Abstract
Protein O-GlcNAcylation, dictated by cellular UDP-N-acetylglucosamine (UDP-GlcNAc) levels, plays a crucial role in posttranslational modifications. The enzyme GlcNAc kinase (NAGK, E.C. 2.7.1.59) catalyzes the formation of GlcNAc-6-phosphate, which is a major substrate for the biosynthesis of UDP-GlcNAc. Recent studies have revealed the expression of NAGK in different types of cells especially in neuronal dendrites. Here, by immunocytochemistry (ICC) and immunonucleochemistry (INC) of cultured rat hippocampal neurons, HEK293T and GT1-7 cells, we have showed that NAGK immuno-reactive punctae being present in the nucleoplasm colocalized with small nuclear ribonucleoprotein-associated protein N (snRNPN) and p54NRB, which are speckle and paraspeckle markers, respectively. Furthermore, NAGK IR cluster was also found to be colocalized with GTF2H5 (general transcription factor IIH, polypeptide 5) immuno reactive punctae. In addition, relative localization to the ring of nuclear lamin matrix and to GlcNAc, which is highly enriched in nuclear pore complexes, showed that NAGK surrounds the nucleus at the cytoplasmic face of the nuclear outer membrane. By in situ proximity ligation assay (PLA) we confirmed the colocalization of NAGK with snRNPN in the nucleus and in dendrites, while we also verified the interactions of NAGK with p54NRB, and with GTF2H5 in the nucleus. These associations between NAGK with speckle, paraspeckle and general transcription factor suggest its regulatory roles in gene expression.
Keywords: dendrite, NAGK, paraspeckle, proximity ligation assay, snRNPN, speckle
INTRODUCTION
Sugar kinases, stress-70 proteins, and actin belong to a super family defined by a fold consisting of two domains with the topology beta beta beta alpha beta alpha beta alpha. These enzymes catalyze ATP phosphoryl transfer or hydrolysis coupled to a large conformational change in which the two domains close around the nucleotide (Hurley, 1996). N-acetylglucosamine kinase (GlcNAc kinase or NAGK; E.C. 2.7.1.59), an enzyme of the sugar-kinase/Hsp70/actin super family (Berger et al., 2002), was first identified in 1970 (Datta, 1970). NAGK is a prominent enzyme in amino sugar metabolism where it catalyzes the conversion of GlcNAc to GlcNAc-6-phosphate. This metabolic pathway eventually yields to UDP-GlcNAc, which is utilized in the synthesis of O-/N-glycans and sialic acid. UDP-GlcNAc is also a substrate for O-GlcNAc transferase, which modifies cytosolic and nuclear proteins at serine or threonine residues by adding a single GlcNAc. Furthermore, these O-GlcNAc modifications are associated with regulatory functions ranging from transcription, translation, cell signaling, and stress response to carbohydrate metabolism (Hart et al., 2011; Wells et al., 2003; Zachara et al., 2004).
Murine NAGK is a homodimeric enzyme of 39 kDa subunits (Hinderlich et al., 1998), whereas cDNA sequence of human NAGK encodes a protein of predicted molecular mass 37.4 kDa (Hinderlich et al., 2000). Crystal structures of homodimeric human NAGK revealed that the N-terminal small and C-terminal large domains form a V-shaped structure which acts as an active center for binding GlcNAc and ATP (Weihofen et al., 2006). Recently, our laboratory reported a non-canonical function of NAGK in neuronal dendritogenesis (Lee et al., 2014a). It was found that NAGK is highly expressed in neuronal dendrites, and NAGK overexpression causes significant increase in the complexity of dendritic arborization, such as, increases in the number of primary dendrites and dendritic branches. In contrast, knockdown of NAGK by short hairpin RNA caused significant dendritic degeneration. In a following article, our laboratory further showed that this non-canonical function is not dependent on the kinase activities of NAGK (Lee et al., 2014b). During these studies, predominant NAGK clusters were noticed in the nuclei of all types of cells observed, which included neurons, astrocytes, human embryonic kidney cell-line HEK293T and mouse hypothalamic GT1-7 cell line. Multiple domains exist in nucleoplasm, and the elucidation of the subnuclear domain where NAGK locates would provide insights regarding its nuclear function. Toward this end, we employed immunocytochemistry (ICC), immunonucleochemistry (INC; Moon et al., 2010), and a proximity ligation assay (PLA), to determine the nuclear location of NAGK.
MATERIALS AND METHODS
Antibodies
The following antibodies were used at the indicated dilutions: chicken polyclonal N-acetylglucosamine kinase (cNAGK) (1:1,000; GenWay Biotech, Inc., USA); O-linked N-acetylglucosamine (GlcNAc_RL2) (1:1000; Affinity BioReagents Inc., USA); MAb actin (1:500; Sigma), rabbit polyclonal N-acetylglucosamine kinase antibody (1:500; GeneTex, Inc., USA); mouse monoclonal GlcNAc kinase (1:50; Santa Cruz Biotechnology); mouse monoclonal (MAb) tubulin -subunit (α-Tubulin) (1:2,000; Developmental Studies Hybridoma Bank, University of Iowa, USA); mouse MAb promyelocytic leukaemia (PML) (1:10; PG-M3: sc-966, Santa Cruz Biotechnology); rabbit polyclonal Nucleolin (1:1000; Sigma); rabbit polyclonal snRNPN (1:1000; Sigma); mouse MAb p54NRB (1:1000; BD Biosciences, Inc., USA); rabbit polyclonal lamin B (1:1000; Young In Frontier Inc., Korea); rabbit Anti-p8/TTD-A (GTF2H5) (1:100; Sigma). The fluorescently labeled secondary antibodies Alexa Fluor 488, 568 (Invitrogen, OR) were used to detect the primary antibodies.
Primary culture of rat hippocampal neurons
Hippocampi from embryonic day 19 (E19) Sprague-Dawley rat pups were dissected, dissociated by trypsin treatment and mechanical trituration, and plated onto 12-mm diameter poly-DL-lysine/laminin coated glass coverslips at a density of ∼150 neurons/mm2, as previously described (Brewer et al., 1993). Cells were initially plated in MACS® Neuro Medium (Miltenyi Biotec Inc., USA) supplemented with MACS NeuroBrew®-21 (Miltenyi Biotec Inc.), 45.95 μM glutamate, 500 μM glutamine, 25 μM 2-mercaptoethanol, and 1% penicillin streptomycin, and fed at 4-day intervals after plating with the same media, but without glutamate and 2-mercaptoethanol. GT1-7 and HEK293T cell were grown on poly-DL-lysine coated glass coverslips in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin streptomycin.
Immunocytochemistry (ICC) and immunonucleochemistry (INC)
For ICC, cells were fixed by sequential paraformaldehyde/methanol fixation [incubation in 4% paraformaldehyde in PBS (20 mM sodium phosphate buffer, pH 7.4, 0.9% NaCl) at RT for 10 min, followed by incubation in methanol at −20°C for 20 min (Moon et al., 2007). For INC (Moon et al., 2010), coverslips were placed on a slide glass, and cells were treated with 0.5% (v/v) (for neuronal cell) and 0.1 % (for cell line) NP-40 for 5 min on ice. After rinsing in PBS, cells were fixed as above. Fixed cells were stored in blocking buffer (5% normal goat serum, 0.05% Triton X-100, and 450 mM NaCl in 20 mM Na2PO4 buffer, pH 7.4) at 4°C, and immunostained using the indicated antibodies (Moon et al., 2007).
Yeast two-hybrid
The small domain of NAGK gene [aa 1∼117] was used as bait for screening its binding proteins in the Matchmaker LexA two-hybrid system (Clontech Laboratories, Inc., USA). In brief, the pLexA vector containing the bait was transformed into yeast strain EGY48 carrying the p8op-lacZ gene. A transformed EGY48 yeast strain was transformed with a mouse brain cDNA library (Takeda et al., 2000) and the positive clones were screened according to the manufacturer’s manual. Unique inserts of positive clones were sequenced, and DNA and protein sequence analyses were performed with the BLAST algorithm at the National Center for Biotechnology Information (NCBI).
Proximity ligation assay (PLA)
Generic in situ PLA was performed using a Duolink assay kit (Olink Bioscience, Sweden), according to the manufacturer’s instructions with minor modifications. Briefly, cells were incubated with primary antibodies in preblocking buffer (5% normal goat serum, 0.05% Triton X-100 in Phosphate Buffered Saline, pH 7.4) at 4°C overnight, and then washed three times for 20 min in preblocking buffer at room temperature. For secondary antibodies conjugated with oligonucleotides, PLA probe anti-mouse MINUS and PLA probe anti-rabbit PLUS were diluted in preblocking buffer and applied for 2 h at 37°C inside a humidity chamber. Other aspects of the assay were performed according to the manufacturer’s instructions.
Image acquisition
A Leica Research Microscope DM IRE2 equipped with I3 S, N2.1 S, and Y5 filter systems (Leica Microsystems AG, Germany) and a high-resolution Cool- SNAP™ CCD camera (Photometrics Inc., USA) was used to take phase-contrast and epifluorescence images. Confocal images (1024 × 1024 pixels) were acquired using a 100X oil-immersion lens using a Leica TCS SP2 confocal system with 488, 543, and 633 nm laser lines. Image contrast and brightness were optimized using Adobe Systems Photoshop 7.0 software.
RESULTS
Nuclear expression of NAGK in cultured rat hippocampal neurons
Initially, we applied ICC to investigate the expression of NAGK in cultured rat hippocampal neurons. As revealed by double-labeling with anti-NAGK and anti-tubulin (Fig. 1A-a) or anti-actin (Fig. 1A-b) antibodies, NAGK-immunoreactive (IR) signals were distributed in the somatodendritic domain and nucleus. In dendrites, the distribution of NAGK was very similar to that of tubulin (Fig. 1A-a). Strong IR signals for both NAGK and tubulin were associated with dendritic shafts (asterisks) while the perikaryon was weakly stained. In contrast, NAGK- and actin-IR signals were not well colocalized; actin-IR signals were distributed at the periphery while NAGK-IR signals were concentrated in the shafts of dendrites (Fig. 1A-b). This feature of NAGK distribution in dendrites implies that NAGK plays a role in dendritic shafts in association with microtubules. In addition to dendritic shafts, NAGK-IR signals also formed clusters in the central area of soma (arrows). These NAGK clusters were roughly round with diverse sizes, the larger ones being ∼1–2 μm in diameter. Occasionally in ICC images, small NAGK clusters formed a circle in soma with a diameter close to that of the nucleus (arrowhead in Fig. 1A-b). These features indicate that NAGK is present in the nucleoplasm and in the nuclear envelope.
Fig. 1.
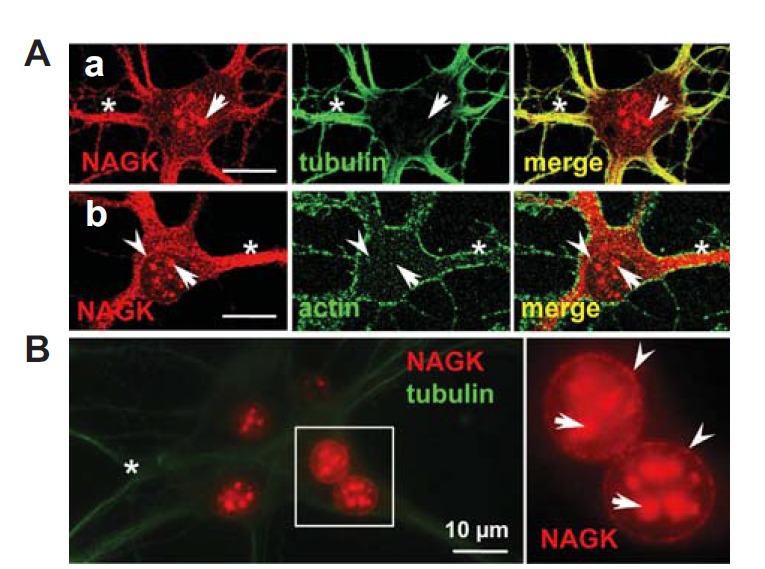
Nuclear expression of NAGK in hippocampal neurons. Cultured rat hippocampal neurons (DIV 21) were double-labeled with indicated antibodies. (A) ICC. Double-labeling with anti-NAGK/-tubulin (a) or with anti-NAGK/actin (b) revealed the expression profile of NAGK was similar to that of tubulin in dendroplasm (asterisks). NAGK clusters in the central area of soma (arrows) and a circular arrangement (arrowheads) were marked. (B) INC. Double-staining ‘naked’ nuclei with anti-NAGK/-tubulin antibodies revealed patch-like NAGK-IR signals in nucleoplasm (arrows) with a circular distribution (arrowheads). The boxed area is enlarged in the inset on the right. Scale bar; 10 μm.
INC visualized the localization of NAGK in the nuclear envelope
The circular distribution of NAGK in soma was not always evident by conventional ICC, probably due to surrounding constituents in soma. Therefore, we applied the INC, which involves the immunostaining of ‘naked’ nuclei after the removal of cytoplasmic components (Moon et al., 2010), to clearly observe circular NAGK IR signals in the soma. Cultured rat hippocampal neurons were subjected to double labeling with antibodies against NAGK and tubulin. The INC process removed most of the cytoplasmic components, including NAGK, as was observed in a fluorescence image (Fig. 1B). Only residual amounts of dendritic tubulin-IR signals, which resisted dissolution due to tight microtubule bundling in neuronal dendrites, were left (Fig. 1B, asterisk). In this image nuclei lit up like moons and the circular distribution of NAGK IR signals was clearly observed around nuclei (arrowheads; Fig. 1B, inset) and large NAGK clusters were observed in nucleoplasm (arrows). These results demonstrate the presence of NAGK in nucleoplasm and its circular distribution along the nuclear envelope.
NAGK localized at the outer membrane of the nuclear envelope
The nuclear envelope is a double unit membrane separated by intermembrane lumen. In most cells, on the inner surface of the nuclear envelope there is a thin fibrous lamina of variable thickness (Fawcett, 1966). This lamina consists of polypeptides called lamin. In thin sections, these form a meshwork of extremely fine filaments that runs parallel to the inner surface of the envelope. The present study indicates that NAGK forms a circle around nuclear envelope. To pinpoint the locations of these NAGK clusters in relation to the nuclear envelope, we performed INC using markers for the inner and outer surfaces of the envelope. For an inner surface marker we used lamin, which forms a thin matrix along the nucleoplasmic face of inner nuclear membrane. For an outer nuclear membrane marker we used GlcNAc, because the unstructured phenylalanine-glycine (FG) repeat regions of the nuclear pore complex are heavily O-GlcNAc-modified, and extend on both the nuclear and cytoplasmic sides of the pore (Li and Kohler, 2014). Therefore, we first investigated whether GlcNAc could serve as a nuclear envelope marker. For this purpose, we double-labeled HEK293T cells with antibodies against GlcNAc and lamin. GlcNAc-IR signals formed clusters (red arrowhead) outside the lamin circles (Fig. 2A, green arrowhead) indicating that these GlcNAc-IR signals were from the nuclear envelope. Having verified that GlcNAc-IR signals represent the nuclear envelope, we double-labeled HEK293T cells and cultured rat hippocampal neurons with anti-NAGK/-lamin (Fig. 2B) or anti-NAGK/-GlcNAc (Fig. 2C) antibodies. The merge images of NAGK- and lamin-IR signals revealed that NAGK clusters lie beyond lamin rings (Fig. 2B, red arrowheads). Notably, in many cases NAGK-IR signals were completely separated from lamin rings (Fig. 2B-b, arrowheads) indicating that the NAGK-IR ring lies well beyond the lamin-IR ring. The merged double-labeled image of NAGK and GlcNAc revealed NAGK rings at the outer margin of GlcNAc-IR rings (Fig. 2C). Therefore, the overall relative positioning from nucleoplasm was inferred to be lamin, GlcNAc, and NAGK. Since lamin forms a nuclear matrix inside the nuclear envelope and GlcNAc moieties protrude from nuclear pores, these results suggest that NAGK is localized at the outer surface of the nuclear envelope.
Fig. 2.
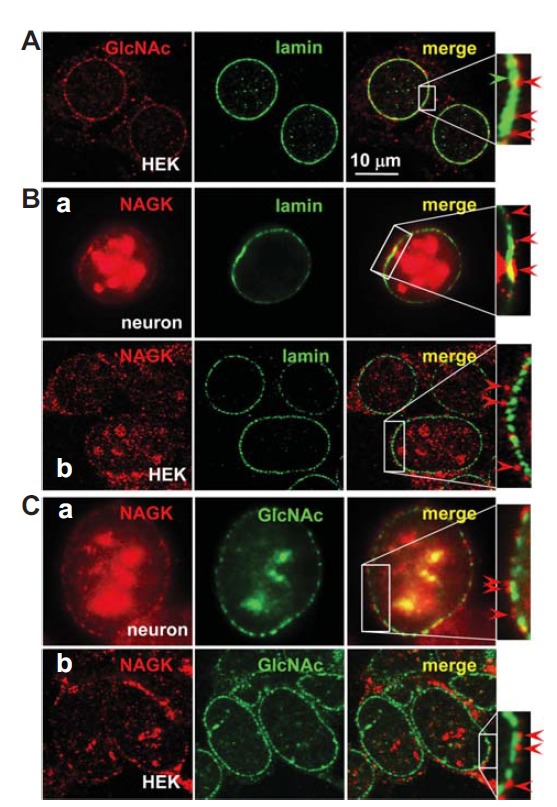
NAGK localized at the outer surface of nuclear membranes. Cultured rat hippocampal neurons (DIV 21) or HEK293T cells were double-labeled with the indicated antibodies. (A) Double–labeling of HEK293T cells with anti-GlcNAc/lamin antibodies showed that the GlcNAc-IR signals lie beyond lamin-IR rings. (B). Double-labeling of hippocampal neurons (a) or HEK293T cells (b) with anti-NAGK/i-lamin antibodies. In both cell types, NAGK-IR rings lie beyond lamin-IR rings. (C). Double-labeling of hippocampal neuron (a) or HEK293T cells (b) with anti-NAGK/GlcNAc antibodies. The NAGK-IR ring was located at the outer marginal ring of GlcNAc-IR signals, which represent the cytoplasmic face of the outer nuclear membrane. Scale bar; 10 μm.
NAGK localized to nuclear speckles and paraspeckles but not to PML body or nucleolus
The nucleoplasm is compartmentalized into highly dynamic sub domains. We found that NAGK formed clusters of various sizes in the nucleoplasm. In order to identify nucleoplasmic NAGK clusters, we double-labeled HEK293T, primary hippocampal or clonal hypothalamic GT1-7 cells with various markers of nucleoplasmic subdomains (Fig. 3). The size, shape, and distribution pattern of NAGK clusters most resembled those of markers for speckle (small nuclear ribonucleoprotein-associated polypeptide N, snRNPN; a paralog of the snRNPB’/B) and paraspeckle (p54NRB). The colocalization of NAGK with snRNPN in neurons (Fig. 3A) and with snRNPB in HEK293T cells (not shown) was almost perfect. The colocalization profile of NAGK with a paraspeckle (p54NRB) marker was varied depending on cell types (Fig. 3B). In neurons, NAGK and p54NRB immunopunctae partially overlapped (Fig. 3B-a), whereas in HEK293T cells, NAGK clusters colocalized partially, as in neurons, with p54NRB clusters in some cells (yellow arrowheads). On the other hand, the two proteins were completely segregated into different domains in other HEK293T cells (Fig. 3B-b, arrows). INC of GT1-7 cells revealed GTF2H5-IR signals overlaid on NAGK clusters (Fig. 3C). NAGK clusters did not colocalize with markers for PML body (Fig. 3D) or nucleolus (nucleolin) (Fig. 3E).
Fig. 3.
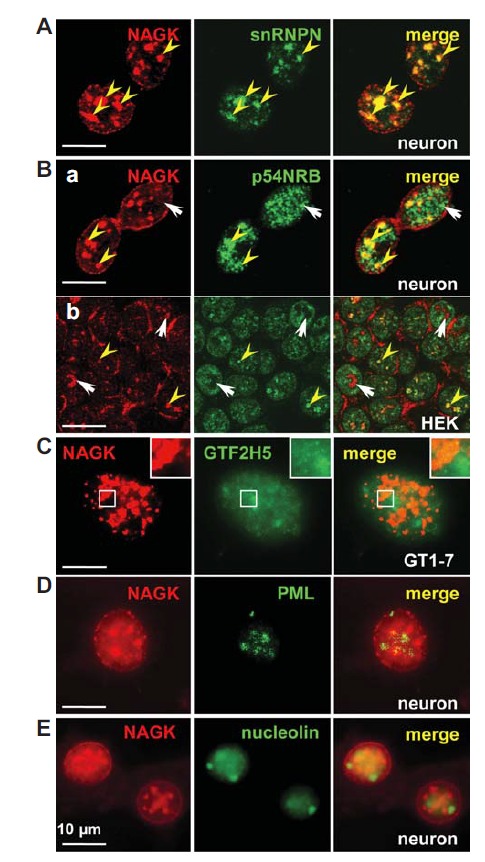
NAGK localization at nuclear subcompartments. Double-labeling of rat hippocampal neurons (DIV 21) with anti-NAGK and various markers for subnuclear domains shows that NAGK colocalized well with snRNPN (a speckle marker; A) and with some select paraspeckles (B-a). Colocalization of NAGK with the paraspeckle marker p54NRB was partial (yellow arrowheads) or absent (arrows) in HEK293T cells (B-b). (C) Immunonucleochemistry of GT1-7 cell showed that General transcription factor 2H5α or TTDA overlay on NAGK-IR clusters, (inset area showing the overlaid portion). NAGK did not colocalize with PML protein (D) or nucleolin (E; a marker for nucleoli). Scale bar; 10 μm
NAGK colocalized with snRNPN also in neuronal dendrites
Considerable evidence supports that pre-mRNA splicing takes place in neuronal dendrites. For example, some auxiliary constituents of spliceosome, like SMN (Jurica and Moore, 2003) and SAM68 (Grange et al., 2004), were found throughout neuronal cytoplasm and to extend to dendrites. Furthermore, the pre-mRNA splicing factors found in dendroplasm were reported to retain the potential to promote pre-mRNA splicing in isolated dendrites (Glanzer et al., 2005). The present and our previous study show that NAGK is highly expressed in neuronal dendrites (Lee at al., 2014a, b). Therefore, we performed ICC to investigate the possibility of NAGK-snRNPN colocalization in dendroplasm. Accordingly, we double-labeled cultured hippocampal neurons (DIV 6) with anti-NAGK and anti-snRNPN antibodies, and resulting fluorescence images showed that both NAGK- and snRNPN-IR punctae were interspersed throughout dendrites (Fig. 4). Merged images revealed the presence of some colocalization of these two proteins (Fig. 4C, arrowheads in inset), suggesting a direct interaction between them.
Fig. 4.
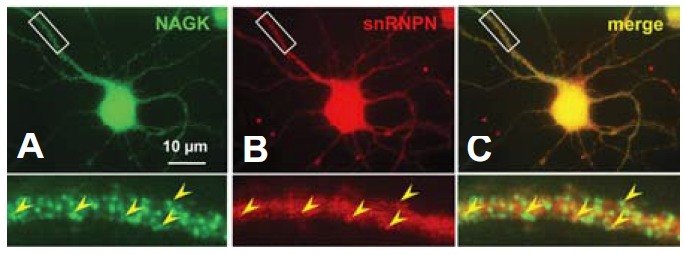
Immunocytochemistry findings showing the distribution of snRNPN immunopunctae and their partial colocalization with NAGK in dendrites. Cultured rat hippocampal neurons (DIV 6) were double-labeled with antibodies against anti-snRNPN and anti-NAGK. Fluorescence images of NAGK- and snRNPN-IR signals are shown with a merged image (A, B, C, respectively). The boxed areas are enlarged at the bottom of the figure. The colocalization of snRNPN and NAGK is indicated by arrowheads in dendritic shafts. Scale bar; 10 μm.
NAGK interacted with snRNPN, p54NRB and GTF2H5
Our initial trials to find the proteins interacting with the small domain of NAGK by yeast two-hybrid selections screened out two yeast clones (clone #3 and 5) coding for snRNPN, and two other clones (clone #15 and 58) coding for the general transcription factor IIH subunit 5 (GTF2H5) (Fig. 5A). To confirm the interaction between the two proteins in the cell, we used in situ proximal ligation assays (PLA), a simple means of detecting direct protein-protein interactions using two primary antibodies each directed against one of the targets of interest (Soderberg et al., 2006). For snRNPN, PLA positive signals (red fluorescence) appeared both in soma and dendrites (Fig. 5B-a). INC followed by PLA showed the interaction between NAGK and snRNPN in the nucleoplasm (Fig. 5B-b). In situ NAGK/p54NRB PLA signals were distributed in the nucleus of HEK293T cells (Fig. 5B-c), and INC followed by NAGK/GTF2H5 PLA revealed positive signals in the nucleoplasm of GT1-7 cells (Fig. 5B-d). In the negative control PLA study, where no primary antibodies were added, showed no specific PLA signal both in neurons and GT1-7 nucleus (data not shown). These results indicated that NAGK directly interacts with snRNPN, p54NRB, and GTF2H5.
Fig. 5.
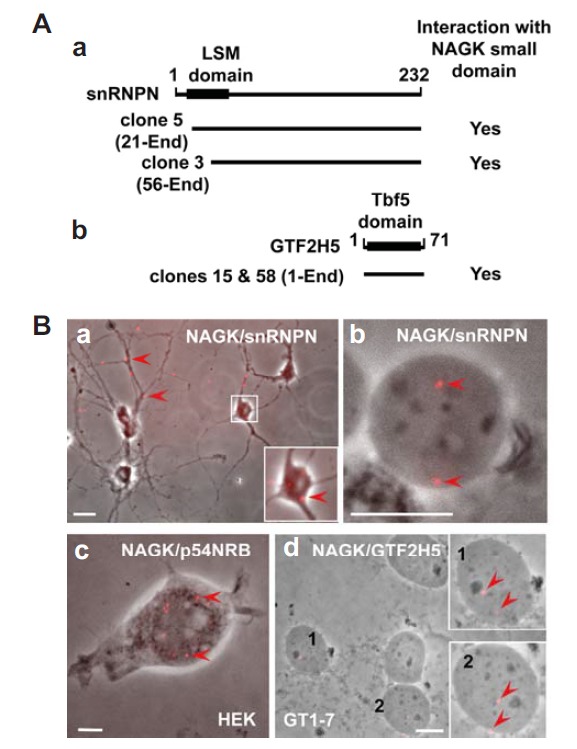
Interaction of NAGK with nuclear proteins. (A) Yeast two hybrid assays using the NAGK small domain as bait identified snRNPN and GTF2H5. The NAGK-interacting domains of snRNPN (a) and GTF2H5 (b) of two different clones for each protein are shown. The positions of like-Sm (LSM) and TTDA subunit of TFIIH basal transcription factor complex (Tbf5) protein domains of each protein were marked on the bars with amino acid numbers. (B) PLA showing interactions of NAGK with nuclear proteins. PLA was performed in rat hippocampal neurons (DIV 11). A low power image shows distribution of NAGK/snRNPN PLA dots (arrowheads) in soma (inset) and dendrites (a). Immunonucleochemistry followed by PLA shows the interaction between NAGK and snRNPN (arrows) in the ‘naked’ neuronal nucleoplasm (b). Scale bars: 5 and 20 μm in a and b, respectively. PLA conducted in HEK293T cells showed interaction between NAGK and paraspeckle protein (p54NRB) in nucleus (c, arrowheads). Scale bar, 5 μm. PLA done in hypothalamic GT1-7 cells showed NAGK/TTDA (GTF2H5) PLA dots in nucleoplasm (arrowheads). Scale bar, 10 μm.
DISCUSSION
In this study, we describe the characteristic distribution patterns of NAGK in different sub nuclear compartments. ICC of cultured hippocampal neurons showed somatodendritic expression of NAGK, and its nuclear expression profile was clearly depicted by INC where an irregular cluster-like expression pattern in nucleoplasm and a circular arrangement along the outer side of the nuclear envelope was found. Double-labeling of ‘naked’ nuclei in INC showed that NAGK was colocalized with snRNPN (a speckle marker protein), p54NRB (a paraspeckle marker protein), and TTDA (GTF2H5). We also found these protein interactions (snRNPN, GTF2H5) by PLA and in yeast two hybrid screening.
Subcompartments of nucleoplasm are involved in the splicing, translation, and transcriptional regulation of genes (Spector, 2001; Zimber et al., 2004). Of these, speckle (also known as interchromatin granule cluster) is a key nuclear subcompartment where splicing and transcription take place simultaneously. Speckle is considered the hub for many pre-mRNA splicing factors, including snRNPs and serine/arginine-rich (SR) proteins (Fu, 1995). Of the snRNPs, snRNAs, such as, U1, U2, U4, U5 and U6 (7), form complexes with B, B′, D, D′, E, F and G snRNP associated proteins. snRNPN is a paralog of the snRNPB’/B gene, which is alternatively spliced to produce SmB or SmB’ spliceosomal core proteins (Gray et al., 1999). snRNPN is expressed in a tissue-specific manner, and is most abundant in rat brain, less so in heart and undetectable in other tissues examined (McAllister et al., 1988). Therefore, it is interpreted that an ancestral duplication of the snRNPB’/B gene gave rise to the snRNPN paralog which evolved new functions in specific tissues. However, our PLA and INC results showed that NAGK interacts with SmB’/B in HEK293T (data not shown) and snRNPN in neurons, which suggests that NAGK has the same function in neuronal and non-neuronal cells.
In addition to pre-mRNA splicing, mounting evidence suggests that speckle also modulates transcription processes. For examples, a subpopulation of the largest subunit of RNA polymerase II molecules is distributed in speckle domains in transcriptionally active nuclei (Bregman et al., 1995). Significant levels of RNA Polymerase II-mediated transcription sites are associated with nuclear speckles (Wei et al., 1999). Depletion of a prototypical SR protein SC35 induced Pol II accumulation within the gene body and attenuated elongation, suggesting speckle proteins play critical roles in transcriptional elongation (Lin et al., 2008). About 80% of pre-mRNA splicing occurs cotranscriptionally, and active spliceosomes localize in situ to regions of decompacted chromatin at the periphery of or within nuclear speckles, whereas the release of post-transcriptionally spliced mRNA from speckles is coupled to the nuclear mRNA export pathway (Girard et al., 2012).
The present study also shows snRNPN/NAGK interaction in neuronal dendrites, indicating involvement of NAGK in local splicing process at dendrites. Splicing in dendrites has been extensively researched since the discovery of protein synthetic machinery including ribosomes, membranous constituents of endoplasmic reticulum, and Golgi apparatus in dendroplasm. Nova proteins (neuron-specific RNA binding proteins) regulate alternative splicing in neurons (Jensen et al., 2000), and immunofluorescence and EM studies of spinal cord motor neurons demonstrated that Nova localizes beneath synaptic contacts in dendrites (Racca et al., 2010), which demonstrates the presence and functionality of pre-mRNA-splicing components in dendrites. For example, splicing factors, SF2/ASF, USAF65, and Sm or SR proteins are located in proximal and distal portions of dendrites and their branch points. Furthermore, reporter pre-mRNAs can be translated in dendritic cytoplasm after dendritic splicing and when isolated dendrites are transfected with chicken delta-crystallin pre-mRNA or luciferase reporter pre-mRNA, splicing junctions clustered at or near expected splice sites (Glanzer et al., 2005). These observations suggest that pre-mRNA-splicing factors in dendroplasm retain the potential to promote pre-mRNA splicing. It is now widely accepted that translation of mRNAs localized to dendrites is regulated in response to neural activity and is required for many forms of long term synaptic plasticity. Thus, it appears NAGK may play a role in the splicing of the pre-mRNAs involved in local dendritic translation. Further research on NAGK is needed to elucidate its role in neuronal dendrites.
In the present study, we found that many NAGK clusters overlapped those of p54RNB, a paraspeckle marker, in neurons and some HEK293T cells. However, in some HEK293T cells, NAGK formed a few large clusters that were completely segregated from p54RNB. Paraspeckles are nuclear bodies that are usually visualized in cultured cell lines as 2∼20 foci during interphase and during all of mitosis except for telophase (Naganuma and Hirose, 2013). At telophase, when the two daughter nuclei are formed, there is no RNA polymerase (Pol) II transcription. When Pol II transcription is inhibited, paraspeckle protein 1 (PSP1) leaves paraspeckles and accumulates at the nucleolus, concentrating in crescent-shaped structures termed “perinucleolar caps” (Fox et al., 2002). p54NRB forms a heterodimer with PSP1 and is targeted to paraspeckles in a RNA-dependent manner whereas its accumulation at perinucleolar caps does not require RNA (Fox et al., 2005; Shav-Tal et al., 2005). We did not study whether NAGK is present in the perinucleolar cap. However, our study showed that NAGK is not present in the nucleolus. Therefore, in cultured HEK293T cells, some cells in Pol II inhibition may exhibit segregation in NAGK and p54NRB signals. Paraspeckles are involved in diverse gene expression associated regulatory events such as regulation of the transcription and pre-mRNA splicing events associated with important physiological events and diseases (Lagier-Tourenne and Cleveland, 2009; Shav-Tal and Zipori, 2002). NAGK may participate in these processes.
Interaction of NAGK with GTF2H5 (TTDA) was confirmed by the yeast two-hybrid assay, INC and PLA. These findings support that NAGK might have a role in gene transcription. GTF2H, a multi subunit complex, is essential for Pol II transcription and Nucleotide Excision Repair (NER). GTF2H5 is a small (8 kDa) protein subunit, of which mutation causes trichothiodystrophy (TTD-A) disease. Under normal condition TTDA maintained equilibrium of free and GTF2H bound states, but shifts into high GTF2H bound condition in presence of DNA lesion by UV-irradiation (Giglia-Mari et al., 2006; Nonnekens et al., 2013). Therefore, NAGK may regulate NER by binding with GTF2H5.
In summary, we show that NAGK interacts with speckle (snRNPN), paraspeckle (p54NRB), GTF2H5 in the nucleoplasm, and also present in outer nuclear membrane. Altogether, our present study implies a diverse but significant non-canonical function of NAGK in the nucleus.
Acknowledgments
We thank Eun-Jung Jung in our laboratory for technical assistance. This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2006116).
REFERENCES
- Berger M, Chen H, Reutter W, Hinderlich S. Structure and function of N-acetylglucosamine kinase. Identification of two active site cysteines. Eur. J. Biochem. 2002;269:4212–4218. doi: 10.1046/j.1432-1033.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Datta A. Studies on hog spleen N-acetylglucosamine kinase. I. Purification and properties of N-acetylglucosamine kinase. Biochim. Biophys. Acta. 1970;220:51–60. doi: 10.1016/0005-2744(70)90228-7. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Miquel C, Theil AF, Mari PO, Hoogstraten D, Ng JM, Dinant C, Hoeijmakers JH, Vermeulen W. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 2006;4:e156. doi: 10.1371/journal.pbio.0040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Will CL, Peng J, Makarov EM, Kastner B. Posttranscriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc. Natl. Acad. Sci. USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange J, Boyer V, Fabian-Fine R, Fredj NB, Sadoul R, Goldberg Y. Somatodendritic localization and mRNA association of the splicing regulatory protein Sam68 in the hippocampus and cortex. J. Neurosci. Res; 2004. pp. 654–666. [DOI] [PubMed] [Google Scholar]
- Gray TA, Smithwick MJ, Schaldach MA, Martone DL, Graves JA, McCarrey JR, Nicholls RD. Concerted regulation and molecular evolution of the duplicated SNRPB′/B and SNRPN loci. Nucleic Acids Res. 1999;27:4577–4584. doi: 10.1093/nar/27.23.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderlich S, Nöhring S, Weise C, Franke P, Stäsche R, Reutter W. Purification and characterization of N-acetylglucosamine kinase from rat liver: comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 1998;252:133–139. doi: 10.1046/j.1432-1327.1998.2520133.x. [DOI] [PubMed] [Google Scholar]
- Hinderlich S, Berger M, Schwarzkopf M, Effertz K, Reutter W. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 2000;267:3301–3308. doi: 10.1046/j.1432-1327.2000.01360.x. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Cho SJ, Moon IS. The non-canonical effect of N-acetyl-D-glucosamine Kinase on the formation of neuronal dendrites. Mol. Cells. 2014a;37:248–256. doi: 10.14348/molcells.2014.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Dutta S, Moon IS. Upregulation of Dendritic Aroborzation by N-acetylglucosamine Kinase is not dependent on its Kinase activity. Mol. Cells. 2014b;37:322–329. doi: 10.14348/molcells.2014.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kohler JJ. Glycosylation of the nuclear pore. Traffic. 2014;15:347–361. doi: 10.1111/tra.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister G, Amara SG, Lerner MR. Tissue-specific expression and cDNA cloning of small nuclear ribonucleoprotein-associated polypeptide N. Proc. Natl. Acad. Sci. USA. 1988;85:5296–5300. doi: 10.1073/pnas.85.14.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon IS, Cho SJ, Jin I, Walikonis R. A simple method for combined fluorescence in situ hybridization and immunocytochemistry. Mol. Cells. 2007;24:76–82. [PubMed] [Google Scholar]
- Moon IS, Lee HS, Park SD, Seog DH. Immunonucleochemistry: a new method for in situ detection of antigens in the nucleus of cells in culture. Cytotechnol. 2010;62:83–93. doi: 10.1007/s10616-010-9266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long noncoding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnekens J, Cabantous S, Slingerland J, Mari PO, Giglia-Mari G. In vivo interactions of TTDA mutant proteins within TFIIH. J. Cell Sci. 2013;126:3278–3283. doi: 10.1242/jcs.126839. [DOI] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front. Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Takeda S, Yamazaki H, Seog DH, Kanai Y, Terada S, Hirokawa N. Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J. Cell Biol. 2000;148:1255–1265. doi: 10.1083/jcb.148.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen WA, Berger M, Chen H, Saenger W, Hinderlich S. Structures of human N-acetylglucosamine kinase in two complexes with N-acetylglucosamine and with ADP/glucose: insights into substrate specificity and regulation. J. Mol. Biol. 2006;364:388–399. doi: 10.1016/j.jmb.2006.08.085. [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol. Life Sci. 2003;60:222–228. doi: 10.1007/s000180300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signaling in health and disease. Cell. Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]


