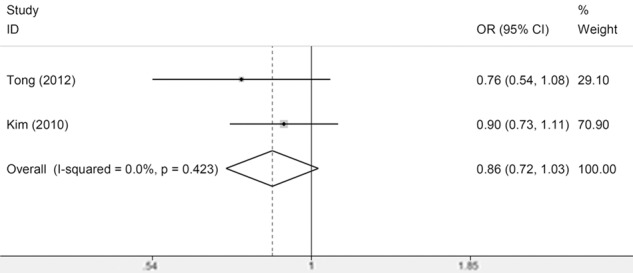
Meta-Analysis of association between FasL-844 polymorphism and susceptibility to leukemia (CC vs TC/TT).
There were some limitations of our meta-analysis. First, there was heterogeneity among studies and the heterogeneity was significant under several models. The heterogeneity was still existed in some subgroups under several models and the heterogeneity (Table 2). So, we could not conclude whether the heterogeneity came from ethnicity or leukemia type in subgroups (ALL in Fas-670A/G polymorphism; Caucasians or AML in Fas-1377G/A polymorphism).Second, studies on the association between Fas polymorphisms and cancer risk mainly focus on solid tumor. The number of studies on hematological malignancy was limited. So the case group and control group were fewer in number, which could increase the likelihood of type I and type II errors. Only 2 studies assessed association between FasL-844T/C polymorphism and leukemia risk were included. Third, although no obvious publication bias was detected by funnel plot or Egger’s test. Fourth, in the subgroup analyses by ethnicity, most studies were from Caucasians and no studies among Africans, suggesting the inapplicability of our results for these populations. Fifth, several risk factors are related to hematological malignancies, such as age, sex, family history, environmental factors, cancer stage, viral and bacterial infections, toxic chemistry, smoking status, and so on. Our meta-analysis didn’t discuss these information due to lack of original information. Sixth, leukemia contains different types while our meta-analysis mainly included AML and ALL. Seventh, our meta-analysis was limited to language; the included published studies were all in English. In spite of these limitations, our meta-analysis had several advantages. First, the quality of the case-control studies included in our meta-analysis was satisfactory and met our inclusion criteria. Second, we did not detect any publication bias, suggesting that the whole pooled result was unbiased. Third, our study is the first meta-analysis assesses the Fas/FasL polymorphisms and leukemia risk.
This meta-analysis suggests that there was no association between Fas/FasL polymorphisms and susceptibility to leukemia except the association between Fas-1377G/A polymorphism and ALL risk. The A allele of Fas-1377G/A was suggested as a protective factor in ALL risk. However, further studies with large sample size are needed to further assess the association between Fas/FasL and susceptibility to leukemia, especially on FasL-844T/C polymorphism.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant No. 81372249, No. 81300431), Foundation of the Ministry of Education of China for Returned Scholars, Research Fund for the Doctoral Program of Higher Education of The Ministry of national education China (Grant No. 20114433110012), Guangdong Provincial Department of education, science and technology innovation project (Grant No. 2012KJCX0025), Key Project of Science and Technology of Guangzhou city (12C22121595), Natural Science Foundation of Guangdong Province, China (Grant No. S2013040014449), National Training Programs of Innovation for Undergraduates of Southern Medical University (Grant No. 201312121001).
Disclosure of conflict of interest
None.
References
- 1.Malek S. Molecular biomarkers in chronic lymphocytic leukemia. Adv Exp Med Biol. 2013;792:193–214. doi: 10.1007/978-1-4614-8051-8_9. [DOI] [PubMed] [Google Scholar]
- 2.Satoh Y, Matsumura I, Tanaka H, Harada H, Harada Y, Matsui K, Shibata M, Mizuki M, Kanakura Y. C-terminal mutation of RUNX1 attenuates the DNA-damage repair response in hematopoietic stem cells. Leukemia. 2012;26:303–11. doi: 10.1038/leu.2011.202. [DOI] [PubMed] [Google Scholar]
- 3.Inazawa J, Itoh N, Abe T, Nagata S. Assignment of the human Fas antigen gene (Fas) to 10q24.1. Genomics. 1992;14:821–2. doi: 10.1016/s0888-7543(05)80200-9. [DOI] [PubMed] [Google Scholar]
- 4.Behrmann I, Walczak H, Krammer PH. Structure of the human APO-1 gene. Eur J Immunol. 1994;24:3057–62. doi: 10.1002/eji.1830241221. [DOI] [PubMed] [Google Scholar]
- 5.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–92. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholz M, Cinatl J. Fas/FasL interaction: a novel immune therapy approach with immobilized biologicals. Med Res Rev. 2005;25:331–42. doi: 10.1002/med.20025. [DOI] [PubMed] [Google Scholar]
- 7.Huang QR, Morris D, Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol. 1997;34:577–82. doi: 10.1016/s0161-5890(97)00081-3. [DOI] [PubMed] [Google Scholar]
- 8.Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, Roddam PL, Skibola CF, Smith MT, Morgan GJ. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63:4327–30. [PubMed] [Google Scholar]
- 9.Wu J, Siddiqui J, Nihal M, Vonderheid EC, Wood GS. Structural alterations of the FAS gene in cutaneous T-cell lymphoma (CTCL) Arch Biochem Biophys. 2011;508:185–91. doi: 10.1016/j.abb.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HJ, Jin XM, Kim HN, Lee IK, Park KS, Park MR, Jo DY, Won JH, Kwak JY, Kim HJ, Choi JS, Juhng SW, Choi C. Fas and FasL polymorphisms are not associated with acute myeloid leukemia risk in Koreans. DNA Cell Biol. 2010;29:619–24. doi: 10.1089/dna.2010.1032. [DOI] [PubMed] [Google Scholar]
- 11.Gormus U, Ergen A, Yaylim-Eraltan I, Yilmaz H, Turna A, Bozkurt N, Isbir T. Fas-1377 A/G polymorphism in lung cancer. In Vivo. 2007;21:663–6. [PubMed] [Google Scholar]
- 12.Wang M, Wu D, Tan M, Gong W, Xue H, Shen H, Zhang Z. FAS and FAS ligand polymorphisms in the promoter regions and risk of gastric cancer in Southern China. Biochem Genet. 2009;47:559–68. doi: 10.1007/s10528-009-9264-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Zheng Z, Yu W, Lin H, Cui B, Cao F. Polymorphisms of the FAS and FASL genes and risk of breast cancer. Oncol Lett. 2012;3:625–628. doi: 10.3892/ol.2011.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunobiki O, Ueda M, Toji E, Yamamoto M, Akashi K, Sato N, Izuma S, Torii K, Tanaka I, Okamoto Y, Noda S. Genetic Polymorphism of Cancer Susceptibility Genes and HPV Infection in Cervical Carcinogenesis. Patholog Res Int. 2011;2011:364069. doi: 10.4061/2011/364069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lima L, Morais A, Lobo F, Calais-da-Silva FM, Calais-da-Silva FE, Medeiros R. Association between FAS polymorphism and prostate cancer development. Prostate Cancer Prostatic Dis. 2008;11:94–8. doi: 10.1038/sj.pcan.4501002. [DOI] [PubMed] [Google Scholar]
- 16.Tong N, Zhang L, Sheng X, Wang M, Zhang Z, Fang Y, Xue Y, Li J, Zhang Z. Functional polymorphisms in FAS, FASL and CASP8 genes and risk of childhood acute lymphoblastic leukemia: a case-control study. Leuk Lymphoma. 2012;53:1360–6. doi: 10.3109/10428194.2011.654117. [DOI] [PubMed] [Google Scholar]
- 17.Valibeigi B, Amirghofran Z, Golmoghaddam H, Hajihosseini R, Kamazani FM. Fas Gene Variants in Childhood Acute Lymphoblastic Leukemia and Association with Prognosis. Pathol Oncol Res. 2013 doi: 10.1007/s12253-013-9705-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Geng P, Li J, Ou J, Xie G, Wang N, Xiang L, Sa R, Liu C, Li H, Liang H. Association of fas -1377 g/a polymorphism with susceptibility to cancer. PLoS One. 2014;9:e88748. doi: 10.1371/journal.pone.0088748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Xing GH, Fan CC. Association between the FAS rs2234767G/A polymorphism and cancer risk: a systematic review and meta-analysis. DNA Cell Biol. 2014;33:320–7. doi: 10.1089/dna.2013.2273. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, He B, Li R, Pan Y, Gao T, Deng Q, Sun H, Song G, Wang S. Association of the polymorphisms in the Fas/FasL promoter regions with cancer susceptibility: a systematic review and meta-analysis of 52 studies. PLoS One. 2014;9:e90090. doi: 10.1371/journal.pone.0090090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 22.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 24.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farre L, Bittencourt AL, Silva-Santos G, Almeida A, Silva AC, Decanine D, Soares GM, Alcantara LC Jr, Van Dooren S, Galvão-Castro B, Vandamme AM, Van Weyenbergh J. Fas 670 promoter polymorphism is associated to susceptibility, clinical presentation, and survival in adult T cell leukemia. J Leukoc Biol. 2008;83:220–2. doi: 10.1189/jlb.0407198. [DOI] [PubMed] [Google Scholar]
- 27.Rozenfeld-Granot G, Toren A, Amariglio N, Brok-Simoni F, Rechavi G. Fas gene polymorphisms in chronic myeloid leukemia. Exp Hematol. 2001;29:228–33. doi: 10.1016/s0301-472x(00)00623-8. [DOI] [PubMed] [Google Scholar]
- 28.Rollinson S, Allan JM, Law GR, Roddam PL, Smith MT, Skibola C, Smith AG, Forrest MS, Sibley K, Higuchi R, Germer S, Morgan GJ. High-throughput association testing on DNA pools to identify genetic variants that confer susceptibility to acute myeloid leukemia. Cancer Epidemiol Biomarkers Prev. 2004;13:795–800. [PubMed] [Google Scholar]


