Abstract
The CD14-159 C/T polymorphism has been implicated in susceptibility to acute pancreatitis (AP), but the results were inconclusive. The present meta-analysis aimed to explore the correlation between CD14-159 C/T polymorphism and AP risk. All eligible case-control studies published up to November 10th, 2014 were identified by searching PubMed, Web of Science, CNKI, and WanFang databases. Two reviewers independently identified the literature according to inclusion and exclusion criteria. Meta-analysis was performed using RevMan 5.2 and Stata 12.0 software. A total of five studies comprising 1211 cases and 932 controls were included. Overall, no significant association between CD14-159 C/T polymorphism and AP risk was found under all four genetic models [CT + TT vs CC: OR = 1.09, 95% CI (0.91, 1.31); TT vs CT + CC: OR = 1.04, 95% CI (0.83, 1.29); CT vs CC: OR = 1.08, 95% CI (0.89, 1.32); TT vs CC: OR = 1.15, 95% CI (0.88, 1.49)]; In stratification analysis by disease severity, we also failed to detect any association between CD14-159C/T polymorphism and the risk of mild AP (MAP) or severe AP (SAP); In subgroup analysis by ethnicity, similar results were observed in Asian and European populations. This meta-analysis suggested that the CD14-159C/T polymorphism is not associated with the susceptibility of acute pancreatitis.
Keywords: CD14, acute pancreatitis, polymorphism, meta-analysis
Introduction
Acute pancreatitis (AP) is a common disease that normally runs a benign course in the majority of patients. However, 25% to 30% of patients experience a severe attack with a high mortality rate [1]. Although gallstones, heavy alcohol consumption and overeating are generally considered the main risk factors for AP, the exact etiology underlying AP is still unclear. Recently, variants in several innate immunity genes have been identified as biologically plausible candidates for effects on AP, such as CD14.
CD14 is expressed on the surface of monocytes, macrophages, and neutrophils as membrane CD14 (mCD14) and in the serum as soluble CD14 (sCD14) and its expression may be partially regulated at the genetic level [2]. The CD14 gene is localized on chromosome 5q31.1, in a region shown to be linked to type 2 T lymphocyte (Th2) prevalent phenotypes, which encodes a receptor protein that binds to lipopolysaccharide (LPS), its primary ligand, and interacts with co-receptors toll-like receptor 4 (TLR4) and lymphocyte antigen 96 (LY96) [3,4]. There are several polymorphism sites in the CD14 gene, and a well-studied common single nucleotide polymorphism (SNP) in the promoter region of CD14, -159C/T (rs2569190, also known as CD14-260C/T), has been associated with increased CD14 expression in vitro and in the serum of children [5].
Recently, the CD14-159C/T polymorphism is investigated extensively to the susceptibility of AP [6-11]. However, the results remain controversial. Therefore, we conduct a meta-analysis to evaluate the association between the CD14-159C/T polymorphism and AP risk.
Materials and methods
Search strategy
A literature research was conducted using PubMed, Web of Science, CNKI and WanFang databases up to November 10th, 2014 without language restrictions. Relevant studies were identified using the terms: [‘cluster of differentiation 14 or CD14’] AND [‘genetic polymorphism or polymorphisms or SNP’] AND [‘acute pancreatitis or AP or mild acute pancreatitis or MAP or severe acute pancreatitis or SAP’]. The search was restricted to humans. Additional studies were identified by a hand search of references of original or review articles on this topic.
Inclusion criteria and exclusion criteria
Studies were included if they met the following criteria: (1) studies that evaluated the association between the CD14-159C/T polymorphism and acute pancreatitis risk, (2) in a case-control study design, and (3) had detailed genotype frequency of cases and controls or could be calculated from the article text. Studies were excluded when they were: (1) not case-control study, (2) review, comment or editorial articles, (3) insufficient data, and (4) repetitive studies.
Data extraction
Two investigators independently extracted data and reached consensus on all of the items. If they generated different results, disagreements were discussed and resolved by a third investigator. Data extracted from the selected articles included: the first author’s name, year of publication, country of origin, ethnicity of study population, genotyping methods, source of control, number of cases and controls and evidence of Hardy-Weinberg equilibrium (HWE) in controls.
Statistical analysis
The risk of AP associated with CD14-159C/T polymorphism was estimated for each study by odds ratio (OR) and 95% confidence interval (95% CI). Four different ORs were calculated: the dominant model (CT + TT vs. CC), the recessive model (TT vs. CT + CC), heterozygote comparison (CT vs. CC), and homozygote comparison (TT vs. CC). The χ2-test-based Q statistic test and I2 statistic were used to assess the between-study heterogeneity [12]. When a significant Q test (P > 0.1) or I2 < 50% indicated homogeneity across studies, the fixed effects model was used [13], or else the random effects model was used [14]. HWE among controls for each study was examined by χ2 test. We performed stratification analyses on ethnicity (Asian, European) and severity of AP (MAP, SAP). Analysis of sensitivity, after removing the studies deviating from HWE, was performed to evaluate the stability of the results. The potential publication bias was examined by Begg’ funnel plot and Egger’s regression test [15,16]. P < 0.05 was regarded as statistically significant.
Statistical analyses were performed using the Cochrane Collaboration RevMan 5.2 and STATA package version 12.0 (Stata Corporation, College Station, Texas).
Results
Study characteristics
The search strategy retrieved 114 potentially relevant studies. According to the inclusion criteria, 6 studies [6-11] with full-text were included in this meta-analysis and 108 studies were excluded. Among those 6 publications, we excluded one study [11] because they did not present detailed genotyping information. Therefore, 5 eligible case-control studies, included 1211 cases and 932 controls, met the inclusion criteria. The characteristics of selected studies are summarized in Table 1. Of the 5 eligible studies, all were written in English. Two ethnicities were addressed: 3 studies [7,8,10] on Asian populations and 2 studies [6,9] on Caucasians. The distribution of genotypes in the controls was consistent with the HWE for all selected studies, except for two studies [7,10].
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Country | Ethnicity | Genotype methods | Source of controls | Sample size (case/control) | Case | Control | HWE (P value) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| CC | CT | TT | CC | CT | TT | ||||||||
| Balog [6] | 2005 | Hungary | European | RT-PCR | PB | 77/71 | 22 | 35 | 20 | 21 | 39 | 11 | 0.309 |
| Chao [7] | 2005 | China | Asian | PCR-RFLP | HB | 177/117 | 84 | 82 | 11 | 49 | 62 | 6 | 0.015 |
| Masamune [8] | 2010 | Japan | Asian | PCR-RFLP | PB | 346/319 | 71 | 172 | 103 | 69 | 143 | 107 | 0.106 |
| Tukiainen [9] | 2008 | Finland | European | MALDI-TOF MS | PB | 396/309 | 147 | 182 | 67 | 133 | 129 | 47 | 0.095 |
| Zhang [10] | 2005 | China | Asian | PCR-RFLP | PB | 215/116 | 128 | 59 | 28 | 71 | 32 | 13 | 0.004 |
RT-PCR: real-time polymerase chain reaction; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; MALDI-TOF MS: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; PB: population-based; HB: hospital-based.
Quantitative data synthesis
There were no inter-study heterogeneity among overall studies of the CD14-159C/T polymorphism in all four genetic models (dominant model: I2 = 0%, P = 0.59; recessive model: I2 = 4%, P = 0.38; CT vs. CC: I2 = 0%, P = 0.49; TT vs. CC: I2 = 0%, P = 0.75). Therefore, we used the fixed-effects model that generated wider CIs. Overall, no significant associations between the CD14-159C/T polymorphism and AP risk were found [dominant model: OR = 1.09, 95% CI (0.91, 1.31); recessive model: OR = 1.04, 95% CI (0.83, 1.29); CT vs. CC: OR = 1.08, 95% CI (0.89, 1.32); TT vs. CC: OR = 1.15, 95% CI (0.88, 1.49)] (Figure 1).
Figure 1.
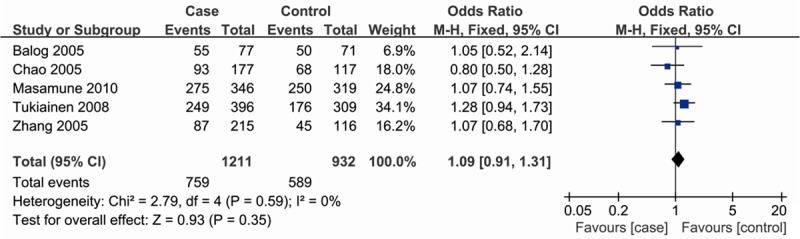
Meta-analysis of the association between CD14-159C/T polymorphism and susceptibility to AP (dominant model).
In the subgroup analysis by severity, similar results were observed in either MAP or SAP (Figure 2; Table 2). Stratification based on ethnicity, we also failed to detect any association in both Asian and European populations (Table 2).
Figure 2.
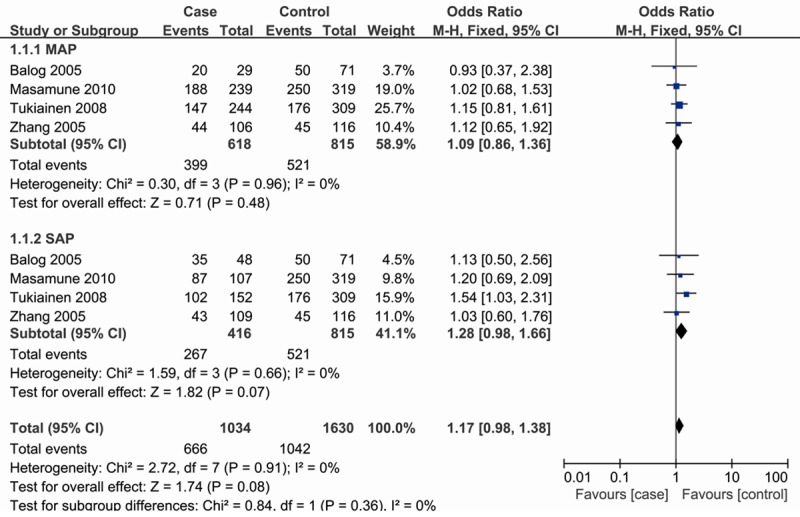
Subgroup analysis by severity of odds ratios for association between CD14-159C/T polymorphism and risk of AP (dominant model).
Table 2.
Pooled analysis for the associations between the CD14-159C/T polymorphism and risk of AP
| Models | Variables | N* | Test of association | Test of heterogeneity | Publication bias | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| OR (95% CI) | Z | P | I2 | P | Egger’s test (P value) | |||
| Dominant model | Total | 5 | 1.09 (0.91, 1.31) | 0.93 | 0.35 | 0 | 0.59 | 0.296 |
| Severity | ||||||||
| MAP | 4 | 1.09 (0.86, 1.36) | 0.71 | 0.48 | 0 | 0.96 | ||
| SAP | 4 | 1.28 (0.98, 1.66) | 1.82 | 0.07 | 0 | 0.66 | ||
| Ethnicity | ||||||||
| Asian | 3 | 0.99 (0.77, 1.26) | 0.10 | 0.92 | 0 | 0.58 | ||
| European | 2 | 1.24 (0.94, 1.64) | 1.52 | 0.13 | 0 | 0.62 | ||
| Recessive model | Total | 5 | 1.04 (0.83, 1.29) | 0.31 | 0.76 | 4% | 0.38 | 0.127 |
| Severity | ||||||||
| MAP | 4 | 0.94 (0.73, 1.22) | 0.44 | 0.66 | 0 | 0.41 | ||
| SAP | 4 | 1.16 (0.86, 1.56) | 0.96 | 0.34 | 10% | 0.34 | ||
| Ethnicity | ||||||||
| Asian | 3 | 0.92 (0.69, 1.22) | 0.60 | 0.55 | 0 | 0.58 | ||
| European | 2 | 1.26 (0.88, 1.81) | 1.25 | 0.21 | 20% | 0.26 | ||
| CT vs CC | Total | 5 | 1.08 (0.89, 1.32) | 0.77 | 0.44 | 0 | 0.49 | 0.150 |
| Severity | ||||||||
| MAP | 4 | 1.10 (0.86, 1.41) | 0.76 | 0.44 | 0 | 0.97 | ||
| SAP | 4 | 1.26 (0.95, 1.68) | 1.61 | 0.11 | 0 | 0.40 | ||
| Ethnicity | ||||||||
| Asian | 3 | 1.00 (0.77, 1.30) | 0.03 | 0.98 | 0 | 0.43 | ||
| European | 2 | 1.20 (0.89, 1.61) | 1.19 | 0.23 | 0 | 0.34 | ||
| TT vs CC | Total | 5 | 1.15 (0.88, 1.49) | 1.02 | 0.31 | 0 | 0.75 | 0.471 |
| Severity | ||||||||
| MAP | 4 | 1.05 (0.77, 1.42) | 0.29 | 0.77 | 0 | 0.67 | ||
| SAP | 4 | 1.32 (0.92, 1.88) | 1.51 | 0.13 | 0 | 0.77 | ||
| Ethnicity | ||||||||
| Asian | 3 | 1.01 (0.71, 1.42) | 0.03 | 0.97 | 0 | 0.84 | ||
| European | 2 | 1.36 (0.91, 2.03) | 1.51 | 0.13 | 0 | 0.58 | ||
Number of comparison;
MAP: mild acute pancreatitis; SAP: severe acute pancreatitis.
Sensitivity analyses
We examined the influence of these studies on the pooled OR by repeating the meta-analysis while excluding the study that was not in HWE. The estimated pooled ORs were not materially altered [dominant model: OR = 1.18, 95% CI (0.94, 1.47); recessive model: OR = 1.01, 95% CI (0.79, 1.29); CT vs. CC: OR = 1.19, 95% CI (0.94, 1.51); TT vs. CC: OR = 1.14, 95% CI (0.85, 1.53)], indicating that our results were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the potential publication bias in the available literature. The shape of funnel plots did not reveal any evidence of funnel plot asymmetry (Figure 3). Egger’s test also showed that there was no statistical significance for the evaluation of publication bias (dominant model: P = 0.296; recessive model: P = 0.127; CT vs. CC: P = 0.150; TT vs. CC: P = 0.471).
Figure 3.
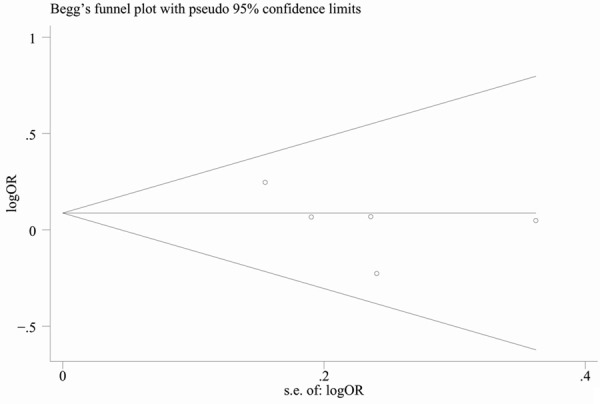
Begg’s funnel plot for publication bias (dominant model).
Discussion
AP is a common disorder that severely affects human health. Many candidate genes were reported to be associated to AP risk, such as TNF-α, IL, ACE, GSTs, TLRs and CD14, and there have been reported polymorphisms associated with AP risk in these candidate genes [17]. Yin YW [18] reported the TNF-α gene-308A/G polymorphism is not associated with AP risk. Later on, another meta-analysis [19] conducted by Yin YW suggested the IL-8-251 T/A polymorphism, but not the IL-1b, IL-6 and IL-10 polymorphisms, is associated with an increased risk of AP. As for CD14, it is a pattern-recognition receptor that plays a central role in innate immunity and directs the adaptive immune responses [20]. As a co-receptor of TLRs, CD14 acts primarily by transferring LPS and other bacterial ligands from circulating LPS-binding protein to the TLR4/MD-2 signaling complex [21].
The -159C/T polymorphism in CD14 gene has been investigated the association with many diseases, such as inflammatory bowel disease [22], cancer [23], alcoholic liver disease [24], allergic rhinitis [25]. To our knowledge, this is the first meta-analysis which comprehensively assessed the association between CD14-159C/T polymorphism and AP risk. The current meta-analysis including 5 case-control studies and 2143 subjects were conducted to explore the association of the CD14-159C/T polymorphism with AR. Overall, no evidence has indicated that the CD14-159C/T polymorphism was associated with the susceptibility to AP. That is to say, the CD14-159C/T genotype distribution between AP and control group was no significant difference. In the subgroup analysis by ethnicity and severity, similarly, we did not detect any association between CD14-159C/T polymorphism and AP risk in Asians, Europeans, MAP and SAP. The results may be caused by the following reasons: (1) the association between the -159C/T polymorphism of CD14 gene and AP is indeed unrelated; (2) interactions with other genetic variants are the possible reasons; and (3) gene-environmental factors (such as lifestyle) may also affect the results.
It would be hard to interpret results, if significant heterogeneity were present. In this study, we did not find any obvious heterogeneity across studies. In addition, studies have reported that deviation from HWE might reflect the presence of genotyping errors, population stratification and selection bias in the controls [26]. Therefore, individual studies not in HWE were excluded to assess the stability of our results. However, we also failed to detect any association between CD14-159 C/T polymorphism and AP risk after exclusion of two studies [7,10], which further confirmed the results.
Some limitations of this meta-analysis should be addressed. First, because of incomplete raw data or publication limitations, some relevant studies could not be included in our analysis. Second, our results were based on unadjusted estimates, while lacking of the information for the date analysis may cause serious confounding bias. Third, the number of published studies was not large enough, especially in subgroup analysis (e.g. etiology). Thus, we may fail to explore the real association.
In conclusion, this meta-analysis suggested that the CD14-159C/T polymorphism is not associated with AP risk. However, considering the limitations in our study, large and well-designed studies are warranted to validate our findings.
Disclosure of conflict of interest
None.
References
- 1.Neoptolemos JP, Raraty M, Finch M, Sutton R. Acute pancreatitis: the substantial human and financial costs. Gut. 1998;42:886–891. doi: 10.1136/gut.42.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 3.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 4.Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 5.Leung TF, Tang NL, Sung YM, Li AM, Wong GW, Chan IH, Lam CW. The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol. 2003;14:255–260. doi: 10.1034/j.1399-3038.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 6.Balog A, Gyulai Z, Boros LG, Farkas G, Takács T, Lonovics J, Mándi Y. Polymorphism of the TNF-alpha, HSP70-2, and CD14 genes increases susceptibility to severe acute pancreatitis. Pancreas. 2005;30:e46–50. doi: 10.1097/01.mpa.0000153329.92686.ac. [DOI] [PubMed] [Google Scholar]
- 7.Chao YC, Chu HC, Chang WK, Huang HH, Hsieh TY. CD14 promoter polymorphism in Chinese alcoholic patients with cirrhosis of liver and acute pancreatitis. World J Gastroenterol. 2005;11:6043–6048. doi: 10.3748/wjg.v11.i38.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masamune A, Kume K, Kikuta K, Watanabe T, Hirota M, Satoh K, Kanno A, Suzuki N, Kakuta Y, Shimosegawa T. -651C/T promoter polymorphism in the CD14 gene is associated with severity of acute pancreatitis in Japan. J Gastroenterol. 2010;45:225–33. doi: 10.1007/s00535-009-0163-2. [DOI] [PubMed] [Google Scholar]
- 9.Tukiainen E, Kylänpää ML, Puolakkainen P, Kemppainen E, Halonen K, Orpana A, Methuen T, Salaspuro M, Haapiainen R, Repo H. Polymorphisms of the TNF, CD14, and HSPA1B genes in patients with acute alcohol-induced pancreatitis. Pancreas. 2008;37:56–61. doi: 10.1097/MPA.0b013e31815d9bad. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DL, Zheng HM, Yu BJ, Jiang ZW, Li JS. Association of polymorphisms of IL and CD14 genes with acute severe pancreatitis and septic shock. World J Gastroenterol. 2005;11:4409–4413. doi: 10.3748/wjg.v11.i28.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman SH, Salter G, Holmfield JH, Larvin M, McMahon MJ. Soluble CD14 receptor expression and monocyte heterogeneity but not the C-260T CD14 genotype are associated with severe acute pancreatitis. Crit Care Med. 2004;32:2457–2463. doi: 10.1097/01.ccm.0000148008.99716.9c. [DOI] [PubMed] [Google Scholar]
- 12.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 13.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 16.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss FU, Simon P, Mayerle J, Kraft M, Lerch MM. Germline mutations and gene polymorphism associated with human pancreatitis. Endocrinol Metab Clin North Am. 2006;35:289–302. doi: 10.1016/j.ecl.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Yin YW, Hu AM, Sun QQ, Liu HG, Wang Q, Zeng YH, Xu RJ, Hou ZZ, Zhang SJ. Association between tumor necrosis factor-alpha gene -308A/G polymorphism and the risk of acute pancreatitis: a meta-analysis. J Surg Res. 2012;178:409–414. doi: 10.1016/j.jss.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Yin YW, Sun QQ, Feng JQ, Hu AM, Liu HL, Wang Q. Influence of interleukin gene polymorphisms on development of acute pancreatitis: a systematic review and meta-analysis. Mol Biol Rep. 2013;40:5931–5941. doi: 10.1007/s11033-013-2700-6. [DOI] [PubMed] [Google Scholar]
- 20.Pugin J, Heumann ID, Tomasz A, Kravchenko W, Akamatsu Y, Nishijima M, Glauser MP, Tobias PS, Ulevitch RJ. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 21.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes Infect. 2002;4:903–914. doi: 10.1016/s1286-4579(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Hu J, Fan R, Zhou J, Zhong J. Association between CD14 gene C-260T polymorphism and inflammatory bowel disease: a meta-analysis. PLoS One. 2012;7:e45144. doi: 10.1371/journal.pone.0045144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Guo X, Yu S, Song J, Zhang J, Cao Z, Wang J, Liu M, Dong W. Association between CD14 Gene Polymorphisms and Cancer Risk: A Meta-Analysis. PLoS One. 2014;9:e100122. doi: 10.1371/journal.pone.0100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng T, Zhang CL, Han XY, Zhao S, Xie KQ. Association between CD14-159C>T polymorphisms and the risk for alcoholic liver disease: a meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:1183–1189. doi: 10.1097/MEG.0b013e3283612ff1. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Wang J. Association of CD14 gene-159C/T polymorphism with allergic rhinitis risk: a meta-analysis. Eur Arch Otorhinolaryngol. 2014;271:1601–1607. doi: 10.1007/s00405-013-2793-5. [DOI] [PubMed] [Google Scholar]
- 26.Boccia S, De Feo E, Gallì P, Gianfagna F, Amore R, Ricciardi G. A systematic review evaluating the methodological aspects of meta-analyses of genetic association studies in cancer research. Eur J Epidemiol. 2010;25:765–775. doi: 10.1007/s10654-010-9503-z. [DOI] [PubMed] [Google Scholar]


