Abstract
Degradation of chlorophyll (Chl) by Chl catabolic enzymes (CCEs) causes the loss of green color that typically occurs during senescence of leaves. In addition to CCEs, STAYGREEN1 (SGR1) functions as a key regulator of Chl degradation. Although sgr1 mutants in many plant species exhibit a stay-green phenotype, the biochemical function of the SGR1 protein remains elusive. Many recent studies have examined the physiological and molecular roles of SGR1 and its homologs (SGR2 and SGR-LIKE) in Chl metabolism, finding that these proteins have different roles in different species. In this review, we summarize the recent studies on SGR and discuss the most likely functions of SGR homologs.
Keywords: abiotic stress, chlorophyll catabolic enzymes, chlorophyll degradation, STAYGREEN (SGR), SGR-LIKE (SGRL)
INTRODUCTION
Mendel’s green cotyledon trait in pea (Pisum sativum) has been widely used to demonstrate the principle of inheritance and the single recessive gene responsible for the non-yellowing trait has intrigued scientists for many years. In 2006, Armstead and colleagues finally identified the gene controlling Mendel’s green cotyledon trait (Armstead et al., 2006), and it is now named STAYGREEN1 (also termed STAY-GREEN1; SGR1) or NONYELLOWING1 (NYE1) in Arabidopsis (Cha et al., 2002; Park et al., 2007; Ren et al., 2007). At about the same time, different groups also independently identified SGR1 homologs in Arabidopsis thaliana (Ren et al., 2007) and rice (Oryza sativa; Jiang et al., 2007; Park et al., 2007), and subsequent work isolated and characterized SGR1 homologs in various plant species, including tomato (Solanum lycopersicum; Barry et al., 2008), bell pepper (Capsicum annuum; Barry et al., 2008), tall fescue (Festuca arundinacea; Wei et al., 2011), Medicago truncatula (Zhou et al., 2011), and soybean (Glycine max; Fang et al., 2014) (Fig. 1A).
Fig. 1.
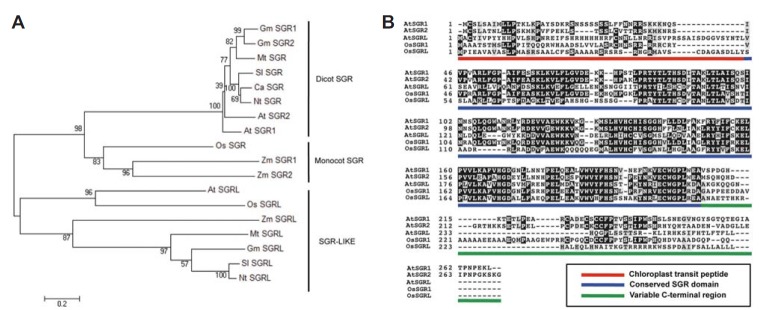
Phylogenetic tree and domain structure of SGR homologs in plants. (A) The phylogenic tree was constructed using MEGA 5.1 (http://www.megasoftware.net/megamac.php). The evolutionary relationship was inferred using the Neighbor-joining method. Numbers at branch points represent bootstrap values of 100 replicate trees. The accession numbers of SGRs and SGRLs in the GenBank, AGI (Arabidopsis Gene Index) or TGI (The Gene Index; TC) are as follows: At SGR1 (Arabidopsis thaliana), At4g22920; At SGR2, At4g11910; At SGRL, At1g44000; Ca SGR (pepper, Capsicum annuum), EU414631, AAY98500.1, ACB56586; Os SGR (rice, Oryza sativa), Os09g36200, AY850134; Os SGRL, Os04g59610, AK105982; Gm SGR1 (soybean, Glycine max), AY850141; Gm SGR2, AY850142; Gm SGRL, TC216309; Mt SGR (Medicago truncatula), BF633258; Mt SGRL, TC182595+GD185310; Nt SGR (tobacco, Nicotiana tabacum), ABY19382; Nt SGRL, TC129900; Sl SGR (tomato, Solanum lycopersicum), DQ100158; SI SGRL, TC118764; Zm SGR1 (maize, Zea mays), AY850136; Zm SGR2, AY850137; Zm SGRL, TC503941. (B) Domain structure of Arabidopsis and rice SGR homologs. Chloroplast transit peptides (Red line), the conserved SGR domains (Blue line), and the variable C-terminal regions (Green line) are shown.
Early analysis indicated that SGR1 might function in chlorophyll (Chl) degradation, as: (i) sgr1 knockout mutants exhibit a common stay-green phenotype in several plant species, (ii) sgr1 mutants display a type C nonfunctional (cosmetic) stay-green phenotype, which affects Chl degradation, but not other aspects of senescence (Thomas and Howarth, 2000), and (iii) rice SGR1 physically interacts with light-harvesting complex subunits of photosystem II (LHCII) in vitro and in vivo (Park et al., 2007). Indeed, Park et al. (2007) examined whether SGR1 has enzymatic properties typical of Chl catabolic enzymes (CCEs) in the chloroplast, but found that SGR does not bind to Chl nor does it have chlorophyllase activity. This indicates that SGR1 does not function as a CCE in the chloroplast.
The function of other SGR homologs is another missing piece of the puzzle in the study of SGR1 (Fig. 1B). Previous phylogenetic analysis classified the SGR family of higher plants into two groups (Barry et al., 2008; Hörtensteiner, 2009; Sakuraba et al., 2014b). One group comprises the genuine SGR subfamily, and mutations in these SGR genes cause a type C nonfunctional stay-green phenotype, as described above. In the other group, the SGR-LIKE (SGRL) subfamily, the C-terminal sequence of SGRL proteins differs considerably from that of the SGR subfamily proteins (Hörtensteiner, 2009). To our knowledge, however, all higher plants contain at least one SGRL, indicating that SGRL has essential functions in green plants, probably acting in Chl breakdown. Furthermore, some plant species contain two or more SGRs; for example, Arabidopsis has SGR1 and SGR2, in addition to one SGRL (Barry et al., 2008; Sakuraba et al., 2014b). The physiological roles of these SGR homologs had remained unclear. However, after 2010, several studies have addressed the physiological and molecular roles of SGRs and revealed their functions.
SGR1, CCES, AND LHCII INTERACT DURING LEAF SENESCENCE
Many studies have examined the functions of SGR1 and CCEs in dark-induced senescence (Kusaba et al., 2007; Meguro et al., 2011; Park et al., 2007; Pružinská et al., 2003; Sakuraba et al., 2012; Schelbert et al., 2009). Although SGR1 interacts with the subunits of LHCII in vivo (Park et al., 2007), how SGR activates Chl degradation during leaf senescence has remained unclear. Work in Arabidopsis and rice has identified six plastid-localized CCEs: Non-yellowing coloring1 (NYC1) (Horie et al., 2009; Kusaba et al., 2007) and NYC1-Like (NOL) (Sato et al., 2009) encoding two different Chl b reductases, 7-Hydroxymethyl chlorophyll a reductase (HCAR) (Meguro et al., 2011), Pheophytin pheophorbide hydrolase (PPH) (Morita et al., 2009; Schelbert et al., 2009), Pheophorbide a oxygenase (PAO) (Pružinská et al., 2003; Tanaka et al., 2003), and Red chlorophyll catabolite reductase (RCCR) (Pružinská et al., 2007). SGR1 physically interacts with all six CCEs (Sakuraba et al., 2012; 2013). Moreover, SGR1 and six CCEs specifically interact with LHCII proteins, but not with other photosystem components, such as LHCI or photosystem core proteins. These results indicate that SGR1, CCEs, and LHCII form dynamic, multi-protein complexes for Chl degradation during senescence. These multi-protein complexes may also be required for Chl detoxification in non-senescing green tissues. Colored intermediates of Chl breakdown, pheophorbide a (Pheide a) and red Chl catabolite (RCC), are potentially phototoxic, because deficiency of PAO or RCCR in pre-senescent leaves leads to accelerated cell death in Arabidopsis (Hirashima et al., 2009; Mach et al., 2001; Pružinská et al., 2003; 2007), maize (Gray et al., 1997; 2002), and rice (Tang et al., 2011). Thus, the SGR1-CCE-LHCII complex likely allows the metabolic channeling of potentially phototoxic Chl breakdown intermediates to minimize the risk of photodynamism caused by Chl breakdown intermediates during natural or dark-induced senescence (Fig. 2), and SGR1 has an important role in recruiting CCEs to LHCII (Sakuraba et al., 2012).
Fig. 2.
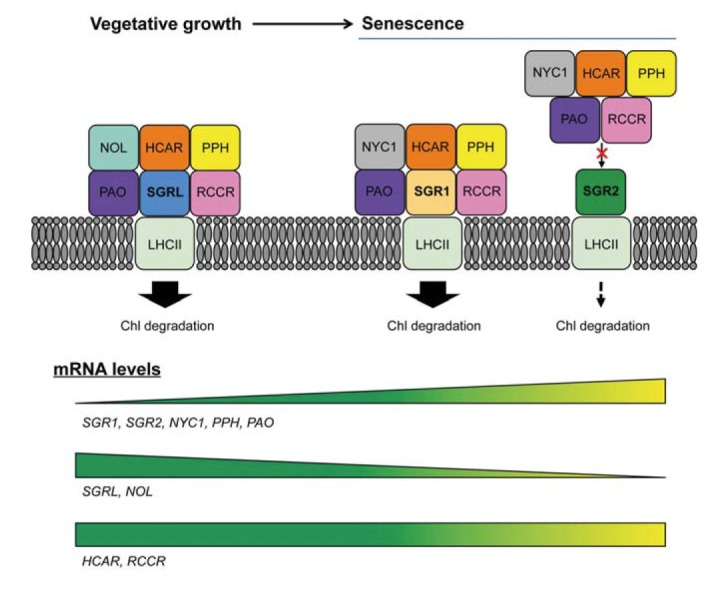
Divergent functions of Arabidopsis SGR1, SGR2, and SGRL proteins. In presenescent leaves during vegetative growth, SGRL and NOL transcripts are much abundant than SGR1 and NYC1 transcripts. Thus, under abiotic/biotic stress conditions, SGRL and NOL are probably the main components of the SGRL-CCEs-LHCII complex. During leaf senescence, SGR1 forms a dynamic, multi-protein complex with CCEs on LHCII to activate Chl degradation. However, SGR2 negatively regulates Chl degradation. Like SGR1 and SGRL, SGR2 can interact with LHCII. However, SGR2 does not interact with most CCEs. Thus, SGR2 likely interrupts the formation of the SGR1-CCE or SGRL-CCE complexes, leading to adjustment of Chl degradation rate in senescing chloroplasts. SGR, STAYGREEN; SGRL, SGR-LIKE; NYC1, NON-YELLOW COLORING1; NOL, NYC1-LIKE; Chl, chlorophyll; CCE, Chl catabolic enzyme; LHCII, light-harvesting complex II.
SGR1 AND SGRL FUNCTION IN STRESS-INDUCED LEAF YELLOWING
Chl breakdown also occurs in response to several abiotic and biotic stresses, in addition to senescence (Lim et al., 2007). Arabidopsis SGR1 and SGRL overexpressors exhibit an early leaf yellowing phenotype, while sgr1/nye1-1 and sgrl mutants display a stay-green phenotype under abiotic stress conditions, including high salinity, osmotic stress (mannitol), and drought stress conditions (Sakuraba et al., 2014a; 2014b; 2014c, Fig. 3). Furthermore, one of the Arabidopsis sgr1 alleles, termed noc1, develops less severe disease symptoms (leaf chlorosis and/or necrosis) than wild type after infection with the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 or the fungal pathogen Alternaria brassicicola (Mecey et al., 2011). Thus, SGR1, SGRL, and CCEs also function in leaf yellowing induced by biotic and abiotic stress.
Fig. 3.

Distinct responses of three SGR overexpressors to salt stress. WT (Col-0) and overexpressors of SGR1 (SGR1-OX), SGR2 (SGR2-OX), and SGRL (SGRL-OX) were grown for 3 weeks under long-day conditions (16-hour light/day), and then 200 mM NaCl solution was supplied for 10 days. SGR, STAYGREEN; DST, days of salt treatment.
SGRs and CCEs may also form a multi-protein complex for Chl degradation under biotic and abiotic stress conditions, similar to their interactions during senescence. Interestingly, transcript levels of SGR1, NYC1, PPH, and PAO dramatically increase during senescence, but transcripts of SGRL and NOL are more abundant in pre-senescent leaves during vegetative growth (Sakuraba et al., 2014b). Furthermore, SGRL interacts more strongly with NOL than with other CCEs (Sakuraba et al., 2014c). Thus, the composition of the SGRL-CCE complexes likely differs, with SGRL and NOL as the main components of the SGRL-CCE complex during stress-induced leaf yellowing and SGR1 and NYC1 as the main components during senescence (Fig. 2). The transcripts of HCAR and RCCR remain at nearly constant levels throughout development, including during natural senescence (Sakuraba et al., 2013), indicating that HCAR and RCCR participate in the SGR-CCE complex during both stress- and senescence-induced leaf yellowing (Fig. 2).
DISTINCT RELATIONSHIP OF TWO SGR HOMOLOGS IN SOYBEAN AND ARABIDOPSIS
As described above, different plant species have different numbers of SGR homologs. For example, rice and barley (Hordeum vulgare) have just one SGR homolog, but Arabidopsis and soybean have two SGR homologs, SGR1 and SGR2 (Sakuraba et al., 2014b). Recent discoveries have shed light on the differing physiological functions and relationships of these pairs of SGR homologs. In soybean, the leaves and seed embryos of d1 d2 double mutants show a significant stay-green phenotype (Chao et al., 1995; Guiamet et al., 1991) and recent work showed that D1 and D2 encode GmSGR1 and GmSGR2, respectively (Fang et al., 2014). The GmSGR1 and GmSGR2 homologs resulted from one of the two whole-genome duplication events that occurred in soybean 13 million years ago. Interestingly, d1 leaves showed a much weaker stay-green phenotype than the d1 d2 leaves, and the d1 seeds turned yellow normally, indicating that the two GmSGRs function redundantly.
In contrast with the two GmSGRs, the two Arabidopsis SGRs have different functions during leaf yellowing. An Arabidopsis SGR2 overexpressor line shows a stay-green phenotype and sgr2 null mutants exhibit a leaf yellowing phenotype under dark-induced senescence and abiotic stress conditions (Sakuraba et al., 2014b; Fig. 3). By contrast, SGR1 overexpressor shows early leaf yellowing and sgr1-1/nye1-1 mutants exhibit a stay-green phenotype, opposite phenotypes to those of SGR2 overex- pressors and sgr2 mutants. SGR2 has a much weaker physical interaction with CCEs compared with the interaction of SGR1 with CCEs. Because SGR2 also can interact with LHCII, an excess of SGR2 may interrupt the formation of the SGR1-CCEs-LHCII complex. Furthermore, SGR1 and SGR2 can interact with each other to form homo- and heterodimers. These two interesting characteristics of SGR2, (i) its lower capacity to interact with CCEs and (ii) its ability to form heterodimers with SGR1, may cause its negative effect on Chl degradation during leaf senescence. Arabidopsis SGR1 and SGR2 likely acquired their antagonistic functions to balance Chl catabolism in senescing chloroplasts, thus preventing inappropriate Chl degradation (Fig. 2). Indeed, rice SGR1 and SGRL overexpressors showed a premature leaf-yellowing phenotype (Jiang et al., 2011; Rong et al., 2013) and Arabidopsis SGR1 and SGRL overexpressors showed a normal phenotype during vegetative growth (Sakuraba et al., 2012, 2014c). These findings suggest that this difference is partly caused by the existence of SGR2 in Arabidopsis. Thus, SGR homologs are not always positive regulators of Chl degradation, and this depends on the independent evolutionary processes that occurred in each species.
SGRS FUNCTION IN SEED DEGREENING AND MATURATION
Proper seed maturation requires prompt degradation of Chl because any Chl remaining in mature seeds has severe, negative effects on seed storability and longevity (Clerkx et al., 2003; Johnson-Flanagan and Spencer, 1994). Although Arabidopsis sgr1 (SALK_070891) and sgr2 (SALK_003830) mutants produce normal yellow seeds, sgr1 sgr2 double mutants produce green seeds, indicating that Arabidopsis SGR1 and SGR2 function redundantly in seed degreening (Delmas et al., 2013). Knockout mutants of the abscisic acid (ABA) signaling-associated transcription factor ABA INSENSITIVE3 (ABI3) also produce green seeds (Clerkx et al., 2003; Koornneef et al., 1989) and electrophoretic mobility shift assays revealed that ABI3 directly binds to the promoters of SGR1 and SGR2 (Delmas et al., 2013). Furthermore, ectopic expression of SGR1 and SGR2 in abi3 knockout mutants rescues the phenotype from green to yellow seeds, indicating that SGR1 and SGR2 have pivotal roles in seed degreening during maturation. Arabidopsis nyc1 mutant seeds also contain green pigments (Nakajima et al., 2012), indicating that NYC1 also functions in seed degreening. Zhang et al. (2014) recently reported that SGR1 and SGR2 also affect the biosynthesis of tocopherol, a major component of Arabidopsis seeds (Sattler et al., 2004), during seed development; for example, the seeds of SGR1-overexpressing plants had significantly lower tocopherol contents. Thus, these results indicate that SGR1 and SGR2 have important roles in balancing Chl degradation and tocopherol biosynthesis during seed maturation.
Some aspects of seed degreening remain unclear. The seeds of pph mutants do not show a green phenotype, although the pph leaves show a stay-green phenotype (Shelbert et al., 2009). We also confirmed that two other mutants of CCEs, hcar and pao, have a stay-green phenotype but do not produce green seeds (unpublished). Furthermore, some CCE genes, such as HCAR, NOL, and RCCR have lower expression levels during seed maturation than during other developmental stages. Thus, it is not yet clear whether these CCEs function in seed degreening in Arabidopsis. Also, SGR2 acts as a negative regulator of leaf yellowing (Sakuraba et al., 2014b), but it promotes seed degreening. Thus, these results strongly suggest that Chl degradation mechanisms differ during seed degreening and leaf senescence.
OTHER INTERESTING FUNCTIONS OF SGR
Ripening of tomato fruits involves accumulation of carotenoids, including lycopene and β-carotene, along with a concomitant decrease in Chl levels (Fraser et al., 1994). A recent study indicated that tomato SGR1 (Solanum lycopersicum SGR1; SlSGR1) balances these two major events of fruit ripening. In SlSGR1 RNAi fruits, three kinds of carotenoids (β-carotene, lycopene, and phytoene) accumulate to higher levels than in non-transgenic tomato fruits (Luo et al., 2013), indicating that SlSGR1 affects the regulation of carotenoid accumulation in ripening tomato fruits. Furthermore, SlSGR1 physically interacts with the carotenoid biosynthesis enzyme phytoene synthase 1 (PSY1), which has an important role in tomato fruit nutrition (Fraser et al., 1994; 2004). Based on these results, the authors proposed that the physical interaction of SlSGR1 and PSY1 leads to a decrease in PSY1 activity and thus causes reduced lycopene accumulation. This study showed the physical relationship between SGR1 and PSY1 in tomato fruits, prompting the intriguing question of whether SGR1 interacts with PSY1 or other carotenoid biosynthesis enzymes to regulate carotenoid accumulation in other tissues, such as pre-senescent leaves, or in other plant species. During leaf senescence, the level of carotenoids declines as Chl is degraded (Biswal, 1995), indicating the activation of carotenoid degradation pathways and inhibition of carotenoid biosynthesis. Thus, SGR1 may have pivotal functions in the regulation of carotenoid accumulation during leaf senescence.
All SGR family proteins in higher plants are predicted to localize to chloroplasts (Barry et al., 2008), indicating that they likely function in plastids, most likely in Chl degradation. However, in addition to its role in senescing chloroplasts, Medicago truncatula SGR (MtSGR) also functions in nodule senescence in roots (Zhou et al., 2011). MtSGR expression is higher in senescing nodules than any other organs, including senescent leaves. Furthermore, the nodules of Medicago sgr mutants (termed NF2089) show significant down-regulation of several nodule senescence-associated genes, indicating that MtSGR affects nodule senescence in legumes. Nodule cells contain other plastid types, such as leucoplasts, and it would be an intriguing idea (and likely) that SGRs do not only occur in chloroplasts, but generally occur in plastids where they execute their diverse functions. Thus, their role in fruit carotenoid biosynthesis likely involves chromoplasts. Functional analyses of MtSGR in nodule senescence of Medicago and SlSGR1 in fruit ripening of tomato will expand our understanding of the potential roles of SGRs in physiological processes beyond Chl degradation.
TRANSCRIPTIONAL REGULATORY NETWORK OF SGR EXPRESSION
Although most studies of SGR have examined its biochemical and physiological functions, recent work in Arabidopsis and rice has identified several transcription factors that directly promote SGR expression. As described above, ABI3 binds to the canonical B3 domain-binding RY motif (CATGCA) in the promoters of SGR1 and SGR2 during seed degreening (Delmas et al., 2013). Recent work showed that Phytochrome Interacting Factor 4 (PIF4) and PIF5 strongly activate SGR1 expression during dark-induced senescence (Sakuraba et al., 2014b). In the PIF4/PIF5-dependent leaf senescence cascade, ABI5 and ENHANCED EM LEVEL (EEL), direct targets downstream of PIF4/PIF5, bind to the ABA-Responsive Element (ABRE) motif (YACGT) in the SGR1 promoter (Sakuraba et al., 2014d). Notably, ABI3, ABI5, and EEL are ABA signaling-associated transcription factors (Giraudat et al., 1992; Jakoby et al., 2002), and ABRE-Binding Factor 2/3/4 (ABF2/3/4) bZIPs also induced SGR1 expression (Fujita et al., 2009), indicating that ABA signaling has an important role in the activation of SGR1 expression and Chl degradation. Interestingly, NYC1 is also directly activated by ABA-signaling transcription factors, including ABI5 and EEL (Sakuraba et al., 2014d) during dark-induced senescence and ABF4 during seed degreening (Nakajima et al., 2012). Furthermore, the OsNAP transcription factor, which is an ABA signaling-associated gene, directly activates the transcription of both SGR1 and NYC1 in rice (Liang et al., 2014). Thus, several ABA-signaling transcription factors likely function to co-activate SGR1 and NYC1, and this close transcriptional relationship between SGR1 and NYC1 is important for Chl degradation during both leaf senescence and seed degreening.
FUTURE RESEARCH ON THE FUNCTIONS OF SGRS
Recent functional studies of SGR1 and its homologs revealed that SGR1 can interact with various chloroplast proteins, including LHCII proteins, all known CCEs, and PSY1 (Luo et al., 2013; Sakuraba et al., 2012; 2013). However, the amino acids in SGR that affect these physical interactions with other proteins remain to be identified. Park et al. found that V99 is important for OsSGR1 function because the rice sgr mutant has a V99M missense mutation, but this amino acid substitution does not affect the interaction of OsSGR1 with LHCII (Park et al., 2007). More detailed analyses, such as the combination of site-directed mutagenesis and protein-protein interaction assays, will reveal more about the biochemical function of SGR.
SGR1 could interact with other, unknown proteins. The most intriguing candidate for a potential SGR1-interactor is metal-chelating substance (MCS), which is considered to be required for the removal of the Mg atom of Chl a during Chl breakdown, but remains to be identified (Hörtensteiner and Krautler, 2011). Because all of the six known CCEs physically interact with SGR1 (Sakuraba et al., 2012; 2013), MCS likely also interacts with SGR1 and other CCEs as an essential factor in the multi-protein complexes. Another candidate SGR1-interactor is Thylakoid Formation 1 (THF1), a homolog of psb29 identified in the cyanobacterium Synechocystis sp PCC6803 (Wang et al., 2004). THF1 is closely associated with Chl breakdown because both Arabidopsis and rice thf1 mutants exhibit a stay-green phenotype (Huang et al., 2013; Yamatani et al., 2013). Furthermore, THF1 physically interacts with all LHCII subunit proteins (Lhcb1 to Lhcb6; Huang et al., 2013). Thus, it is possible that THF1 is near SGR1 and CCEs in LHCII, possibly by physical interactions, during Chl breakdown. The subunits of chloroplast proteases are also candidate SGR-interacting proteins. Chl and photosystem apoproteins should be degraded at almost the same stage, and chloroplast proteases likely degrade apoproteins. It is well known that chloroplast-localized FtsH protease participates in the degradation of D1 protein, a subunit of photosystem II, under stress conditions (Kato and Sakamoto, 2009). Glutamyl endopeptidase, a chloroplast-localized serine protease, is proposed to be involved in the degradation of Lhcb1 (Forsberg et al., 2005). Furthermore, the transcripts of several subunits of Clp protease are most abundant in the chloroplasts of senescing leaves (Olinares et al., 2011). This indicates that Clp protease also has an important role in degradation of chloroplast proteins, possibly including CCEs, during leaf senescence. Indeed, Arabidopsis Clp protease is involved in destabilization of the Chl biosynthetic enzyme chlorophyllide a oxygenase (Nakagawara et al., 2007). Collectively, further proteomic analyses, such as mass spectrometric analysis of pull-down fractions, using epitope-tagged SGR1 protein, will provide good approaches to find as-yet-unidentified SGR1-interacting proteins.
Acknowledgments
This work was carried out with the support of “Next-Generation BioGreen21 Program for Agriculture & Technology Development (Project No. PJ01106301)”, Rural Development Administration, Republic of Korea.
REFERENCES
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. Cross-species identification of Mendel’s/locus. Science. 2007;315:73. doi: 10.1126/science.1132912. [DOI] [PubMed] [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008;147:179–187. doi: 10.1104/pp.108.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B. Carotenoid catabolism during leaf senescence and its control by light. J. Photochem. Photobiol. B, Biol. 1995;30:3–13. [Google Scholar]
- Cha KW, Lee YJ, Koh HJ, Lee BM, Nam YM, Paek NC. Isolation, characterization, and mapping of the stay green mutant in rice. Theor. Appl. Genet. 2002;104:526–532. doi: 10.1007/s001220100750. [DOI] [PubMed] [Google Scholar]
- Chao WS, Liu V, Thomson WW, Platt K, Walling LL. The impact of chlorophyll-retention mutations, d1d2 and cyt-G1, during embryogeny in soybean. Plant Physiol. 1995;107:253–262. doi: 10.1104/pp.107.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJ, Vries HB, Ruys GJ, Groot SP, Koornneef M. Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol; 2003. pp. 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JGB, McCourt P, Samuel MA. ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA. 2013;110:e3888–e3894. doi: 10.1073/pnas.1308114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Li C, Li W, Wang Z, Zhou Z, Shen Y, Wu M, Wu Y, Li G, Kong LA, et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J. 2014;77:700–712. doi: 10.1111/tpj.12419. [DOI] [PubMed] [Google Scholar]
- Forsberg J, Strom J, Kieselbach H, Larsson K, Alexciev A, Engstrom A, Akerlund HE. Protease activities in the chloroplast capable of cleaving an LHCII N-terminal peptide. Physiol. Plant. 2005;123:21–29. [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression) Plant Physiol. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004;43:228–265. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell. 1997;89:25–31. doi: 10.1016/s0092-8674(00)80179-8. [DOI] [PubMed] [Google Scholar]
- Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS. Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol. 2002;130:1894–1907. doi: 10.1104/pp.008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiamet JJ, Schwartz E, Pichersky E, Nooden LD. Characterization of cytoplasmic and nuclear mutations affecting chlorophyll and chlorophyll-binding proteins during senescence in soybean. Plant Physiol. 1991;96:227–231. doi: 10.1104/pp.96.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima M, Tanaka R, Tanaka A. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:719–729. doi: 10.1093/pcp/pcp035. [DOI] [PubMed] [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009;284:17449–17456. doi: 10.1074/jbc.M109.008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009;14:155–162. doi: 10.1016/j.tplants.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Krautler B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta. 2011;1807:977–988. doi: 10.1016/j.bbabio.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J. Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol. Plant. 2013;6:1673–1691. doi: 10.1093/mp/sst069. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007;52:197–209. doi: 10.1111/j.1365-313X.2007.03221.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Chen Y, Li M, Xu X, Wu G. Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 2011;253:375–387. doi: 10.1111/j.1744-7909.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- Johnson-Flanagan AM, Spencer MS. Ethylene production during development of mustard (Brassica juncea) and canola (Brassica napus) seed. Plant Physiol. 1994;106:601–606. doi: 10.1104/pp.106.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. Protein quality control in chloroplast: a current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 2009;146:463–469. doi: 10.1093/jb/mvp073. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10013–10018. doi: 10.1073/pnas.1321568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Luo Z, Zhang J, Li J, Yang C, Wang T, Ouyang B, Li H, Giovannoni J, Ye Z. A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013;198:442–452. doi: 10.1111/nph.12175. [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. U.S.A. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecey C, Hauck P, Trapp M, Pumplin N, Plovanich A, Yao J, He SY. A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 2011;157:1965–1974. doi: 10.1104/pp.111.181826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell. 2011;23:3442–3453. doi: 10.1105/tpc.111.089714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009;59:940–952. doi: 10.1111/j.1365-313X.2009.03919.x. [DOI] [PubMed] [Google Scholar]
- Nakagawara E, Sakuraba Y, Yamasato A, Tanaka R, Tanaka A. Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J. 2007;49:800–809. doi: 10.1111/j.1365-313X.2006.02996.x. [DOI] [PubMed] [Google Scholar]
- Nakajima S, Ito H, Tanaka R, Tanaka A. Chlorophyll b reductase plays an essential role in maturation and storability of Arabidopsis seeds. Plant Physiol. 2012;160:261–273. doi: 10.1104/pp.112.196881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Kim J, van Wijk KJ. The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta. 2011;1807:999–1011. doi: 10.1016/j.bbabio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–1664. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Tanner G, Anders I, Roca M, Hörtensteiner S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15259–15264. doi: 10.1073/pnas.2036571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Anders I, Aubry S, Schenk N, Tapernoux-Luthi E, Muller T, Krautler B, Hörtensteiner S. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell. 2007;19:369–387. doi: 10.1105/tpc.106.044404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007;144:1429–1441. doi: 10.1104/pp.107.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong H, Tang Y, Zhang H, Wu P, Chen Y, Li M, Wu G, Jiang H. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013;170:1367–1373. doi: 10.1016/j.jplph.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell. 2012;24:507–518. doi: 10.1105/tpc.111.089474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Kim YS, Yoo SC, Hörtensteiner S, Paek NC. 7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem. Biophys. Res. Commun. 2013;430:32–37. doi: 10.1016/j.bbrc.2012.11.050. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Lee SH, Kim YS, Park OK, Hörtensteiner S, Paek NC. Delayed degradation of chlorophylls and photosynthetic proteins in Arabidopsis autophagy mutants during stress-induced leaf yellowing. J. Exp. Bot. 2014a;65:3915–3925. doi: 10.1093/jxb/eru008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Park SY, Kim YS, Wang SH, Yoo SC, Hörtensteiner S, Paek NC. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol. Plant. 2014b;7:1288–1302. doi: 10.1093/mp/ssu045. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kim D, Kim YS, Hörtensteiner S, Paek NC. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014c;588:3830–3837. doi: 10.1016/j.febslet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G. Phytochrome-interacting transcription factors PIF4 and PIF5 induced leaf senescence in Arabidopsis. Nat. Commun. 2014d;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009;57:120–131. doi: 10.1111/j.1365-313X.2008.03670.x. [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell. 2009;21:767–785. doi: 10.1105/tpc.108.064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 2003;44:1266–1274. doi: 10.1093/pcp/pcg172. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011;168:1952–1959. doi: 10.1016/j.jplph.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ. Five ways to stay green. J. Exp. Bot. 2000;51:329–337. doi: 10.1093/jexbot/51.suppl_1.329. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL. Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 2004;136:3594–3604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Guo Y, Kuai B. Isolation and characterization of a chlorophyll degradation regulatory gene from tall fescue. Plant Cell Rep. 2011;30:1201–1207. doi: 10.1007/s00299-011-1028-8. [DOI] [PubMed] [Google Scholar]
- Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, et al. NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll - protein complexes during leaf senescence. Plant J. 2013;74:652–662. doi: 10.1111/tpj.12154. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu T, Ren G, Hörtensteiner S, Zhou Y, Cahoon EB, Zhang C. Chlorophyll degradation: the tocopherol biosynthesis-related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol. 2014;166:70–79. doi: 10.1104/pp.114.243709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Pislariu C, Nakashima J, Fu C, Jiang Q, Quan L, Blancaflor EB, Tang Y, Bouton JH, et al. From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011;157:1483–1496. doi: 10.1104/pp.111.185140. [DOI] [PMC free article] [PubMed] [Google Scholar]


