Synopsis
Salmonella, Shigella, and Yersinia cause a well-characterized spectrum of disease in humans, ranging from asymptomatic carriage to hemorrhagic colitis and fatal typhoidal fever. These pathogens are responsible for millions of cases of food-borne illness in the U.S. each year, with substantial costs measured in hospitalizations and lost productivity. In the developing world, illness caused by these pathogens is not only more prevalent, but is also associated with a greater case-fatality rate. Classical methods for identification rely on selective media and serology, but newer methods based on mass spectrometry and PCR show great promise for routine clinical testing.
Keywords: Salmonella, Shigella, Yersinia, Enteric, Gram-negative bacilli, gastroenteritis
Introduction
In this review, we discuss three enteric pathogens: Salmonella, Shigella, and Yersinia. These important members of Enterobacteriaceae are responsible for significant morbidity and mortality, causing diarrhea and a spectrum of associated symptoms from mild to severe in most parts of the world. In this review, we cover infection and epidemiology, taxonomic classification, collection, transport, and storage of specimens, culture techniques, molecular detection methods, susceptibility testing, and treatment. Discussions that pertain to individual organisms are organized into individual sections starting with Salmonella, followed by Shigella, then Yersinia. Topics common to all three (such as collection, transport and storage of specimens, and molecular multiplex methods of detection) are discussed as they first occur, denoted as such in paragraph headings. Throughout, priority of discussion is placed on newer techniques and data, with less emphasis on details of classical methods available in reference textbooks.
Salmonella Introduction
Members of the genus Salmonella cause a well-characterized spectrum of disease in humans, ranging from asymptomatic carriage to fatal typhoidal fever. In the developed world, food-borne acute gastroenteritis and enterocolitis are the most common forms of Salmonella infection, with an estimated 1.2 million annual cases of non-typhoidal Salmonellosis occurring in the U.S.1–3 Though relatively uncommon in the U.S., typhoid, paratyphoid, and enteric fever constitute a very serious global public health problem, with 25 million new infections and >200,000 deaths occurring annually.4, 5
Salmonella is a member of the Enterobacteriaceae, originally characterized by their ability to metabolize citrate as a sole carbon source and lysine as a nitrogen source, as well as their ability to produce hydrogen sulfide.6 However, classical biochemical testing alone does not unambiguously distinguish key pathogenic members of this genus and modern classification relies instead on serology and increasingly on molecular methods.
Salmonella Disease Manifestations
Infection with Salmonella typically follows two very different disease courses, depending on whether the infecting Salmonella strain is a typhoidal or non-typhoidal serovar. Infection with non-typhoidal serovars ordinarily presents as diarrhea associated with fever and abdominal cramping 12–72 hours after infection.7 In most cases in healthy individuals, this infection runs a self-limited course over 4–7 days but, in susceptible hosts, certain non-typhoidal strains of Salmonella may spread systemically to other sites in the body. Though this is more common in those with compromised immune systems or underlying medical conditions (e.g., sickle cell anemia), systemic spread of non-typhoidal Salmonella strains may be seen in otherwise healthy individuals as well.
In contrast to infection with non-typhoidal Salmonella, infection with typhoidal strains (primarily serovars Typhi and Paratyphi) presents as a systemic, often serious, disease. After invading through intestinal mucosa, typhoidal strains disseminate through a transient primary bacteremia that may occur without diarrhea.5 Following hematogenous dissemination, some individuals will develop typhoid fever, which involves high temperature (>39° C), vomiting, and headache, sometimes with complications that include neurologic involvement, intestinal perforation and death.5
Salmonella Taxonomic classification
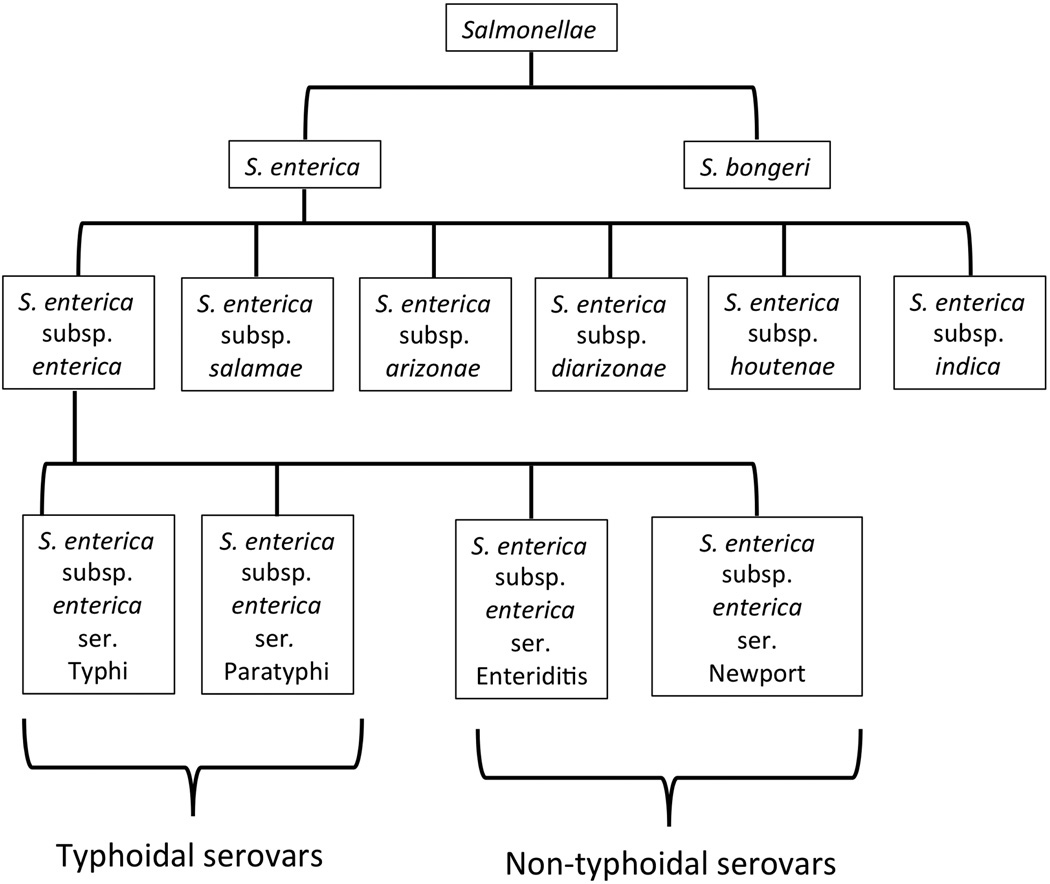
The classification of the salmonellae has a complicated history, resulting in part from multiple independent investigators using phenotypic, serologic, and genotypic methods to characterize phylogenetic relationships within the genus, and in part from disagreements on nomenclature. The most recent consensus defines a classification scheme that recognizes two principle species of Salmonella: S. enterica and S. bongori (Figure 1). In this scheme, S. enterica is further classified into six subspecies: Subspecies I, or S. enterica subsp. enterica; Subspecies II, or S. enterica subsp. salamae; Subspecies IIIa, or S. enterica subsp. arizonae; Subspecies IIIb, or S. enterica subsp. diarizonae; Subspecies IV, or S. enterica subsp. houtenae; and Subspecies VI, or S. enterica subsp. indica.8–10 Recent sequence analysis has shed light on the genetic relationships within this genus and has largely supported the above classification scheme.4, 11–20
Figure 1.
Relationships within the Salmonella genus, including species, subspecies, and serovar designations are illustrated. Note that serovars do not have official taxonomic status. Four representative serovars are shown for S. enterica subsp. enterica. Only the most common typhyoidal serovars, and representative non-typhoidal serovars are shown. There are greater than 2500 serovars in total, with the most common disease-causing serovars belonging to S. enterica subsp. Enterica.
The seven principle members of the Salmonella genus can be further subtyped by serologic methods, based on three antigens: O, H, and Vi. The serologic typing scheme identifies >2500 serovars.21 The resolution provided by serologic typing methods has proved valuable to epidemiologic tracking of isolates in outbreaks. Given that S. enterica subsp. enterica strains constitute the majority (as much as 99.5%) of isolates cultured from humans and other warm-blooded animals, it is perhaps not surprising that the majority of disease causing serovars belong to this subspecies.6, 21 In contrast, S. bongori and the other members of S. enterica are more commonly isolated from cold-blooded animals and environmental sources, and Salmonellosis caused by serovars representing these other species is relatively rare, though infections do occur.
Collection, Transport and Storage of Specimens for Detection of Salmonella/Shigella/Yersinia
For laboratory diagnosis in cases of gastrointestinal disease, fecal specimens should be collected at the early stages of illness, ideally before antibiotics have been initiated.6, 22 Whole stools are the preferred specimen for culture, and examination of multiple specimens may improve the recovery of Salmonella/Shigella/Yersinia.23 Consultation with the laboratory may be required where specimen rejection rules do not allow for serial cultures. Most commonly used pH-buffered stool transport media are compatible with recovery of Salmonella/Shigella/Yersina, though Cary-Blair transport medium is preferred by many labs, due to its compatibility with other common stool pathogens.6, 22
Fecal specimens should be examined immediately on receipt, or stored at 4° C if plating for culture or inoculation of broth will be delayed for greater than 1–2 hours following collection.6, 22 However, it should be noted that refrigeration of specimens containing Shigella in non-pH-buffered transport media may decrease recovery in culture.22 In cases of suspected systemic spread, as with typhoidal Salmonella, cultures from other sources (blood, bone marrow, lymph node, and bone biopsy) that may be submitted should be collected and transported according to standard procedures appropriate to these specimen types.
Salmonella Culture and Isolation
Stool culture is the most common source from which non-typhoidal serovars of Salmonella are recovered. Non-typhoidal strains of Salmonella may also be recovered from blood and tissue (lymph node, bone marrow, etc.) in cases with systemic spread. Typhoidal strains may be more easily isolated from cultures of extra-intestinal sites than from fecal cultures.6
Salmonella may be cultured on a variety of solid media. Typically, two selective and differential media, one of which is highly selective, are inoculated with the stool specimen. Hektoen and xylose-lysine-deoxycholate (XLD) agars are highly selective and both detect H2S production, facilitating identification of Salmonella species. More highly selective agars, including Salmonella-Shigella, bismuth sulfite, and brilliant green agars may inhibit some strains of Salmonella sp., and thus are often used in combination with a less selective agar.22 For this reason, a less selective differential enteric medium, such as MacConkey or eosin methylene blue, and a nonselective medium, such as 5% sheep blood agar may be inoculated in addition as part of the stool culture work up, depending on lab preference.22 However, the growth of fecal flora on non-selective agars may obscure Salmonella colonies that are present in low numbers.
Stool may be inoculated into enrichment broths in addition to plated cultures. Enrichment broths serve to allow the growth of Salmonella while suppressing the growth of normal fecal flora, and thereby can improve recovery yield. Two commonly used enrichment broths are tetrathionate broth and selenite broth.22 Once isolated, Salmonella should be sub-cultured using standard techniques to obtain colonies for identification and susceptibility testing (if indicated), as well as for submission to the local public health laboratory.
Identification Methods for Salmonella
Once Salmonella has been isolated in culture, there are a variety of methods available for identification and classification.6 Definitive identification ordinarily relies on a combination of phenotypic and serologic methods. The widely-used Kauffmann-White serologic typing scheme is based on the LPS O antigen, the H1 and H2 flagellar antigens, and the Vi antigen.21 While the O and H1 antigens are detectable in almost all strains of Salmonella, the H2 antigens are present only in certain strains and the Vi antigen is found predominantly in typhoidal strains.6 It should be noted that Vi, though useful for detection of serovar Typhi, may also be expressed in Citrobacter sp. and therefore the Vi antigen alone cannot be used for definitive identification.6
O/H/Vi typing by the Kauffmann-White scheme yields greater than 2500 serovars, which may be designated by antigenic formulae expressed with the following convention: O antigen(s), Vi antigen if present: phase 1 H antigens(s):phase 2 H antigen(s) if present 6, 21. In the Kauffman-White scheme, the >1500 serovars from S. enterica subsp. enterica receive names; those from the other subspecies of S. enterica and S. bongeri are referred to only by formulae.21 By convention serovar names are capitalized and not italicized, and it is not necessary to include the enterica subspecies designation when referring to the serovar, as only subspecies enterica serovars are named. Thus both S. enterica subsp. enterica ser. Enteriditis and S. enterica ser. Enteriditis are correct designations 21. It is important to note that serovars do not have taxonomic status and should not be confused with species or subspecies of Salmonella.
As stated above, the majority of identified serovars and the majority of human pathogens belong to S. enterica subsp. enterica. Some serovars, such as Typhi and Paratyphi are largely restricted to humans. Other serovars are generally restricted to particular zoonotic reservoirs and only occasionally cause human infection. However, infections caused by these serovars may be severe with systemic spread. These species include Derby and Choleraesuis (pig-adapted), Saintpaul (poultry-adapted), and Dublin (bovine-adapted).6 Yet other serovars, particularly Typhimurium and Enteritidis pass between human and zoonotic reservoirs with ease, perhaps accounting for their high prevalence. CDC Survey data from 2011 suggests that the three most commonly reported serovars in the U.S. during the reporting period were Enteritidis, Typhimurium, and Newport.24 Non-typhoidal Salmonellosis was traditionally associated with meat and poultry products, but more recently outbreaks have been caused increasingly by produce.1–3
A number of techniques now exist for screening of primary specimens and enrichment broths before isolated colonies are available for the above methods. Selenite enrichment broth cultures may be screened by the Wellcolex Color Salmonella (Remel Inc., Lenexa, KS), a serologic agglutination-based method.25 There are important limitations in sensitivity and specificity that one must be aware of with the Wellcolex test; in particular, expression of Vi antigen in Citrobacter sp. and may lead to false positives with this assay and not all Salmonella sp. are identified by this test.26
Antimicrobial Susceptibility Testing for Salmonella
Current CLSI guidelines 27 state that routine susceptibility testing is indicated for typhoidal Salmonella serovars (Typhi and Paratyphi A-C) from all sites, and for non-typhoidal serovars from extra-intestinal sites. Fecal isolates should be tested for susceptibility to ampicillin, a fluoroquinolone, and trimethoprim-sulfamethoxazole. For extra-intestinal isolates of Salmonella, a third-generation cephalosporin should be tested and reported additionally, and chloramphenicol may be tested and reported if requested. It should be noted that 1st and 2nd generation cephalosporins, cephamycins and aminoglycosides may appear active in vitro against Salmonella, but are not effective clinically and should not be reported as susceptible.27
Current CLSI breakpoints for ciprofloxacin and levofloxacin for Salmonella are lower than for other Enterobacteriaceae.27 CLSI recommends that laboratories unable to implement the current (lowered) MIC breakpoints for fluoroquinolone testing should use nalidixic acid to test for reduced fluoroquinolone susceptibility. Strainsresistant to nalidixic acid may be associated with clinical fluoroquinolone failures. Salmonella strains producing ESBL beta-lactamases and NDM-1 type carbapenemases have been reported, as well as S. enterica ser. Typhi isolates with chromosomally-integrated multi-drug resistance islands.28–30 It is therefore important for the laboratory and clinicians to be aware of the possibility of multi-drug- and carbapenem-resistant isolates.
Identification of Salmonella/Shigella/Yersinia by Mass Spectrometry
Molecular methods, including mass spectrometry and PCR-based multiplex panels have been developed for the detection of enteric bacteria, and some laboratories are beginning to incorporate these techniques. These newer methods have the potential to improve our ability to provide rapid and accurate bacterial identification, but they have limitations that are important to understand. Commercial matrix assisted laser desorption ionization – time of flight mass spectrometry (MALDI-TOF MS) instruments can provide rapid identifications of Salmonella and Yersinia, and limited identification of Shigella.31, 32 Both the Bruker Biotyper and Biomerieux Vitek MS MALDI-TOF MS systems are FDA approved for in vitro diagnostic use to identify cultured isolates of Y. enterocolitica and Y. pseudotuberculosis (species level identification) and Salmonella (genus level identification), but not Shigella. The Biomerieux Vitex MS system carries a manufacturer’s note that confirmatory testing is recommended when identifications of Salmonella are made. Neither system is FDA approved for species-level or serovar-level classifications of Salmonella. However, recent literature suggests that MALDI-TOF mass spectrometry-based systems may have the power to distinguish among subspecies and some Salmonella serovars.33 As this technology is new and experience is still relatively limited, caution is appropriate, as illustrated by a recent report of a patient with both bacteremia and gastrointestinal symptoms for whom an isolate of Y. pseudotuberculosis was misidentified as Y. pestis using a MALDI-TOF MS method. This highlights the need for comprehensive MALDI-TOF MS spectral databases in combination with careful validations and appropriate additional confirmatory testing for this group of less commonly seen bacteria.34
Pulsed field gel electrophoresis (PFGE) has been used extensively for epidemiologic typing of strains of Salmonella. Recently, a variety of molecular methods have been developed for distinguishing among strains of Salmonella/Shigella/Yersina, including multi-locus sequence typing approaches, and PCR-based approaches.35–42 Whole genome sequencing-based approaches have also been used, facilitated by published reference genomes.13, 14, 43–45
PCR-Based Molecular Methods to Detect Salmonella/Shigella/Yersinia
Recently, a number of PCR-based multiplex GI pathogen identification panels have been marketed for use with primary stool specimens. These include the bioMerieux Biofire Filmarray system,46 BD MAX system,47 Luminex xTAG system,48, 49 Savyon Diagnostics system,48 and Genetic Signatures system.50 These panels allow rapid identification of Salmonella, Shigella, and Yersinia from primary stool specimens, and offer substantially improved turnaround time on primary laboratory diagnosis compared with culture-based methods. Recovery of isolates from culture is still required for taxonomic classification and susceptibility testing. It should be noted that none of the commercially available multiplex PCR panels include Y. pseudotuberculosis in the target list.
Though experience with these new panels is still relatively limited, a number of recent publications have begun to shed light on their performance. Wessels et al. found that in comparison with conventional diagnostic methods, the Luminex xTAG panel reported almost twice as many pathogen identifications.51 However, not all of these positive results were confirmed by independent PCR assays, including one Shigella sp. and four Salmonella sp. out of a total of 83 positives reported by the instrument. The authors therefore recommended that confirmatory testing be performed before reporting Salmonella with this method. A study of Luminex xTAG ASR reagents demonstrated sensitivity for Salmonella of 92% and for Shigella of 93%.52 Conclusions regarding the sensitivity of this assay for detecting Yersinia sp. were limited by the inclusion of only 3 isolates.52 A study comparing the Biofire FilmArray and Luminex xTAG systems demonstrated higher positivity rate with both assays compared to routine methods; however, presumed false positives were observed, consistent with other studies.46 The Genetics Signatures EasyScreen system tested in Australia showed very good correlation of results between conventional and molecular methods for Salmonella, but evaluation of performance for the detection of Shigella and Yersinia was limited by the small numbers of isolates of these genera included in the study.50 Adequate validation of these multiplexed panels can be quite difficult due to the number of pathogens that must be tested, and the need to verify all presumed false positives with another equivalently-sensitive method. Significant testing and experience should be accumulated before a lab considers replacing traditional methods with a single molecular method.
Another approach under investigation is PCR electrospray ionization mass spectrometry (PCR-ESI-MS). Pierce et al. evaluated a PCR-ESI-MS-based assay for testing of enteric pathogens in food samples that employed broad-range oligonucleotide primers targeted to highly conserved regions in the genomes of target organisms 53. Salmonella was detected in samples tested by this method with high sensitivity, but Shigella was detected in only 81% of cases. As with MALDI-TOF MS-based methods, PCR-ESI-MS-based assays are very dependent on the completeness of the database. In this study, identification of bacteria from food samples required the use of selective enrichment media and additional incubation time. Further work is needed to incorporate this method into routine use by food safety investigators.53
Shigella Epidemiology, Disease Manifestations & Treatment
Symptoms of Shigella infection include fever, malaise, watery diarrhea, cramping abdominal pain, and myalgia. The incubation period is 1–4 days, and the illness often resolves in 5–7 days. After 2–3 days, the volume and frequency of diarrhea may decrease to be replaced with blood and mucus in feces (dysentery), along with straining. Some individuals may not have symptoms, but can still transmit the bacteria to others.54 Although not as frequent as gastrointestinal disease, there are multiple reports of bacteremia and other extraintestinal infections.55–57 Complications of Shigella infection include hemolytic uremic syndrome, and reactive arthritis.58
The worldwide incidence of shigellosis has been reported to be approximately 165 million cases, but the mortality has decreased substantially over the past three decades.59–61 Though the causes of this decrease in shigellosis-associated mortality are likely to be multifactorial, it has occurred along with a decrease in prevalence of S. dysenteriae type 1.61 Infection with S. dysenteriae type 1 carries relatively high mortality in the developing world, as demonstrated by the case fatality rate of 5–15% in Africa and Central America. There are an estimated 500,000 cases of shigellosis per year in the U.S. with 38 deaths.62
The only known natural reservoirs of Shigella are humans and large primates. Shigellosis is highly infectious, with a minimum inoculum of only 10–100 organisms required to cause disease.63 Outbreaks often occur in the summer months. Transmission is through the fecal-oral route, with spread occurring person-to-person, and through consumption of water or food contaminated with feces from infected individuals. Not surprisingly, shigellosis seen more commonly under conditions that facilitate spread of bacteria through the fecal-oral route, such as in daycare centers or in areas without indoor plumbing. Outbreaks have additionally been described among people in custodial institutions, orthodox Jews, international travelers, and men who have sex with men.64–68 Asymptomatic infection is associated with the spread of disease and prolongation of outbreaks due to silent transmission. Severe intestinal and extra-intestinal manifestations can occur with all four species of Shigella, but are most common with S. dysenteriae type 1, due in part to the production of Shiga toxin.69 Infections due to S. dysenteriae are often acquired by international travel and are often multidrug resistant.70 Shigellosis due to S. boydii is uncommon, and limited to the Indian subcontinent.71 S. sonnei is endemic in the United States and other developed countries. Infections associated with S. sonnei tend to be mild or asymptomatic.
Antimicrobial therapy has been demonstrated to decrease the duration, transmission, and severity of symptoms. A decision to treat is based on severity of symptoms, and on the desire to reduce spread, balanced against the goal to reduce the development of more resistant isolates. In the U.S., shigellosis is reportable to state-level public health labs, and Shigella resistance is tracked by the National Antimicrobial Resistance Monitoring System for Enteric Bacteria, a collaboration of multiple U.S. government agencies.
Shigella Laboratory Identification
Shigella, a member of the Enterobacteriaceae family, is classified into four serologic subgroups: S. dysenteriae (Group A), S. flexneri (Group B), S. boydii (Group C), and S. sonnei (Group D). Though many authors have treated these subgroups of Shigella as distinct taxonomic species, Escherichia coli and Shigella are very similar genetically (80–90%), and the argument has been made that almost all Shigella strains could be considered a biotype of E. coli.72 S. boydii 13 is the exception and has been reclassified as Escherichia albertii.73, 74 Because of their substantial genetic similarity, distinguishing Shigella from E. coli often presents a challenge for the clinical microbiology laboratory. However, though they are genetically similar, Shigella and non-Shiga toxin-producing strains of E. coli demonstrate different clinical behavior, and their distinction in the microbiology laboratory facilitates their continued epidemiological tracking as distinct entities.
To optimize detection of Shigella in stool, samples should be plated on MacConkey and either xylose-lysine-deoxycholate, Hektoen enteric, or deoxycholate citrate agar. Colonies are bluish-green on Hektoen agar and do not have the black center seen with Salmonella, as Shigella do not produce H2S. Shigella do not ferment lactose and xylose and are relatively inert biochemically. Some strains of S. sonnei are exceptions and may ferment lactose. Most isolates do not produce gas, except some S. flexneri.6, 75 Appropriate colonies can be characterized further using Kligler iron or triple sugar iron agar. Lysine decarboxylase tests are typically negative for Shigella. Groups A, B, and C have similar biochemical characteristics, but S. sonnei has ornithine decarboxylase activity and beta-galactosidase activity.75
Serologic typing should be performed on Shigella isolates. Subgroup A (S. dysenteriae) has 17 serotypes, subgroup B (S. flexneri) has 14 serotypes further divided into subserotypes, subgroup C (S. boydii) has 19 serotypes, and subgroup D (S. sonnei) is a single serotype.76, 77 Confirmation of identification and serotype should be performed at a reference lab or public health lab for isolates from extraintestinal infections, or when the primary lab does not have full serotyping capabilities. Enteroinvasive E. coli are evolutionary intermediates between E. coli and Shigella.78 Some of the enteroinvasive E. coli (EIEC) may cross-react with Shigella anti-sera. It should be noted that serological testing of the serum of an infected individual is generally not useful for diagnosis.
Active or typical E. coli strains can be distinguished from Shigella by several tests, including lysine decarboxylase, motility, gas production, acetate utilization, and mucate and lactose fermentation. However, inactive strains of E. coli can have overlapping reactions with Shigella. Inactive strains of E. coli that are nonmotile, anerogenic biotypes are called Alkalescens-Dispar bioserotypes. In order to address this issue, different laboratories have developed different algorithms for the work-up of E coli and Shigella in stool specimens; however, many labs do not employ procedures to distinguish E. coli from Shigella when isolated from other body sites.
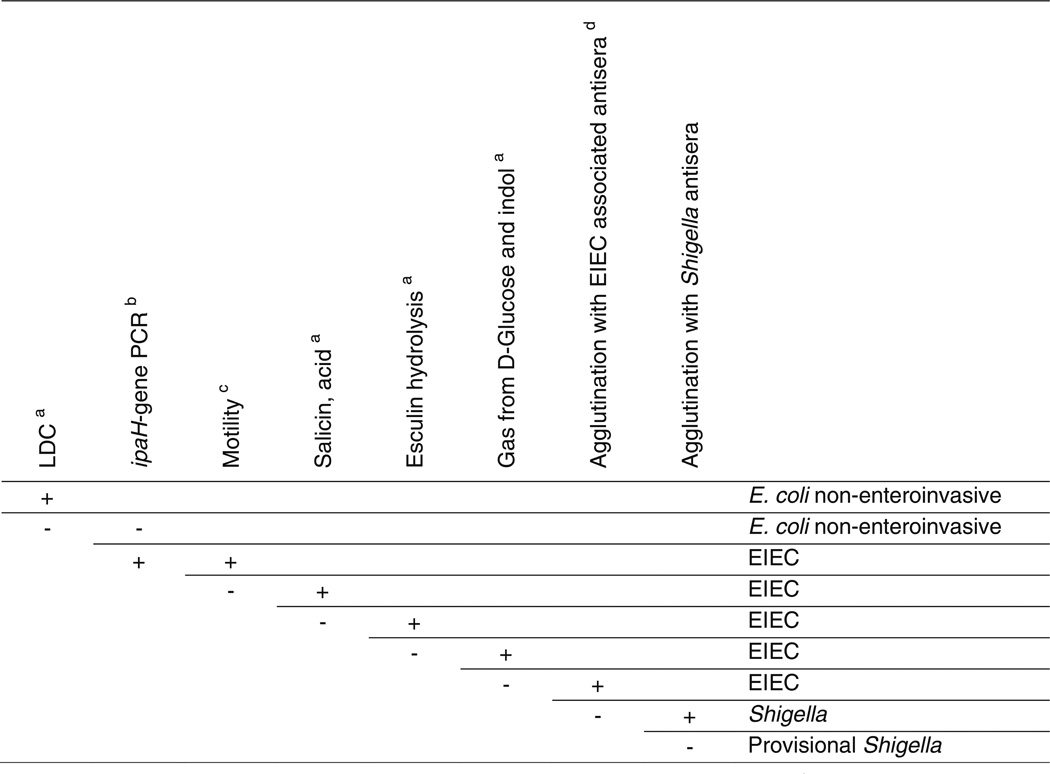
Given that E. coli is a much more common isolate to recover from most sterile site cultures than Shigella, and given that many labs restrict procedures to distinguish Shigella from E. coli to stool specimens, a Shigella occurring in a sterile site specimen may be misidentified as an E. coli. This is especially likely if identification is dependent on an automated system, as commercial systems such as the Vitek 2 have a high rate of misidentification.79 There are a number of reports examining API20E strips for the identification of Shigella, with varying reports of accuracy.80–83 If a Shigella isolate from an extra-intestinal site were misidentified as an E. coli, the reflex susceptibility performed and reported would be that appropriate for an E. coli isolate. However, the CLSI guidelines differ for the two organisms, with first and second generation cephalosporins, cephamycins, and aminoglycosides excluded from testing for Shigella isolates, as false in vitro susceptibility my occur.27 As a consequence, a Shigella misidentified from a site such as blood, could mistakenly be treated with an ineffective agent (e.g. aminoglycoside), with possible clinical failure. Furthermore, such a case could be missed in the tracking of Shigella incidence, unless Shigella was isolated from a concurrent stool specimen. An algorithm to distinguish E. coli and Shigella is presented in Figure 2.72
Figure 2.
Key for differentiation of Shigella, enteroinvasive Escherichia coli (EIEC), and noninvasive Escherichia coli. (From van den Beld MJ, Reubsaet FA. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 2012;31(6):899–904; with permission)
- Based on Edwards and Ewing’s identification of Enterobacteriaceae, 4th edition, 1986 and/or Cowan and Steel’s manual for the identification of medical bacteria, 3rd edition, 1993.
- Performed with a standard PCR protocol, with primers designed to amplify a part of the conserved region of ipaH7.8, as described by Buysse et al., Microb. Pathog. 19(5):335–349.
- Incubated for 24 h in BHI-medium at 37°C.
- Known O:H serotypes of EIEC according to Bergey’s manual of Systematic Bacteriology, 2nd edition, volume 2, The Proteobacteria, Part B The Gammaproteobacteria.
Given the long-standing difficulty in distinguishing Shigella from E. coli, some newer methods have been evaluated. Khot and Fisher applied ClinProTools software from Bruker Daltonics to develop and algorithm to distinguish Shigella from E. coli using MALDI-TOF mass spectrometry. The 3% rate of misidentification these authors achieved is better than that seen with automated biochemical instruments. However, given the still significant error rates, such an approach does not obviate the need for traditional biochemical approaches.84
Guidelines have been written to test all stool of all patients with acute, community-acquired diarrhea for Shiga-toxin producing E. coli, because selective testing strategies such as testing only bloody stool, or stool from children, or during summer months, fail to detect all positive samples.85 Due to low prevalence, not all hospitals have chosen to test all such stool samples. The Shiga toxin produced by S. dysenteriae type 1 is very similar to the Shiga toxin 1 produced by some enterohemorrhagic E. coli, so some rapid assays may detect the toxin from either E. coli or this specific Shigella species.86–88 Shigellas that do not produce the toxin should not be detected with those assays. It should be noted that Shiga toxin-encoding genes can be detected in healthy volunteers by PCR of fecal samples, so correlation with clinical symptoms and culture of an isolate are essential for diagnosis when using this type of molecular assay.89
Shigella Susceptibility Testing
Shigella solates should be tested against ampicillin, a fluoroquinolone, and trimethoprim-sulfamethoxazole.27 There is significant resistance to ampicillin and trimethoprim-sulfamethoxazole.68, 90 In Asia, resistance has been reported to ciprofloxacin, pivmecillinam, and azithromycin, with some emerging resistance to third-generation cepholosporins and additional drugs.91–96 A recent report from studies in Bangladesh show an increase in resistance to ciprofloxacin from 0% in 2004 to 44% in 2010.97
Shigella Virulence Mechanism
Shigella pathogenesis involves translocation through ileal and colonic M cells, uptake by macrophages, basolateral invasion of epithelial cells, and dissemination within the mucosa.98, 99 Shigella has a large virulence plasmid, encoding a Type III-secretion system and a set of secreted proteins. The system results in the injection of virulence factors into host colonic epithelial cells, leading to damage to the epithelial lining, as well as proteins that antagonize the adaptive and innate immune response.98, 100 Shigella virulence is enhanced by the presence of enterotoxins, which is separate from the Shiga toxin produced in limited strains.101 The genes encoding the Shiga toxin genes are located in the genome of heterogeneous lambdoid prophages. These are highly mobile genetic elements that play a role in horizontal gene transfer and genome diversification.102 Shiga toxin is made by the Shigella dysenteriae type I strains and E. coli O157:H7 among other E. coli shiga toxin-producing strains. Shiga-like toxin II, also seen in some E. coli strains is 56% homologous with Shiga toxin (or Shiga-like toxin I).103
Shigella Vaccine
The declining incidence of shigellosis with increasing age suggests that natural immunity develops, implying that vaccines eliciting this natural response may be effective. There is currently no licensed Shigella vaccine, but there are multiple vaccines are in preclinical stages or clinical trials, including live attenuated, killed whole-cell, conjugate, and subunit vaccines.104 Vaccines have been designed against the O antigens, and also conjugate vaccines have been developed using several different conserved antigenic molecules.101 Data are limited regarding the immune parameter that correlates with protective immunity.77 More studies are needed before we will know if a Shigella vaccine will prove effective.
Yersinia Epidemiology & Disease Manifestations
Yersinia are zoonotic agents distributed worldwide and there are both pathogenic and nonpathogenic strains.105 Yersinia species associated with human disease include Y. pseudotuberculosis, Y. enterocolitica, and Y. pestis. Of these clinically significant species, we will discuss only Y. enterocolitica and Y. pseudotuberculosis, as this review is focused on enteric pathogens. Disease due to Y. enterocolitica manifests as terminal ileitis, lymphadenitis, and acute enterocolitis due to ingestion of contaminated food or water. Symptoms typically develop 4–7 days after exposure and may last 1–3 weeks. Complications include skin rash, joint pains, and bacteremia.106 Right-sided abdominal pain in some cases of Y. enterocolitica infection may be confused with appendicitis. Illness may include erythema nodosum, polyarthritis, and less commonly septicemia or endocarditis.107–110 Reactive arthritis is an uncommon complication, but may be seen in especially in immunocompromised patients and those with the HLA-B27 allele. Associated disorders also include inflammatory bowel disease and autoimmune thyroid disorders. Iron overload due to underlying conditions, such as hereditary hemochromatosis or beta thalassemia, or treatment with deferoxamine, leads to an increased susceptibility to septicemia manifestations.105, 111
Infections with Y. enterocolitica are more common in the winter months and often seen in young children. There is one culture-confirmed case of Y. enterocolitica per 100,000 individuals each year in the United States. The CDC tracks foodborne disease with the surveillance network, FoodNet. The source of infection can include contaminated prepackaged deli meats, undercooked pork, unpasteurized milk, or untreated water. Preparation of chitterlings (large intestines of pigs), a dish for holiday meals, has been associated with multiple cases of infection in infants, due to caregivers handling of the contaminated food112–117 This organism is able to multiply at refrigerated temperatures, contributing to its role in food products and transfusion-related infections.118
Y. pseudotuberculosis is endemic in a variety of animals, including fowl. Y. pseudotuberculosis usually produces a self-limited disease. The infection can manifest as mesenteric lymphadenitis, and may also be confused with appendicitis.119 Septicemic illness is rare and if seen, would most likely occur in someone with underlying disorders that increase susceptibility to severe infection.120–122 Y. pseudotuberculosis has been reported as a foodborne pathogen.123, 124
Yersinia Laboratory Identification
Yersinia is a gram-negative bacillus in the Enterobacteriacae family. There are 11 species of Yersina, but only three are clearly human pathogens.75 Genome sequencing reveals that Y. pestis and Y. pseudotuberculosis are closely related, but significantly different than Y. enterocolitica.125 Yersinia can appear small and coccobacillary in Gram-stained smears. It exhibits bipolar staining described as a safety pin shape on Giemsa staining. Yersinia grow on blood, chocolate, and MacConkey agar, but may be overgrown by other organisms due to slow growth. Yersinia can form pinpoint colonies on both blood agar and MacConkey agar in 24 hours, particularly Y. pseudotuberculosis. Yersinia are catalase positive, oxidase negative and ferment glucose. Yersinia species have an alkaline over acid pattern on Kligler Iron Agar and are nonmotile at 36°C, but motile at 22°C. Optimal growth is observed at 25–32°C. Longer incubation reveals grey-white, 1–2 mm, convex colonies. Y. enterocolitica appears as small, lactose-negative colonies on MacConkey in 48 hours. Selective media, such as cefsulodin-irgasan-novobiocin (CIN) and incubation at lower temperatures can enhance detection. Colonies appear as a bull’s-eye with a red center on CIN agar, although some other bacteria will also give this appearance. Use of eosin methylene blue agar and triple-sugar iron agar may not result in a clear distinction from other coliforms. Y. enterocolitica incubated at room temperature will be more biochemically active and better identification at this temperature may be obtained with some systems such as API 20E, because commercial systems such as Vitek may fail to identify these organisms under routine conditions.126, 127
Y. pseudotuberculosis is a pleomorphic gram-negative bacillus that can grow at temperatures ranging from 4°C to 43°C, with optimal temperatures between 25–28°C.128 Enteric Yersinia species can be distinguished biochemically; ornithine decarboxylase, sucrose, and sorbitol are all positive for Y. enterocolitica and negative for Y. pseudotuberculosis. Enzyme-linked immunosorbent assays and lateral-flow assays exist, but are not commonly used for routine labs, instead being used primarily in research.
Yersinia Serogroups
Of the six biovars of Y. enterocolitica (1A, 1B, 2, 3, 4), , five are pathogenic for humans.129 It has been proposed that biotype 1A strains may represent a potential group of emerging pathogens. They can be identified in clinical specimens, but clear association with human disease has not been established.130 Serology has been used in investigations of plague infections caused by Y. pestis, in cases when an organism was not recovered in culture. However, serology for Y. enterocolitica and Y. pseudotuberculosis is somewhat limited by cross-reactions due to infections with other bacterial species, and due to human exposure to nonpathogenic Yersinia species.
Yersinia Virulence
One virulence factors of Yersinia is yersiniabactin , a siderophore iron uptake system present in virulent Yersinia, including Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biotype 1B. The genes involved in the biosynthesis, transport, and regulation of yeriniabactin are located on a mobile genetic element, the high-pathogenicity island. This mobile element has contributed to the horizontal spread of yersiniabactin genes into other Enterobacteriacae. In addition, Yersina species contain alternative iron siderophore scavenging systems, with some strains containing all three systems, yersiniabactin, pseudochelin, and yersiniachelin.131
Y. pseudotuberculosis has a virulence plasmid, pYV, that carries a type III secretion system that forms a needle structure on the bacterial surface for the injection of Yersinia outer membrane proteins (Yops) into target host cells. This system results in disruption of both the innate and adaptive immune response, inhibiting phagocytosis.128
Yersinia Susceptibility Testing
Some Y. enterocolitica have chromosomally encoded beta lactamases, conferring resistance to ampicillin, cephalothin, and carbenicillin. Treatment of Y. enterocolitica may include an aminoglycoside, doxycycline, trimethoprim-sulfamethoxazole, a third-generation cephalosporin, or a fluoroquinolone.106 There is significant resistance to fluoroquinolones in some regions, due to mutation of the gyr A gene and efflux mechanisms.132 Y. pseudotuberculosis infections do not usually require treatment, but bacteremia may be treated with ampicillin, tetracycline, or streptomycin. The organism is usually susceptible to extended-specturm cephalosporins, aminoglycosides, chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole.133
Chronic disease associated with Salmonella/Shigella/Yersinia Infections
An examination of cases of gastroenteritis due to Salmonella, Campylobacter, Shigella, or Yersinia in US military personnel lead to a report of a higher incidence of chronic health sequelae than some previous reports.134 Pathogen-specific increases were observed with associated increased risk of irritable bowel disease, dyspepsia, constipation, and gastroesophogeal reflux disease, with a relative risk of 13.1 for inflammatory bowel disease following Yersinia infection. Three separate metaanalyses have shown a significantly increased risk of irritable bowel syndrome following gastroenteritis due to Shigella.135–137
Key points.
The global disease burden from Salmonella/Shigella/Yersinia, organisms reportable to public health departments, is quite high, with significant but fewer infections in the developed world.
Although there is a broad spectrum of disease associated with these pathogens, infections are self-limiting in the majority of cases. Antimicrobial therapy is ordinarily indicated only for severe gastrointestinal and systemic disease.
Inactive strains of E. coli can be difficult to distinguish from Shigella strains, using multiple laboratory methods.
Molecular methods such as PCR and mass spectrometry are being used more routinely for identification of these organisms, but both methods have limitations.
There is active investigation in the areas of vaccine development and characterization of virulence mechanisms.
Box 1. Prevention of Yersinia Infections.
Avoid raw or undercooked pork.
Avoid unpasteurized milk and milk products.
Wash hands with soap and water before preparing food or eating, and after contact with animals or raw meat.
Avoid contact with young children while preparing raw chitterlings.
Clean all kitchen boards and utensils after preparing raw meat.
Dispose of animal feces in a sanitary manner.
(From Yersinia. National Center for Emerging and Zoonotic Infectious Diseases. Centers for Disease Control and Prevention (CDC). Available at: http://www.cdc.gov/nczved/divisions/dfbmd/diseases/yersinia/)
Box 2. Barriers to development of a Shigella Vaccine.
We have a limited understanding of protective immunity for Shigella.
There are any serotypes of Shigella.
There is a lack of an adequate animal model.
Concerns of producing reactive arthritis in vaccine trial recipients.
Concerns of gastrointestinal tract side effects or chronic conditions.
Challenges in manufacturing process.
Some promote funding for clean water and sanitation over vaccine studies.
From Barry EM, Pasetti MF, Sztein MB, et al. Progress and pitfalls in Shigella vaccine research. Nature reviews Gastroenterology & hepatology 2013;10(4):245–255; with permission.
SELF ASSESSMENT.
-
Which of the following statements is true regarding Salmonella?
S. bongeri is a common cause of Salmonellosis in humans.
Typhus is caused by a serovar of S. enterica subsp. Indica.
Serovars of Salmonella have official taxonomic status similar to the named Salmonella subspecies.
Typhoidal strains of Salmonella are more likely to cause systemic disease than non-typhoidal strains.
Unpasteurized milk products are the most common source of human Salmonella infection in the U.S.
-
Which of the following statements is true regarding Shigella?
Diarrhea from Shigella infection is almost always bloody.
All patients with Shigella infection should receive antibiotics promptly to avoid sequelae.
The black-centered colony on Hektoen agar is characteristic for Shigella.
Two of the four groups of Shigella species typically carry the shiga toxin gene.
Inactive strains of E. coli are very difficult to distinguish from Shigella with multiple laboratory identification systems.
-
Which of the following statements is true regarding Yersinia?
Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis all have equally similar genomes.
It is common for patients with Y. enterocolitica to have bacteremia as a complication.
There is currently no role for MALDI-TOF MS in the identification of Yersinia due to the poor performance in published studies.
There are multiple reports of chronic gastrointestinal diseases as sequelae of Yersinia infections.
There are 50 serotypes of Yersinia known to be pathogenic in humans.
Answers to multiple choice questions
D
E
D
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patrick ME, Adcock PM, Gomez TM, et al. Salmonella enteritidis infections, United States, 1985–1999. Emerging infectious diseases. 2004;10(1):1–7. doi: 10.3201/eid1001.020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden CR. Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2006;43(4):512–517. doi: 10.1086/505973. [DOI] [PubMed] [Google Scholar]
- 3.Hanning IB, Nutt JD, Ricke SC. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne pathogens and disease. 2009;6(6):635–648. doi: 10.1089/fpd.2008.0232. [DOI] [PubMed] [Google Scholar]
- 4.Holt KE, Thomson NR, Wain J, et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC genomics. 2009;10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annual review of microbiology. 2014;68:317–336. doi: 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- 6.Nataro JP, Bopp CA, Fields PI, Kaper JB, Strockbine NA, Escherichia, Shigella, Salmonella . In: Manual of Clinical Microbiology. 10th ed. Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Vol. 1. Washington, D.C: ASM Press; 2011. pp. 603–626. [Google Scholar]
- 7. www.cdc.gov/salmonella/general/
- 8.Le Minor LaP M. Dsignation of Salmonella enterica sp. nov., nom. rev., as the Type and Only Species of the Genus Salmonella. International Journal of Systematic Bacteriology. 1987;37(4):465–468. [Google Scholar]
- 9.Euzeby JP. Revised Salmonella nomenclature: designation of Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987 sp. nov., nom. rev. as the neotype species of the genus Salmonella Lignieres 1900 (approved lists 1980), rejection of the name Salmonella choleraesuis (Smith 1894) Weldin 1927 (approved lists 1980), and conservation of the name Salmonella typhi (Schroeter 1886) Warren and Scott 1930 (approved lists 1980) Request for an opinion. International journal of systematic bacteriology. 1999;49(Pt 2):927–930. doi: 10.1099/00207713-49-2-927. [DOI] [PubMed] [Google Scholar]
- 10.Tindall BJ, Grimont PA, Garrity GM, et al. Nomenclature and taxonomy of the genus Salmonella. International journal of systematic and evolutionary microbiology. 2005;55(Pt 1):521–524. doi: 10.1099/ijs.0.63580-0. [DOI] [PubMed] [Google Scholar]
- 11.Reeves MW, Evins GM, Heiba AA, et al. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. Journal of clinical microbiology. 1989;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards RA, Olsen GJ, Maloy SR. Comparative genomics of closely related salmonellae. Trends in microbiology. 2002;10(2):94–99. doi: 10.1016/s0966-842x(01)02293-4. [DOI] [PubMed] [Google Scholar]
- 13.McClelland M, Sanderson KE, Spieth J, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413(6858):852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 14.Parkhill J, Dougan G, James KD, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413(6858):848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 15.Porwollik S, McClelland M. Lateral gene transfer in Salmonella. Microbes and infection / Institut Pasteur. 2003;5(11):977–989. doi: 10.1016/s1286-4579(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 16.Chiu CH, Tang P, Chu C, et al. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic acids research. 2005;33(5):1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker S, Dougan G. The genome of Salmonella enterica serovar Typhi. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45(Suppl 1):S29–S33. doi: 10.1086/518143. [DOI] [PubMed] [Google Scholar]
- 18.Litrup E, Torpdahl M, Malorny B, et al. Association between phylogeny, virulence potential and serovars of Salmonella enterica. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2010;10(7):1132–1139. doi: 10.1016/j.meegid.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Lan R, Reeves PR, Octavia S. Population structure, origins and evolution of major Salmonella enterica clones. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2009;9(5):996–1005. doi: 10.1016/j.meegid.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Jacobsen A, Hendriksen RS, Aaresturp FM, et al. The Salmonella enterica pangenome. Microbial ecology. 2011;62(3):487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimont PADaW F. Antigenic Formulae of the Salmonella Serovars. WHO Collaborating Centre for Reference and Research on Salmonella. (9th edition.) 2007:1–166.
- 22.Gilligan PH, Janda JM, Karmali MA, Miller MJ. Laboratory Diagnosis of Bacterial Diarrhea. Cumitech. 1992;12A [Google Scholar]
- 23.Ethelberg S, Olsen KE, Gerner-Smidt P, et al. The significance of the number of submitted samples and patient-related factors for faecal bacterial diagnostics. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2007;13(11):1095–1099. doi: 10.1111/j.1469-0691.2007.01816.x. [DOI] [PubMed] [Google Scholar]
- 24. www.cdc.gov/ncezid/dfwed/pdfs/salmonella-annual-report-2011-508c.pdf.
- 25.Bouvet PJ, Jeanjean S. Evaluation of two colored latex kits, the Wellcolex Colour Salmonella Test and the Wellcolex Colour Shigella Test, for serological grouping of Salmonella and Shigella species. Journal of clinical microbiology. 1992;30(8):2184–2186. doi: 10.1128/jcm.30.8.2184-2186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scientific T. Wellcolex Color Salmonella Rapid Latex Agglutination Test. Package Insert. Queried on 11/22/2014, Available at: http://www.thermoscientific.com/en/product/wellcolex-color-salmonella-rapid-latex-agglutination-test-kit.html.
- 27.CLSI. M100-S24: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. 2014 M100-S24. [Google Scholar]
- 28.Gonzalez-Lopez JJ, Piedra-Carrasco N, Salvador F, et al. ESBL-Producing Salmonella enterica Serovar Typhi in Traveler Returning from Guatemala to Spain. Emerging infectious diseases. 2014;20(11):1918–1920. doi: 10.3201/eid2011.140525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irfan S, Khan E, Jabeen K, et al. Clinical isolates of Salmonella enterica serovar Agona producing NDM-1 metallo-beta-lactamase: First report from Pakistan. Journal of clinical microbiology. 2014 doi: 10.1128/JCM.02396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiou CS, Alam M, Kuo JC, et al. Chromosome-mediated multidrug resistance in Salmonella enterica serovar Typhi. Antimicrobial agents and chemotherapy. 2014 doi: 10.1128/AAC.04081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng J, Fu L, Wang R, et al. Comparison of MALDI-TOF MS, gene sequencing and the Vitek 2 for identification of seventy-three clinical isolates of enteropathogens. Journal of thoracic disease. 2014;6(5):539–544. doi: 10.3978/j.issn.2072-1439.2014.02.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparbier K, Weller U, Boogen C, et al. Rapid detection of Salmonella sp. by means of a combination of selective enrichment broth and MALDI-TOF MS. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2012;31(5):767–773. doi: 10.1007/s10096-011-1373-0. [DOI] [PubMed] [Google Scholar]
- 33.Martiny D, Busson L, Wybo I, et al. Comparison of the Microflex LT and Vitek MS systems for routine identification of bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of clinical microbiology. 2012;50(4):1313–1325. doi: 10.1128/JCM.05971-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerome P, Le Fleche P, Blouin Y, et al. Yersinia pseudotuberculosis ST42 (O:1) Strain Misidentified as Yersinia pestis by Mass Spectrometry Analysis. Genome announcements. 2014;2(3) doi: 10.1128/genomeA.00435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugliese N, Circella E, Pazzani C, et al. Validation of a seminested PCR approach for rapid detection of Salmonella enterica subsp. enterica serovar Gallinarum. Journal of microbiological methods. 2011;85(1):22–27. doi: 10.1016/j.mimet.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Akiba M, Kusumoto M, Iwata T. Rapid identification of Salmonella enterica serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by multiplex PCR. Journal of microbiological methods. 2011;85(1):9–15. doi: 10.1016/j.mimet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Tankouo-Sandjong B, Sessitsch A, Liebana E, et al. MLST-v, multilocus sequence typing based on virulence genes, for molecular typing of Salmonella enterica subsp. enterica serovars. Journal of microbiological methods. 2007;69(1):23–36. doi: 10.1016/j.mimet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Duan R, Liang J, Shi G, et al. Homology analysis of pathogenic Yersinia species Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis based on multilocus sequence typing. Journal of clinical microbiology. 2014;52(1):20–29. doi: 10.1128/JCM.02185-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallik S, Virdi JS. Genetic relationships between clinical and non-clinical strains of Yersinia enterocolitica biovar 1A as revealed by multilocus enzyme electrophoresis and multilocus restriction typing. BMC microbiology. 2010;10:158. doi: 10.1186/1471-2180-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruekit S, Wangchuk S, Dorji T, et al. Molecular characterization and PCR-based replicon typing of multidrug resistant Shigella sonnei isolates from an outbreak in Thimphu, Bhutan. BMC research notes. 2014;7:95. doi: 10.1186/1756-0500-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voskresenskaya E, Savin C, Leclercq A, et al. Typing and clustering of Yersinia pseudotuberculosis isolates by restriction fragment length polymorphism analysis using insertion sequences. Journal of clinical microbiology. 2014;52(6):1978–1989. doi: 10.1128/JCM.00397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YW, Watanabe H, Phung DC, et al. Multilocus variable-number tandem repeat analysis for molecular typing and phylogenetic analysis of Shigella flexneri. BMC microbiology. 2009;9:278. doi: 10.1186/1471-2180-9-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X, Shariat N, Driebe EM, et al. Comparative analysis of subtyping methods against a whole genome sequencing standard in Salmonella enterica serotype Enteritidis. Journal of clinical microbiology. 2014 doi: 10.1128/JCM.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonnell J, Dallman T, Atkin S, et al. Retrospective analysis of whole genome sequencing compared to prospective typing data in further informing the epidemiological investigation of an outbreak of Shigella sonnei in the UK. Epidemiology and infection. 2013;141(12):2568–2575. doi: 10.1017/S0950268813000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sangal V, Holt KE, Yuan J, et al. Global phylogeny of Shigella sonnei strains from limited single nucleotide polymorphisms (SNPs) and development of a rapid and cost-effective SNP-typing scheme for strain identification by high-resolution melting analysis. Journal of clinical microbiology. 2013;51(1):303–305. doi: 10.1128/JCM.02238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khare R, Espy MJ, Cebelinski E, et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. Journal of clinical microbiology. 2014;52(10):3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson NW, Buchan BW, Ledeboer NA. Comparison of the BD MAX enteric bacterial panel to routine culture methods for detection of Campylobacter, enterohemorrhagic Escherichia coli (O157), Salmonella, and Shigella isolates in preserved stool specimens. Journal of clinical microbiology. 2014;52(4):1222–1224. doi: 10.1128/JCM.03099-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry MD, Corden SA, Howe RA. Evaluation of the Luminex xTAG Gastrointestinal Pathogen Panel and the Savyon Diagnostics Gastrointestinal Infection Panel for the detection of enteric pathogens in clinical samples. Journal of medical microbiology. 2014;63(Pt 11):1419–1426. doi: 10.1099/jmm.0.074773-0. [DOI] [PubMed] [Google Scholar]
- 49.Dunbar SA, Ritchie VB, Hoffmeyer MR, et al. Luminex((R)) multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods in molecular biology. 2015;1225:1–27. doi: 10.1007/978-1-4939-1625-2_1. [DOI] [PubMed] [Google Scholar]
- 50.Siah SP, Merif J, Kaur K, et al. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology. 2014;46(1):53–59. doi: 10.1097/PAT.0000000000000022. [DOI] [PubMed] [Google Scholar]
- 51.Wessels E, Rusman LG, van Bussel MJ, et al. Added value of multiplex Luminex Gastrointestinal Pathogen Panel (xTAG(R) GPP) testing in the diagnosis of infectious gastroenteritis. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20(3):O182–O187. doi: 10.1111/1469-0691.12364. [DOI] [PubMed] [Google Scholar]
- 52.Navidad JF, Griswold DJ, Gradus MS, et al. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. Journal of clinical microbiology. 2013;51(9):3018–3024. doi: 10.1128/JCM.00896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierce SE, Bell RL, Hellberg RS, et al. Detection and Identification of Salmonella enterica, Escherichia coli, and Shigella spp. via PCR-electrospray ionization mass spectrometry: isolate testing and analysis of food samples. Applied and environmental microbiology. 2012;78(23):8403–8411. doi: 10.1128/AEM.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shigella: CDC website. http://wwwcdcgov/shigella/ and http://wwwcdcgov/pulsenet/pathogens/shigellahtml.
- 55.Papasian CJ, Enna-Kifer S, Garrison B. Symptomatic Shigella sonnei urinary tract infection. Journal of clinical microbiology. 1995;33(8):2222–2223. doi: 10.1128/jcm.33.8.2222-2223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Appannanavar SB, Goyal K, Garg R, et al. Shigellemia in a post renal transplant patient: a case report and literature review. Journal of infection in developing countries. 2014;8(2):237–239. doi: 10.3855/jidc.3000. [DOI] [PubMed] [Google Scholar]
- 57.Markham KB, Backes C, Jr., Samuels P. Bacteremia and intrauterine infection with Shigella sonnei in a pregnant woman with AIDS. Archives of gynecology and obstetrics. 2012;286(3):799–801. doi: 10.1007/s00404-012-2310-x. [DOI] [PubMed] [Google Scholar]
- 58.Shigella and enteroinvasive Escherichia coli. In: Acheson DWK, Keusch GT, editors; Blaser PDS MJ, Ravdin JI, Greenberg HB, Guerrant RL, editors. Infections of the Gastrointestinal Tract. New York, NY: Raven Press; 1995. pp. 763–784. [Google Scholar]
- 59.Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bulletin of the World Health Organization. 1999;77(8):651–666. [PMC free article] [PubMed] [Google Scholar]
- 60.Bardhan P, Faruque AS, Naheed A, et al. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerging infectious diseases. 2010;16(11):1718–1723. doi: 10.3201/eid1611.090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van de Verg LL, Venkatesan MM. Editorial commentary: a Shigella vaccine against prevalent serotypes. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2014;59(7):942–943. doi: 10.1093/cid/ciu471. [DOI] [PubMed] [Google Scholar]
- 62.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States--major pathogens. Emerging infectious diseases. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DuPont HL, Levine MM, Hornick RB, et al. Inoculum size in shigellosis and implications for expected mode of transmission. The Journal of infectious diseases. 1989;159(6):1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 64.Shigella sonnei outbreak among men who have sex with men--San Francisco, California, 2000–2001. MMWR Morbidity and mortality weekly report. 2001;50(42):922–926. [PubMed] [Google Scholar]
- 65.Day care-related outbreaks of rhamnose-negative Shigella sonnei--six states, June 2001–March 2003. MMWR Morbidity and mortality weekly report. 2004;53(3):60–63. [PubMed] [Google Scholar]
- 66.Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers--Kansas, Kentucky, and Missouri, 2005. MMWR Morbidity and mortality weekly report. 2006;55(39):1068–1071. [PubMed] [Google Scholar]
- 67.Arvelo W, Hinkle CJ, Nguyen TA, et al. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant Shigella sonnei: implications for the management of shigellosis outbreaks among children. The Pediatric infectious disease journal. 2009;28(11):976–980. doi: 10.1097/INF.0b013e3181a76eab. [DOI] [PubMed] [Google Scholar]
- 68.Daskalakis DC, Blaser MJ. Another perfect storm: Shigella, men who have sex with men, and HIV. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;44(3):335–337. doi: 10.1086/510591. [DOI] [PubMed] [Google Scholar]
- 69.Khan WA, Griffiths JK, Bennish ML. Gastrointestinal and extra-intestinal manifestations of childhood shigellosis in a region where all four species of Shigella are endemic. PloS one. 2013;8(5):e64097. doi: 10.1371/journal.pone.0064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tauxe RV, Puhr ND, Wells JG, et al. Antimicrobial resistance of Shigella isolates in the USA: the importance of international travelers. The Journal of infectious diseases. 1990;162(5):1107–1111. doi: 10.1093/infdis/162.5.1107. [DOI] [PubMed] [Google Scholar]
- 71.von Seidlein L, Kim DR, Ali M, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS medicine. 2006;3(9):e353. doi: 10.1371/journal.pmed.0030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van den Beld MJ, Reubsaet FA. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2012;31(6):899–904. doi: 10.1007/s10096-011-1395-7. [DOI] [PubMed] [Google Scholar]
- 73.Huys G, Cnockaert M, Janda JM, et al. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. International journal of systematic and evolutionary microbiology. 2003;53(Pt 3):807–810. doi: 10.1099/ijs.0.02475-0. [DOI] [PubMed] [Google Scholar]
- 74.Hyma KE, Lacher DW, Nelson AM, et al. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. Journal of bacteriology. 2005;187(2):619–628. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winn WC, Jr., Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Woods GL. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6th ed. Baltimore, MD: Lippincott, Williams, & Wilkins; 2006. The Enterobacteriacae; pp. 211–302. [Google Scholar]
- 76.Lindberg AA, Karnell A, Weintraub A. The lipopolysaccharide of Shigella bacteria as a virulence factor. Reviews of infectious diseases. 1991;13(Suppl 4):S279–S284. doi: 10.1093/clinids/13.supplement_4.s279. [DOI] [PubMed] [Google Scholar]
- 77.Porter CK, Thura N, Ranallo RT, et al. The Shigella human challenge model. Epidemiology and infection. 2013;141(2):223–232. doi: 10.1017/S0950268812001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campilongo R, Di Martino ML, Marcocci L, et al. Molecular and functional profiling of the polyamine content in enteroinvasive E. coli : looking into the gap between commensal E. coli and harmful Shigella. PloS one. 2014;9(9):e106589. doi: 10.1371/journal.pone.0106589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carroll KC, Borek AP, Burger C, et al. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of staphylococci and enterococci. Journal of clinical microbiology. 2006;44(6):2072–2077. doi: 10.1128/JCM.02636-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aldridge KE, Gardner BB, Clark SJ, et al. Comparison of micro-ID, API 20E, and conventional media systems in identification of Enterobacteriaceae. Journal of clinical microbiology. 1978;7(6):507–513. doi: 10.1128/jcm.7.6.507-513.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O'Hara CM, Rhoden DL, Miller JM. Reevaluation of the API 20E identification system versus conventional biochemicals for identification of members of the family Enterobacteriaceae: a new look at an old product. Journal of clinical microbiology. 1992;30(1):123–125. doi: 10.1128/jcm.30.1.123-125.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peele D, Bradfield J, Pryor W, et al. Comparison of identifications of human and animal source gram-negative bacteria by API 20E and crystal E/NF systems. Journal of clinical microbiology. 1997;35(1):213–216. doi: 10.1128/jcm.35.1.213-216.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson A, McCarter YS, Tetreault J. Comparison of Crystal Enteric/Nonfermenter system, API 20E system, and Vitek AutoMicrobic system for identification of gram-negative bacilli. Journal of clinical microbiology. 1995;33(2):364–370. doi: 10.1128/jcm.33.2.364-370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khot PD, Fisher MA. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of clinical microbiology. 2013;51(11):3711–3716. doi: 10.1128/JCM.01526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gould LH, Bopp C, Strockbine N, et al. Recommendations for diagnosis of shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recommendations and reports : Morbidity and mortality weekly report Recommendations and reports / Centers for Disease Control. 2009;58(RR-12):1–14. [PubMed] [Google Scholar]
- 86.Gavin PJ, Peterson LR, Pasquariello AC, et al. Evaluation of performance and potential clinical impact of ProSpecT Shiga toxin Escherichia coli microplate assay for detection of Shiga Toxin-producing E. coli in stool samples. Journal of clinical microbiology. 2004;42(4):1652–1656. doi: 10.1128/JCM.42.4.1652-1656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mackenzie AM, Lebel P, Orrbine E, et al. Sensitivities and specificities of premier E. coli O157 and premier EHEC enzyme immunoassays for diagnosis of infection with verotxin (Shiga-like toxin)-producing Escherichia coli. The SYNSORB Pk Study investigators. Journal of clinical microbiology. 1998;36(6):1608–1611. doi: 10.1128/jcm.36.6.1608-1611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He X, Patfield S, Hnasko R, et al. A polyclonal antibody based immunoassay detects seven subtypes of Shiga toxin 2 produced by Escherichia coli in human and environmental samples. PloS one. 2013;8(10):e76368. doi: 10.1371/journal.pone.0076368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Urdahl AM, Solheim HT, Vold L, et al. Shiga toxin-encoding genes (stx genes) in human faecal samples. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2013;121(3):202–210. doi: 10.1111/j.1600-0463.2012.02957.x. [DOI] [PubMed] [Google Scholar]
- 90.Ashkenazi S, Levy I, Kazaronovski V, et al. Growing antimicrobial resistance of Shigella isolates. The Journal of antimicrobial chemotherapy. 2003;51(2):427–429. doi: 10.1093/jac/dkg080. [DOI] [PubMed] [Google Scholar]
- 91.Khatun F, Faruque AS, Koeck JL, et al. Changing species distribution and antimicrobial susceptibility pattern of Shigella over a 29-year period (1980–2008) Epidemiology and infection. 2011;139(3):446–452. doi: 10.1017/S0950268810001093. [DOI] [PubMed] [Google Scholar]
- 92.Taneja N, Mewara A, Kumar A, et al. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. The Journal of antimicrobial chemotherapy. 2012;67(6):1347–1353. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 93.Vinh H, Nhu NT, Nga TV, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC infectious diseases. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang W, Luo Y, Li J, et al. Wide dissemination of multidrug-resistant Shigella isolates in China. The Journal of antimicrobial chemotherapy. 2011;66(11):2527–2535. doi: 10.1093/jac/dkr341. [DOI] [PubMed] [Google Scholar]
- 95.Ghosh S, Pazhani GP, Chowdhury G, et al. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. Journal of medical microbiology. 2011;60(Pt 10):1460–1466. doi: 10.1099/jmm.0.032920-0. [DOI] [PubMed] [Google Scholar]
- 96.Jain S, Sharma M, Gupta R, et al. Multidrug resistant Shigella flexneri : a rare case of septicemia in an infant. Journal of clinical and diagnostic research : JCDR. 2014;8(6):DD03–DD04. doi: 10.7860/JCDR/2014/8434.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Azmi IJ, Khajanchi BK, Akter F, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PloS one. 2014;9(7):e102533. doi: 10.1371/journal.pone.0102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Phalipon A, Sansonetti PJ. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunology and cell biology. 2007;85(2):119–129. doi: 10.1038/sj.icb7100025. [DOI] [PubMed] [Google Scholar]
- 99.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clinical microbiology reviews. 2008;21(1):134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ogawa M, Handa Y, Ashida H, et al. The versatility of Shigella effectors. Nature reviews Microbiology. 2008;6(1):11–16. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- 101.Barry EM, Pasetti MF, Sztein MB, et al. Progress and pitfalls in Shigella vaccine research. Nature reviews Gastroenterology & hepatology. 2013;10(4):245–255. doi: 10.1038/nrgastro.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herold S, Karch H, Schmidt H. Shiga toxin-encoding bacteriophages--genomes in motion. International journal of medical microbiology : IJMM. 2004;294(2–3):115–121. doi: 10.1016/j.ijmm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 103.Donohue-Rolfe A, Acheson DW, Keusch GT. Shiga toxin: purification, structure, and function. Reviews of infectious diseases. 1991;13(Suppl 4):S293–S297. doi: 10.1093/clinids/13.supplement_4.s293. [DOI] [PubMed] [Google Scholar]
- 104.Kim YJ, Yeo SG, Park JH, et al. Shigella vaccine development: prospective animal models and current status. Current pharmaceutical biotechnology. 2013;14(10):903–912. doi: 10.2174/1389201014666131226123900. [DOI] [PubMed] [Google Scholar]
- 105.Bottone EJ. Yersinia enterocolitica: the charisma continues. Clinical microbiology reviews. 1997;10(2):257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yersinia: CDC Website. http://wwwcdcgov/nczved/divisions/dfbmd/diseases/yersinia/
- 107.Cover TL, Aber RC. Yersinia enterocolitica. The New England journal of medicine. 1989;321(1):16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 108.Foberg U, Fryden A, Kihlstrom E, et al. Yersinia enterocolitica septicemia: clinical and microbiological aspects. Scandinavian journal of infectious diseases. 1986;18(4):269–279. doi: 10.3109/00365548609032337. [DOI] [PubMed] [Google Scholar]
- 109.Giamarellou H, Antoniadou A, Kanavos K, et al. Yersinia enterocolitica endocarditis: case report and literature review. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 1995;14(2):126–130. doi: 10.1007/BF02111871. [DOI] [PubMed] [Google Scholar]
- 110.van der Heijden IM, Res PC, Wilbrink B, et al. Yersinia enterocolitica: a cause of chronic polyarthritis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1997;25(4):831–837. doi: 10.1086/515535. [DOI] [PubMed] [Google Scholar]
- 111.Adamkiewicz TV, Berkovitch M, Krishnan C, et al. Infection due to Yersinia enterocolitica in a series of patients with beta-thalassemia: incidence and predisposing factors. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1998;27(6):1362–1366. doi: 10.1086/515025. [DOI] [PubMed] [Google Scholar]
- 112.Yersinia enterocolitica gastroenteritis among infants exposed to chitterlings--Chicago, Illinois, 2002. MMWR Morbidity and mortality weekly report. 2003;52(40):956–958. [PubMed] [Google Scholar]
- 113.Abdel-Haq NM, Asmar BI, Abuhammour WM, et al. Yersinia enterocolitica infection in children. The Pediatric infectious disease journal. 2000;19(10):954–958. doi: 10.1097/00006454-200010000-00002. [DOI] [PubMed] [Google Scholar]
- 114.Fredriksson-Ahomaa M, Stolle A, Korkeala H. Molecular epidemiology of Yersinia enterocolitica infections. FEMS immunology and medical microbiology. 2006;47(3):315–329. doi: 10.1111/j.1574-695X.2006.00095.x. [DOI] [PubMed] [Google Scholar]
- 115.Jones TF. From pig to pacifier: chitterling-associated yersiniosis outbreak among black infants. Emerging infectious diseases. 2003;9(8):1007–1009. doi: 10.3201/eid0908.030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee LA, Gerber AR, Lonsway DR, et al. Yersinia enterocolitica O:3 infections in infants and children, associated with the household preparation of chitterlings. The New England journal of medicine. 1990;322(14):984–987. doi: 10.1056/NEJM199004053221407. [DOI] [PubMed] [Google Scholar]
- 117.Lee LA, Taylor J, Carter GP, et al. Yersinia enterocolitica O:3: an emerging cause of pediatric gastroenteritis in the United States. The Yersinia enterocolitica Collaborative Study Group. The Journal of infectious diseases. 1991;163(3):660–663. doi: 10.1093/infdis/163.3.660. [DOI] [PubMed] [Google Scholar]
- 118.Wagner SJ, Friedman LI, Dodd RY. Transfusion-associated bacterial sepsis. Clinical microbiology reviews. 1994;7(3):290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tertti R, Vuento R, Mikkola P, et al. Clinical manifestations of Yersinia pseudotuberculosis infection in children. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 1989;8(7):587–591. doi: 10.1007/BF01968134. [DOI] [PubMed] [Google Scholar]
- 120.Crchova V, Grondin C. [Urinary infection due to Yersinia pseudotuberculosis] La Vie medicale au Canada francais. 1973;2(1):3–5. [PubMed] [Google Scholar]
- 121.Ljungberg P, Valtonen M, Harjola VP, et al. Report of four cases of Yersinia pseudotuberculosis septicemia and a literature review. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 1995;14(9):804–810. doi: 10.1007/BF01690998. [DOI] [PubMed] [Google Scholar]
- 122.Naiel B, Raul R. Chronic prostatitis due to Yersinia pseudotuberculosis. Journal of clinical microbiology. 1998;36(3):856. doi: 10.1128/jcm.36.3.856-856.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nuorti JP, Niskanen T, Hallanvuo S, et al. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. The Journal of infectious diseases. 2004;189(5):766–774. doi: 10.1086/381766. [DOI] [PubMed] [Google Scholar]
- 124.Jalava K, Hakkinen M, Valkonen M, et al. An outbreak of gastrointestinal illness and erythema nodosum from grated carrots contaminated with Yersinia pseudotuberculosis. The Journal of infectious diseases. 2006;194(9):1209–1216. doi: 10.1086/508191. [DOI] [PubMed] [Google Scholar]
- 125.Reuter S, Connor TR, Barquist L, et al. Parallel independent evolution of pathogenicity within the genus Yersinia. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(18):6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Archer JR, Schell RF, Pennell DR, et al. Identification of Yersinia spp. with the API 20E system. Journal of clinical microbiology. 1987;25(12):2398–2399. doi: 10.1128/jcm.25.12.2398-2399.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sharma NK, Doyle PW, Gerbasi SA, et al. Identification of Yersinia species by the API 20E. Journal of clinical microbiology. 1990;28(6):1443–1444. doi: 10.1128/jcm.28.6.1443-1444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wunderink HF, Oostvogel PM, Frenay IH, et al. Difficulties in diagnosing terminal ileitis due to Yersinia pseudotuberculosis. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2014;33(2):197–200. doi: 10.1007/s10096-013-1943-4. [DOI] [PubMed] [Google Scholar]
- 129.Singhal N, Kumar M, Virdi JS. Molecular analysis of beta-lactamase genes to understand their differential expression in strains of Yersinia enterocolitica biotype 1A. Scientific reports. 2014;4:5270. doi: 10.1038/srep05270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Batzilla J, Heesemann J, Rakin A. The pathogenic potential of Yersinia enterocolitica 1A. International journal of medical microbiology : IJMM. 2011;301(7):556–561. doi: 10.1016/j.ijmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 131.Rakin A, Schneider L, Podladchikova O. Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Frontiers in cellular and infection microbiology. 2012;2:151. doi: 10.3389/fcimb.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Capilla S, Ruiz J, Goni P, et al. Characterization of the molecular mechanisms of quinolone resistance in Yersinia enterocolitica O:3 clinical isolates. The Journal of antimicrobial chemotherapy. 2004;53(6):1068–1071. doi: 10.1093/jac/dkh225. [DOI] [PubMed] [Google Scholar]
- 133.Pham JN, Bell SM, Martin L, et al. The beta-lactamases and beta-lactam antibiotic susceptibility of Yersinia enterocolitica. The Journal of antimicrobial chemotherapy. 2000;46(6):951–957. doi: 10.1093/jac/46.6.951. [DOI] [PubMed] [Google Scholar]
- 134.Porter CK, Choi D, Cash B, et al. Pathogen-specific risk of chronic gastrointestinal disorders following bacterial causes of foodborne illness. BMC gastroenterology. 2013;13:46. doi: 10.1186/1471-230X-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dizdar V, Gilja OH, Hausken T. Increased visceral sensitivity in Giardia-induced postinfectious irritable bowel syndrome and functional dyspepsia. Effect of the 5HT3-antagonist ondansetron. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2007;19(12):977–982. doi: 10.1111/j.1365-2982.2007.00988.x. [DOI] [PubMed] [Google Scholar]
- 136.Falth-Magnusson K, Franzen L, Jansson G, et al. Infant feeding history shows distinct differences between Swedish celiac and reference children. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology. 1996;7(1):1–5. doi: 10.1111/j.1399-3038.1996.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 137.Verdu EF, Mauro M, Bourgeois J, et al. Clinical onset of celiac disease after an episode of Campylobacter jejuni enteritis. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 2007;21(7):453–455. doi: 10.1155/2007/169591. [DOI] [PMC free article] [PubMed] [Google Scholar]