Abstract
Interferon gamma (IFN-γ) is a potent proinflammatory cytokine which plays an important role in the antiviral, antiproliferative, and antitumor activities. So our meta-analysis was conducted to evaluate the correlations between common genetic polymorphisms in the IFN-γ gene and susceptibility to cervical cancer. The +874 polymorphism in IFN gene region reportedly affects cancer risk. In order to derive a more precise estimation of the association, eight clinical case-control studies met all the inclusion criteria and were included in this meta-analysis. A total of 2,375 cancer cases and 2,106 controls were involved in this meta-analysis. Overall, no significant association was detected in allelic model (A allele vs. T allele OR=0.97, 95% CI, 0.73~1.28), homozygote comparison (AA vs. TT OR=1.12, 95% CI, 0.68~1.85), heterozygote comparison (AT vs. TT OR=1.43, 95% CI, 0.97~1.61), dominant model (AA+AT vs. TT OR=1.19, 95% CI, 0.87~1.63), nor recessive model (AA vs. AT+TT OR=0.95, 95% CI, 0.64~1.40). Subgroup analysis based on ethnicity, genotyping method, and Hardy–Weinberg equilibrium status. Ethnicity suggested that genetic polymorphisms in the IFN-γ gene were closely correlated with increased cervical cancer risk among Asians (allele model: OR=1.11, 95% CI=0.61~2.02, P<0.001; recessive model: OR=0.98, 95% CI=0.36~2.96, P<0.001; homozygous model: OR=1.43, 95% CI=0.56~3.65, P=0.001; respectively), but not among Mixed (all P>0.05). In conclusion, the current meta-analysis supported that IFN-γ genetic polymorphism may contribute to cervical cancer susceptibility, and further well-designed studies with large sample size are warranted to validate our conclusion.
Keywords: IFN-γ, genetic polymorphism, susceptibility, cervical cancer, meta-analysis
Introduction
Cervical cancer is the second most common malignancy in women worldwide [1], with approximately 80% of cases arising in developing countries. Innate immune deficiency, environmental aggravation, and genetic mutation have been considered as important pathopoiesis factors.
The genetic polymorphisms in cytokine genes have been investigated widely, which could influence cancer predisposition and prognosis by altering the expression level [2,3].The gene that encodes IFN-γ is located on chromosome 12q24 spanning approximately 5.4 kb and consists of four exons with three intervening regions [4]. The human IFN-γ gene shows the presence of a variable-length CA repeat within the first intron of the gene ranging from 12 to 15 repeats [5]. IFN-γ is a pleiotropic cytokine secreted by type-1 helper (Th1) T cells, cytotoxic T cells, and stimulated natural killer cells in response to antigenic stimulation and involved in activation of macrophages and endothelial cells [6]. It can produced various immune cells such as activated CD4+ T cells, cytotoxic CD8+ cells, and activated natural killer cells [7,8]. On one hand, IFN-γ can exert direct anti-proliferative and anti-metabolic effects on a wide variety of tumor cells [9-13]; on the other hand, IFN-γ can inhibit angiogenesis in the tumor [14,15]. Pravica [16] et al. noted a novel single nucleotide polymorphism (SNP), T to A, located at the +874 position from translation start site in the first intron of IFN-γ gene, which coincides with a putative NF-κB binding site that could play a fundamental role in the induction of constitutively high IFN-γ production. Therefore, a meta-analysis of the published studies was performed, with the aim to clarify the relationship between the IFN-γ polymorphism and cervical cancer risk.
Materials and methods
Identification of eligible studies
PubMed, Embase and Web of Science database searches were performed in our meta analysis. In order to search as many related studies as possible, we used different combinations of the following MeSH terms and keywords: ‘Interferon’ or ‘IFN’, ‘polymorphism’ or ‘SNP’, ‘cervical cancer’ or ‘cervical tumor’ (the last search was updated 1 August 2014). The search was limited to English language papers. All reference lists of reviews and retrieved articles were manually screened for further potential studies.
Inclusion and exclusion criteria
The following criteria were used to decision inclusion eligibility: (1) must be a study focused on the correlation of IFN-γ genetic polymorphisms with cervical cancer susceptibility; (2) clinical case-control studies; and (3) providing sufficient data to calculate the odds ratio (OR) and its corresponding 95% confidence interval (CI). Furthermore, studies that did not meet our inclusion criteria were excluded.
Data extraction
Data of eligible studies were extracted by two authors independently and in duplicate according to the predesigned data-collection form. For each article, extracted information contained the following: the first author’s name, publication year, country of origin, the patients’ ethnicities, genotyping method, numbers of cases and controls, and evidence of HWE. The different ethnic descents were categorized as African, Asian, Caucasian and Mixed.
Statistic analysis
Meta-analysis was performed using the STATA statistical software (Version 12.0, Stata Corporation, College Station, TX, USA). The strength of the association between IFNG +874 T/A polymorphism and the risk of cancer was calculated by Odds ratios (OR) with 95% confidence interval (95% CI) in the following five genetic models: allelic model (A allele vs. T allele), homozygote comparison (AA vs. TT), heterozygote comparison (AT vs. TT), dominant model (AA+AT vs. TT), and recessive model (AA vs. AT+TT). The statistical significance of the pooled OR was assessed with the Z test and a P value of <0.05 was considered significant. Chi-square-based Q test was conducted to measure the heterogeneity between eligible studies, and the existence of heterogeneity was considered significant if P<0.10 [17]. When the between-study heterogeneity was absent, a fixed-effect model (the Mantel-Haenszel method) was used to pool the data from different studies [18]; otherwise, a random-effect model (the DerSimonian and Laird method) was applied [19]. To explore the source of heterogeneity among variables such as ethnicity, and HWE status, both subgroup analyses and logistic met regression analyses were performed [20]. Funnel plots and Egger’s linear regression test were applied to investigate publication bias [21].
Results
Study selection and description
For cervical cancer susceptibility related to the IFN-γ +874 T/A polymorphism, 8 studies with 2,375 cancer cases and 2,106 controls were included in the current meta-analysis. We summarized the study characteristics and methodological quality in Table 1. Among these 8 eligible studies, 3 of them were Asian descendants, 1 Caucasian, 1 African, and the remaining 3 studies were conducted in mixed populations.
Table 1.
Characteristics of eligible studies included in the meta-analysis
| Author | Year | Country | Ethnicity | Cancer types | Genotyping methods | Case | Control | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| TT | AT | AA | TT | AT | AA | |||||||
| do Carmo | 2012 | Brazil | Mixed | Cervical cancer | PCR-SSP | 2 | 3 | 4 | 13 | 46 | 17 | Yes |
| Wang | 2011 | China | Asian | Cervical cancer | PCR-SSP | 21 | 84 | 81 | 38 | 96 | 66 | Yes |
| Ivansson | 2010 | Sweden | Caucasian | Cervical cancer | TaqMan | 286 | 650 | 354 | 157 | 371 | 274 | Yes |
| Gangwar | 2009 | India | Asian | Cervical cancer | PCR-ARMS | 23 | 91 | 86 | 45 | 115 | 70 | Yes |
| Kordi | 2008 | India | Asian | Cervical cancer | PCR-ARMS | 24 | 125 | 51 | 25 | 75 | 100 | Yes |
| Govan 1 | 2003 | South Africa | African | Cervical cancer | PCR-ARMS | 5 | 30 | 61 | 7 | 31 | 102 | No |
| Govan 2 | 2003 | South Africa | Mixed | Cervical cancer | PCR-ARMS | 14 | 66 | 85 | 26 | 81 | 158 | No |
| Guzman | 2008 | Brazil | Mixed | Cervical cancer | PCR-SSP | 130 | 99 | 126 | 67 | Yes | ||
Quantitative data synthesis
A total of 8 studies were identified and analyzed, a summary of the meta-analysis findings on the correlations between IFN-γ polymorphisms and the risk of cervical cancer in each individual study was presented in Table 2, and the summary results were summarized in Table 3. In our meta-analysis the random-effects model was performed. The results declared that IFN-γ genetic polymorphisms might be significantly associated with an increased risk of cervical cancer (allele model: OR=0.97, 95% CI=0.73~1.28, P<0.001; recessive model: OR=0.95, 95% CI=0.64~1.40, P<0.001; homozygous model: OR=1.12, 95% CI=0.68~1.85, P<0.001; respectively).
Table 2.
Detailed association of IFNG +874 T/A polymorphism with cancer risk in each individual study
| Author | Ethnicity | Cancer types | A vs. T allele | AA vs. TT | AT vs. TT | AA+AT vs. TT | AA vs. AT+TT |
|---|---|---|---|---|---|---|---|
|
| |||||||
| OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | |||
| do Carmo | Mixed | Cervical cancer | 1.41 (0.52, 3.84) | 1.53 (0.24, 9.67) | 0.42 (0.06, 2.81) | 0.72 (0.13, 3.88) | 2.78 (0.67, 11.5) |
| Wang | Asian | Cervical cancer | 1.47 (1.10, 1.97)b | 2.22 (1.19, 4.15)b | 1.58 (0.86, 2.91) | 1.84 (1.04, 3.28)b | 1.57 (1.04, 2.37)b |
| Ivansson | Caucasian | Cervical cancer | 0.83 (0.73, 0.94)a | 0.71 (0.55, 0.91)a | 0.96 (0.76, 1.21) | 0.85 (0.69, 1.06) | 0.73 (0.60, 0.88)a |
| Gangwar | Asian | Cervical cancer | 1.54 (1.17, 2.04)b | 2.40 (1.33, 4.35)b | 1.55 (0.87, 2.75) | 0.81 (0.46, 1.44) | 1.72 (1.16, 2.56)b |
| Govan 1 | African | Cervical cancer | 0.73 (0.45, 1.17) | 0.84 (0.26, 2.75) | 1.36 (0.39, 4.74) | 0.96 (0.30, 3.11) | 0.65 (0.37, 1.14) |
| Govan 2 | Mixed | Cervical cancer | 0.84 (0.62, 1.15) | 1.00 (0.50, 2.01) | 1.51 (0.73, 3.13) | 1.17 (0.59, 2.32) | 0.72 (0.49, 1.07) |
| Kordi | Asian | Cervical cancer | 0.60 (0.45, 0.80) | 0.53 (0.28, 1.02) | 1.74 (0.93, 3.26) | 1.05 (0.58, 1.91) | 0.34 (0.22, 0.52) |
| Guzman | Mixed | Cervical cancer | –c | –c | –c | –c | 1.43 (0.97, 2.13) |
Significantly associated with decreased cancer risk.
Significantly associated with increased cancer risk.
Cannot be calculated.
Table 3.
Stratification analysis of genetic susceptibility of IFNG +874 T/A polymorphism to cancer risk
| Category | A allele vs. T allele | AA vs. TT | AT vs. TT | AA+AT vs. TT | AA vs. AT+TT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| N | OR (95% CI) | P | N | OR (95% CI) | P | N | OR (95% CI) | P | N | OR (95% CI) | P | N | OR (95% CI) | P | |
| Total | 7 | 0.97 (0.73, 1.28) | <0.001 | 7 | 1.12 (0.68, 1.85) | <0.001 | 7 | 1.25 (0.97, 1.61) | 0.263 | 7 | 1.19 (0.87, 1.63) | 0.068 | 8 | 0.95 (0.64, 1.40) | <0.001 |
| Ethnicity | |||||||||||||||
| Asian | 3 | 1.11 (0.61, 2.02) | <0.001 | 3 | 1.43 (0.56, 3.65) | 0.001 | 3 | 1.61 (1.14, 2.29) | 0.963 | 3 | 1.56 (1.08, 2.24) | 0.292 | 3 | 0.98 (0.35, 2.69) | <0.001 |
| Caucasian | 1 | 0.83 (0.73, 0.94) | - | 1 | 0.71 (0.55, 0.91) | - | 1 | 0.96 (0.76, 1.21) | - | 1 | 0.85 (0.69, 1.06) | - | 1 | 0.73 (0.60, 0.88) | - |
| African | 1 | 0.73 (0.45, 1.17) | - | 1 | 0.84 (0.26, 2.75) | - | 1 | 1.36 (0.38, 4.74) | - | 1 | 0.96 (0.30, 3.11) | - | 1 | 0.65 (0.37, 1.13)c | - |
| Mixed | 2 | 0.88 (0.66, 1.18) | 0.330 | 2 | 1.02 (0.64, 1.62) | 0.672 | 2 | 1.09 (0.37, 3.25) | 0.218 | 2 | 1.20 (0.58, 2.06) | 0.600 | 3 | 1.17 (0.62, 2.22) | 0.021 |
| Genotyping method | |||||||||||||||
| PCR-SSP | 2 | 1.47 (1.11, 1.94) | 0.939 | 2 | 2.14 (1.18, 3.86) | 0.707 | 2 | 1.13 (0.36, 3.49) | 0.194 | 2 | 1.63 (0.88, 3.03) | 0.301 | 3 | 1.53 (1.16, 2.03) | 0.672 |
| TaqMan | 1 | 0.83 (0.73, 0.94) | - | 1 | 0.71 (0.55, 0.91) | - | 1 | 0.96 (0.76, 1.21) | - | 1 | 0.85 (0.69, 1.06) | - | 1 | 0.73 (0.60, 0.88) | - |
| PCR-ARMS | 4 | 0.87 (0.56, 1.37) | <0.001 | 4 | 1.05 (0.50, 2.17) | 0.008 | 4 | 1.58 (1.11, 2.24) | 0.983 | 4 | 1.19 (0.87, 1.85) | 0.464 | 4 | 0.73 (0.36, 1.46) | <0.001 |
| HWE status | |||||||||||||||
| Yes | 6 | 1.05 (0.73, 1.53) | <0.001 | 6 | 1.20 (0.61, 2.34) | <0.001 | 6 | 1.26 (0.91, 1.76 | 0.136 | 6 | 1.23 (0.81, 1.87) | 0.021 | 6 | 1.07 (0.64, 1.79) | <0.001 |
| No | 2 | 0.81 (0.62, 1.04) | 0.615 | 2 | 1.11 (0.68, 1.85) | 0.802 | 2 | 1.47 (0.78, 2.76) | 0.881 | 2 | 1.19 (0.87, 1.63) | 0.77 | 10 | 0.70 (0.51, 0.96) | 0.768 |
A subgroup analysis was carried out to evaluate the impact of the IFN-γ polymorphism on the risk of cervical cancer. Ethnicity stratified analysis indicated that the genetic polymorphisms in the IFN-γ gene were closely linked to the pathogenesis of cervical cancer among Asian populations (allele model: OR=1.11, 95% CI=0.61~2.02, P<0.001; dominant model: OR=1.56, 95% CI=1.08~2.24, P=0.292; heterozygous model: OR=1.61, 95% CI=1.14~2.29, P=0.963; homozygous model: OR=1.43, 95% CI=0.56~3.65, P=0.001; respectively), but not among Mixed (all P>0.05) (Figure 1). We also performed subgroup analyses based on genotyping method and HWE status. Their results indicate that IFN-γ genetic polymorphisms might be implicated in the risk of cervical cancer in the majority of subgroups, except the PCR-SSP and no HWE status subgroups.
Figure 1.
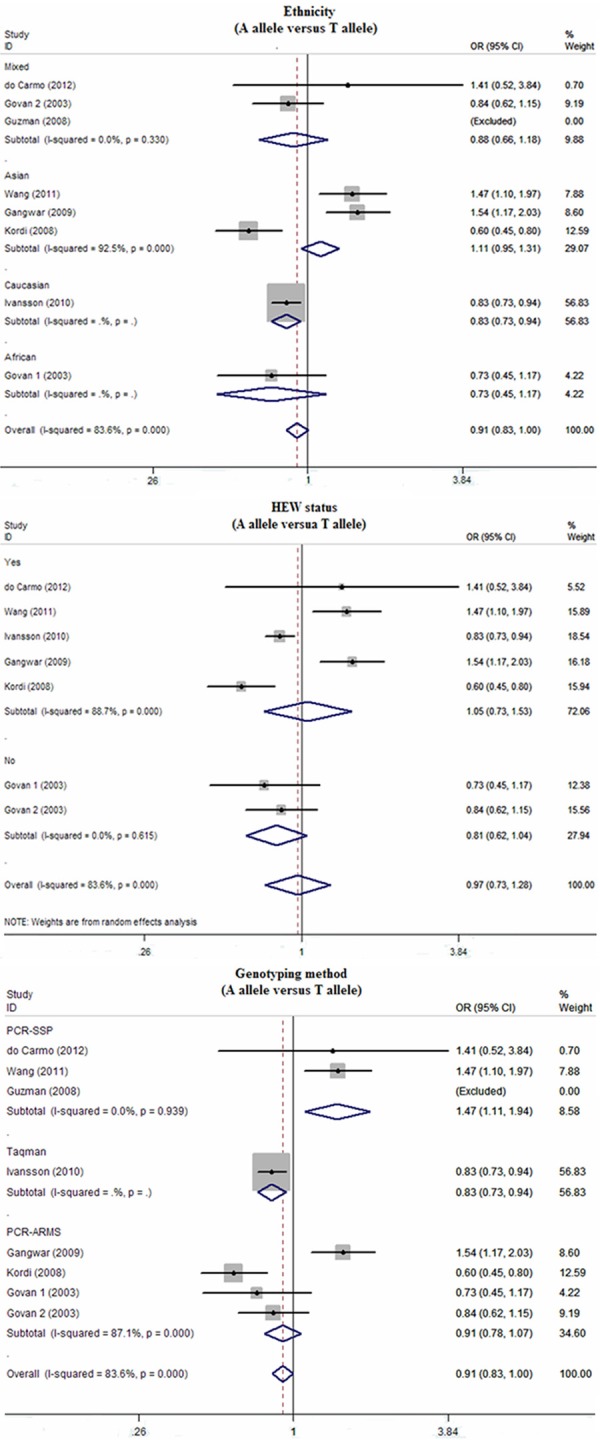
Subgroup analyses by ethnicity, genotyping method, and HEW status of relationships between IFN-γ genetic polymorphisms and susceptibility to cervical cancer.
Sensitivity analysis suggested that no single study could influence the pooled OR (Figure 2). Funnel plots demonstrated evidence of obvious asymmetry (Figure 3). Egger’s test displayed strong statistical evidence of publication bias.
Figure 2.
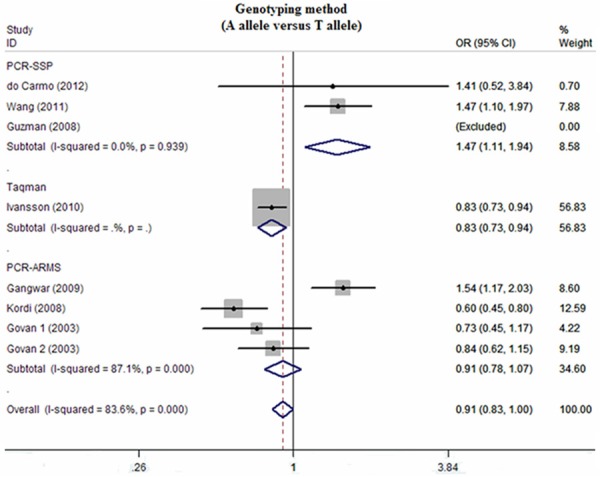
Sensitivity analysis of the summary odds ratio coefficients on the relationships between IFN-γ genetic polymorphisms and susceptibility to cervical cancer.
Figure 3.
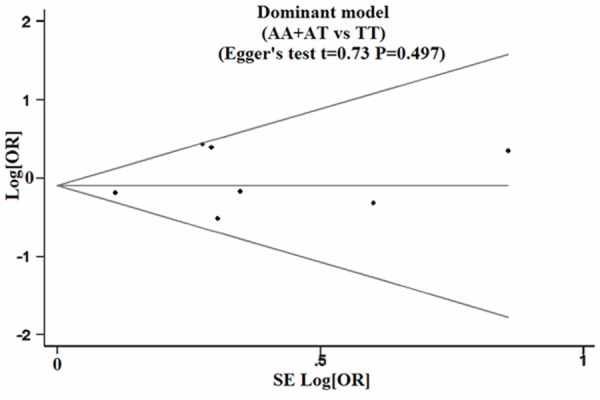
Funnel plot of publication biases on the relationships between IFN-γ genetic polymorphisms and susceptibility to cervical cancer.
Discussion
Cervical cancer is one of the most dangerous causes of health deficiency and death worldwide. Some SNPs of IFN-γ have been reported. The cytokine IFN-γ, an acid-labile protein plays a key role in cervical cancer [8,22-29] and other cancers [30,31]. Among different polymorphisms of IFN-γ, the +874 T/A polymorphism is one of the most widely studied. In our meta-analysis, we evaluated the hypothesis that IFN-γ genetic polymorphisms contribute to susceptibility to cervical cancer. We found that genetic polymorphisms in the IFN-γ gene were significantly correlated with an increased risk of cervical cancer.
This meta-analysis, based on 8 studies, explored the association between IFN-γ +874 T/A and cervical cancer risk, involving about 2,375 cancer cases and 2,106 controls.
To further investigate the relationship between IFN-γ genetic variations and the pathogenesis of cervical cancer, we conducted subgroup analyses based on ethnicity, genotyping method, and HWE status. A stratified analysis by ethnicity was conducted, the findings revealed that there were significant associations between IFN-γ genetic polymorphisms and cervical cancer risk among Asians, but not among Mixed, indicating that ethnic differences may be a potential factor affecting individual’s susceptibility to cervical cancer. Subsequently, we conducted subgroup analysis based on genotyping method revealed that IFN-γ genetic polymorphisms were related to an increased risk of cervical cancer in the PCR-ARMS array subgroups, while no associations were detected in the PCR-SSP subgroups. In the further stratification analysis by HWE status, our results illustrated a positive correlation between IFN-γ genetic polymorphisms and susceptibility to cervical cancer in the HWE status subgroup, but not in the any HWE status size subgroup.
In summary, our findings were consistent with previous studies that IFN-γ genetic polymorphisms may be potential risk factors in the development of cervical cancer, suggesting that these polymorphisms may be of great value for the early diagnosis of cervical cancer.
The current meta-analysis has several limitations that should be pointed out. The first limitation was that the small number of studies was short of sufficient statistical power to assess the correlations between IFN-γ genetic polymorphisms and the pathogenesis of cervical cancer. The second limitation in our meta-analysis failed to obtain original data from the included articles, which may have limited further evaluation of the potential roles of IFN-γ genetic polymorphisms in the development and progression of cervical cancer.
Although our study has several limitations, this is the first meta-analysis focusing on the relationships between IFN-γ genetic polymorphisms and the risk of cervical cancer. Furthermore, we performed a highly sensitive literature search strategy of electronic databases, a manual search of reference lists from relevant studies, and the selection process of eligible articles was based on strict inclusion and exclusion criteria. Importantly, rigorous statistical analysis provided a basis for pooling of information from individual studies.
In conclusion, a strong association was observed between IFN-γ genetic polymorphisms and cervical cancer, and therefore IFN-γ genetic polymorphisms may be valuable as a biomarker. Considering that the quality and quantity of the reviewed articles were limited, larger and well-designed studies should be employed in the future for further confirmation of the association between IFN-γ genetic polymorphisms and cervical cancer.
Disclosure of conflict of interest
None.
References
- 1.Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Jin P, Panelli MC, Marincola FM, Wang E. Cytokine polymorphism and its possible impact on cancer. Immunol Res. 2004;30:181–190. doi: 10.1385/IR:30:2:181. [DOI] [PubMed] [Google Scholar]
- 3.Howell WM, Rose-Zerilli MJ. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr. 2007;137:194S–199S. doi: 10.1093/jn/137.1.194S. [DOI] [PubMed] [Google Scholar]
- 4.Calvo J, Martínez N, Etxagibel A, Calleja S, Sáez-torres C, Sedeño M, Julià R, Muncunill J, Matamoros N, Gayà A. Allelic frequencies of polymorphic variants of cytokine genes (IL1A, IL1B, IL1RN, IL6, IL10, IL12p40, and IFNG) in a Spanish population. Inmunologia. 2002;21:76–86. [Google Scholar]
- 5.Pravica V, Asderakis A, Perrey C, Hajeer A, Sinnott PJ, Hutchinson IV. In vitro production of IFN-gamma correlates with CA repeat polymorphism in the human IFN-gamma gene. Eur J Immunogenet. 1999;26:1–3. doi: 10.1046/j.1365-2370.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- 6.Woodman JP, Dimier IH, Bout DT. Human endothelial cells are activated by IFN-gamma to inhibit Toxoplasma gondii replication. Inhibition is due to a different mechanism from that existing in mouse macrophages and human fibroblasts. J Immunol. 1991;147:2019–2023. [PubMed] [Google Scholar]
- 7.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 8.Billiau A, Matthys P. Interferon-gamma: a historical perspective. Cytokine Growth Factor Rev. 2009;20:97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 10.Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandal M, Bandyopadhyay D, Goepfert TM, Kumar R. Interferon-induces expression of cyclin-dependent kinase-inhibitors p21WAF1 and p27Kip1 that prevent activation of cyclin-dependent kinase by CDK-activating kinase (CAK) Oncogene. 1998;16:217–225. doi: 10.1038/sj.onc.1201529. [DOI] [PubMed] [Google Scholar]
- 12.Buard A, Vivo C, Monnet I, Boutin C, Pilatte Y, Jaurand MC. Human malignant mesothelioma cell growth: activation of janus kinase 2 and signal transducer and activator of transcription 1alpha for inhibition by interferon-gamma. Cancer Res. 1998;58:840–847. [PubMed] [Google Scholar]
- 13.Chen B, He L, Savell VH, Jenkins JJ, Parham DM. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 2000;60:3290–3298. [PubMed] [Google Scholar]
- 14.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 15.Folkman J. Angiogenic zip code. Nat Biotechnol. 1999;17:749. doi: 10.1038/11676. [DOI] [PubMed] [Google Scholar]
- 16.Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol. 2000;61:863–866. doi: 10.1016/s0198-8859(00)00167-1. [DOI] [PubMed] [Google Scholar]
- 17.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang BS, Liu Z, Xu WX, Sun SL. CYP3A5*3 polymorphism and cancer risk: a meta-analysis and meta-regression. Tumour Biol. 2013;34:2357–2366. doi: 10.1007/s13277-013-0783-2. [DOI] [PubMed] [Google Scholar]
- 21.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 22.Kordi Tamandani MK, Sobti RC, Shekari M, Mukesh M, Suri V. Expression and polimorphism of IFN-gamma gene in patients with cervical cancer. Exp Oncol. 2008;30:224–229. [PubMed] [Google Scholar]
- 23.Govan VA, Carrara HR, Sachs JA, Hoffman M, Stanczuk GA, Williamson AL. Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. J Carcinog. 2003;2:3. doi: 10.1186/1477-3163-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.do Carmo Vasconcelos de Carvalho V, de Macêdo JL, de Lima CA, da Conceição Gomes de Lima M, de Andrade Heráclio S, Amorim M, de Mascena Diniz Maia M, Porto AL, de Souza PR. IFN-gamma and IL-12B polymorphisms in women with cervical intraepithellial neoplasia caused by human papillomavirus. Mol Biol Rep. 2012;39:7627–7634. doi: 10.1007/s11033-012-1597-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Zhang C, Walayat S, Chen HW, Wang Y. Association between cytokine gene polymorphisms and cervical cancer in a Chinese population. Eur J Obstet Gynecol Reprod Biol. 2011;158:330–333. doi: 10.1016/j.ejogrb.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecol Oncol. 2010;116:544–548. doi: 10.1016/j.ygyno.2009.10.084. [DOI] [PubMed] [Google Scholar]
- 27.Gangwar R, Pandey S, Mittal RD. Association of interferon-gamma +874A polymorphism with the risk of developing cervical cancer in north-Indian population. BJOG. 2009;116:1671–1677. doi: 10.1111/j.1471-0528.2009.02307.x. [DOI] [PubMed] [Google Scholar]
- 28.Guzman VB, Yambartsev A, Goncalves-Primo A, Silva ID, Carvalho CR, Ribalta JC. New approach reveals CD28 and IFNG gene interaction in the susceptibility to cervical cancer. Hum Mol Genet. 2008;17:1838–1844. doi: 10.1093/hmg/ddn077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kordi Tamandani MK, Sobti RC, Shekari M, Mukesh M, Suri V. Expression and polimorphism of IFN-gamma gene in patients with cervical cancer. Exp Oncol. 2008;30:224–9. [PubMed] [Google Scholar]
- 30.Farhat K, Hassen E, Gabbouj S, Bouaouina N, Chouchane L. Interleukin-10 and interferon-gamma gene polymorphisms in patients with nasopharyngeal carcinoma. Int J Immunogenet. 2008;35:197–205. doi: 10.1111/j.1744-313X.2008.00752.x. [DOI] [PubMed] [Google Scholar]
- 31.Colakogullari M, Ulukaya E, Yilmaztepe Oral A, Aymak F, Basturk B, Ursavas A. The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct. 2008;26:283–290. doi: 10.1002/cbf.1419. [DOI] [PubMed] [Google Scholar]


